Abstract
The degree of white matter (WM) myelination is rather inhomogeneous across the brain. As a consequence, white matter appears differently across the cortical lobes in MR images acquired during early postnatal development. At 1 year old specifically, the gray/white matter contrast of MR images in prefrontal and temporal lobes is limited and thus tissue segmentation results show commonly reduce edaccuracy in these lobes. In this novel work, we propose the use of spatial intensity growth maps (IGM) for T1 and T2 weighted image to compensate for local appearance inhomogeneity. The IGM captures expected intensity changes from 1 to 2 years of age, as appearance inhomogeneity is highly reduced by the age of 24 months. For that purpose, we employ MRI data from a large dataset of longitudinal (12 and 24 month old subjects) MR study of Autism. The IGM creation is based on automatically co-registered images at 12 months, corresponding registered 24 months images, and a final registration of all image to a prior average template. In template space, voxelwise correspondence is thus achieved and the IGM is computed as the coefficient of a voxelwise linear regression model between corresponding intensities at 1-year and 2-years. The proposed IGM shows low regression values of 1–10% in GM and CSF regions, as well as in WM regions at advanced stage of myelination at 1-year. However, in the prefrontal and temporal lobe we observed regression values of 20–25%, indicating that the IGM appropriately captures the expected large intensity change in these lobes due to myelination. The IGM is applied to cross-sectional MRI datasets of 1-year old subjects via registration, correction and tissue segmentation of the corrected dataset. We validated our approach in a small study of images with known, manual “ground truth” segmentations. We furthermore present an EM-like optimization of adapting existing non-optimal prior atlas probability maps to fit known expert rater segmentations.
Index Terms: MRI, tissue segmentation, expectation maximization (EM) algorithm, classification
1. INTRODUCTION
Brain tissue segmentation is a fundamental analysis step to anatomical studies of neurodevelopment. Many methods have been proposed that segment MR images into tissue classes of white matter (WM), gray matter (GM) and cerebrospinal fluid (CSF). The most widely employed approaches are expectation maximization (EM), artificial neural network and fuzzy classification based algorithms [1, 2, 3]. These methods work well on images from subjects older than 2 years of age, as the white matter of the brain has matured enough in order to appear mostly homogenous across the brain. But WM in early postnatal stage undergoes myelination that strongly affects MR appearance. The intensity of not-fully-myelinated WM often appear similar to GM intensity. The progress of myelination in WM is well known be inhomogeneous across the brain by following a pattern of posterior-to-anterior lobes and superior to inferior progression. At 1 year old, the inferior frontal lobes and temporal poles show consequently a reduced WM/GM contrast as compared to other lobes. Not surprisingly, standard tissues segmentation methods, which assume homogeneous within-class appearance across the image, produce incorrect results within the prefrontal and temporal lobes. Commonly, white matter is undersegmented in these lobes. To address this, the addition of a mixed WM/GM class or the use of regional/lobar atlases were proposed [4], though with limited success unless paired longitudinal datasets exist.
The purpose of this study is to develop a novel method for brain tissue segmentation of cross-sectional 1-year old MRI datasets using a novel spatial intensity growth map (IGM). Our IGM captures expected intensity changes from 1 to 2 years of age, as appearance inhomogeneity is highly reduced by the age of 24 months. The IGM is applied to MRI images by deformable registration and subsequent intensity correction. The modified image is then segmented with an EM based tissue segmentation method. The proposed method is then evaluated on 3 T1 weigthed images of 1 year old subjects with manual “ground truth” segmentation.
2. METHODS
2.1. Data
The study population consists of fourteen subjects with longitudinal T1 (160 slices with TR=2400ms, TE=3.16ms, flip angle=8, field of view 224 × 256) and T2 weighted (160 slices with TR=3200ms, TE=499ms, flip angle=120, field of view 256 × 256) MR scans at 12 and 24 months. The subject scans are a selection from scans within the IBIS (Infant Brain Imaging Study) network1 acquired at 4 different sites. The reason that we are randomly selected from the autism and healthy datasets is to avoid being biased segmentation results toward the specific group. All datasets were acquired on 3T Siemens Tim Trio scanners at the same resolution of 1 × 1 × 1mm3. Three additional subjects from the same study were selected for the evaluation study. Using ITK-SNAP2, trained expert researchers determined manual segmentations of WM, GM and CSF of the full T1 image.
2.2. Preprocessing
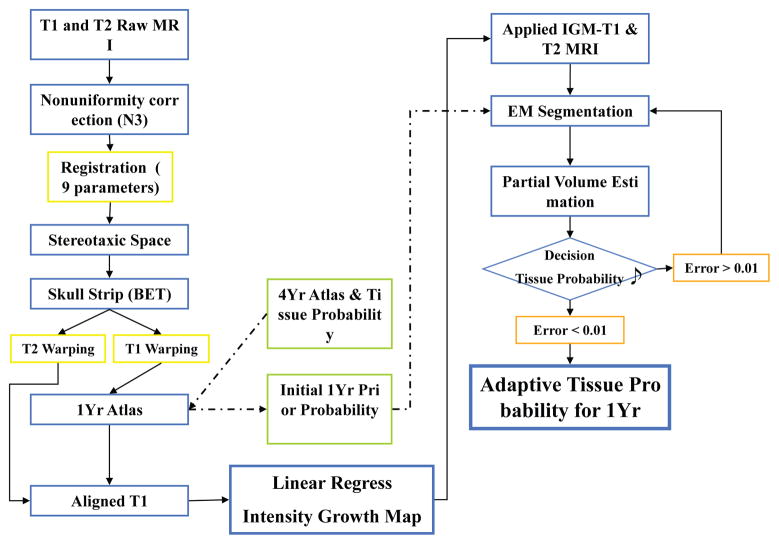
An overview of processing is shown in Fig. 1. All images were first corrected for intensity nonuniformity using N3 [5] resulting from inhomogeneities in the magnetic field. Then we extracted the skull using FSL-BET (Brain Extraction Tool)3 for all subjects [6]. All T2 weighted images were rigidly registered to the corresponding T1 weighted images. Then, the T1 and T2 images at two years of age were mapped into their corresponding intrasubject 1-year old dataset by rigid registration followed by cross-correlation, thin-plate spline based deformable registration [7]. Finally, all T1 MRIs of 1-year subjects were then registered into a prior, common, average template coordinate space by 9 parameter (similarity transform) registration followed by the same deformable registration. The concatenated registration transforms were applied to all the other images, such that all images (T1 weighted and T2 weighted at both 1 and 2 years) are mapped into a common template space. The unbiased, age-appropriate (1-year) atlas template was computed via joint deformable registration that simultaneously minimizes the difference of intensity and transformation [8] from 66 training 1-year old datasets within the IBIS study. Initial probabilistic spatial priors for all tissue classes were determined by co-registering and adapting an existing 4-year old template (see 2.5).
Fig. 1.
Overview of IGM calculation and generation of the optimized tissue probabilities using an iterative EM process.
2.3. Intensity growth map (IGM) generation
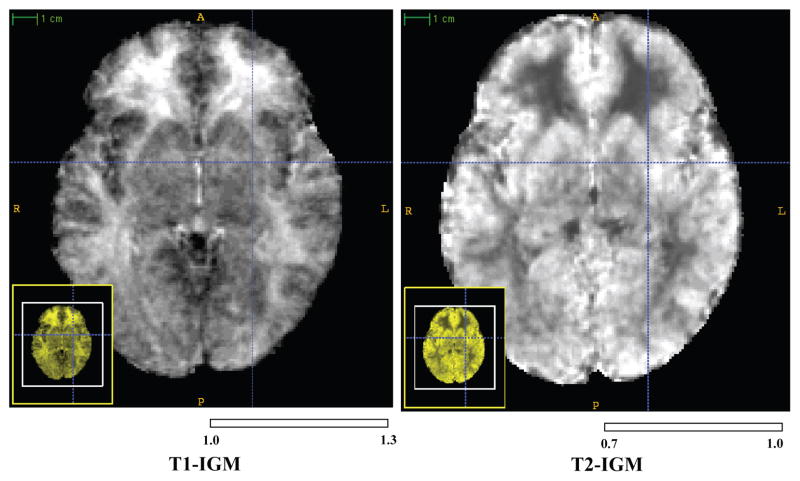
Based on the voxelwise correspondence in the average template space established in the preprocessing, we compute the IGM as the local coefficient map (αi) from a reduced linear regression model (Yi = αi · Xi) between the local intensity at 2-year (Yi) and 1-year (Xi) old data in the each voxel (i). Whereas a monoexponential or second order model would be needed for regression across a longer period or when incorporating multiple time points [9, 10], the selected linear model is a reasonable choice when we focus on a 1-year period from 12 to 24 months of age with only two known timepoints. While the common linear regression model also includes a constant term, we did not consider this constant term, as we assume that the background has the same zero mean intensity at both ages. The resulting IGM map (see Fig. 2) will show high values of areas of high intensity change and low values for those regions of moderate intensity change from 12 to 24 months.
Fig. 2.
IGM shows regression coefficients of about 25% in the prefrontal and temporal lobe regions (ITK-SNAP visualization).
2.4. Enhanced IGM based tissue segmentation
Like most atlas based segmentation approaches, our EM segmentation registers a prior atlas template to a new subject’s T1 or T2 datasets and maps the atlas tissue priors to the subject space. In our approach, we also map the IGM map into the subject image space. The T1 and T2 IGMs are then convoluted with each T1 and T2 weighted image to yield intensity corrected T1 and T2 images as input for an EM based tissue segmentation [11]. Next to the posterior WM, GM and CSF probability maps, our EM segmentation approach also provides a partial volume estimation map (PVE) for each tissue type. Using a thinning based 1D-skeleton of the WM-PVE map binarized at 10%, we further enhance the posterior WM probability setting the WM-PVE-skeleton to 1, as the WM often is thin and more variable in the temporal pole and posterior occipital lobes. This last step does little to the overall volume (less than 1% change), but provides considerable enhancement to any potential cortical thickness analysis following the tissue segmentation.
2.5. Prior optimization
The result of our atlas based brain tissue segmentation is strongly dependent on the prior tissue probability maps defined in the atlas template space. In our application, the initial tissue class priors in the atlas space were determined by deformable registration of an existing 4-year old atlas with known probability priors into the 1-year old atlas. As these mapped probabilistic tissue maps may not be fully appropriate for our 1 year atlas, and we employed an EM-like framework to optimize these maps. The tissue segmentation method (described above) was thereby treated as a black box. As a first step, using the propagated 4-year old probability priors, we computed the segmentation of the 14 subjects already employed in the IGM computation. The resulting posterior maps were mapped back in the atlas space, where they were averaged to represent updated prior probability maps. Then, for all subsequent iterations, we computed the segmentation of three “training” subjects with known manual expert segmentation and compared these to the current iteration’s hard segmentation maps. Difference maps were computed for each tissue class (0 = correct segmentation;+1 = false positive; −1 = false negative), averaged across all three training subjects in the atlas space, and additively applied to the prior probability maps. The optimization iterated until the prior probabilities converged (less than 1% cumulative change in probability across the WM, GM and CSF priors. Convergence was established after 4 iterations.
3. RESULTS
3.1. Intensity growth map
The computed T1 IGM reflects the expected maturation related MR intensity changes between 1-year and 2-year old (see Fig. 2). GM and CSF, which are not related to the myelination process, revealed relatively low change coefficients αi between 1.0 and 1.1 in the T1 image. In WM regions that already underwent considerable myelination, we measured similar coefficient values to those in GM and CSF regions. However, in those WM areas that are known to exhibit a comparatively lower stage of myelination at 1-years of, we observed 20–30% intensity difference between 1-year and 2-year olds (i.e. coefficients(αi) around 1.25). The IGM coefficients for the T2 weighted images provided the same interpretations although it appears inverse when compared to the T1 weighted IGM. In both T1 and T2 IGM, the superior frontal lobe, inferior temporal lobe and temporal pole changed the most. IGM corrected MR images of 1 year old subjects appear visually with the appearance of a 2 year old’s MR image.
3.2. Adaptive one-year-old tissue probability atlas and experimental results
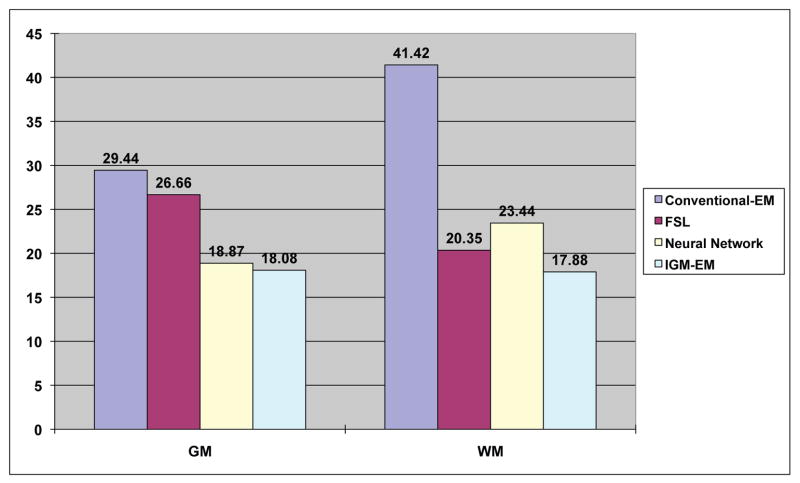
Fig. 3 displays how under-estimations occurred in the inferior temporal lobe and frontal lobe using a conventional EM segmentations method on the uncorrected 1-year old data with known ground truth. In addition over-estimation also occured in the superior frontal lobe due to the low contrast in that region. However, when we applied our proposed IGM-EM based method, we obtained more accurate segmentations of the WM and GM, especially in the low contrast areas of the inferior temporal and superior frontal lobe. We validated the accuracy of the IGM-EM segmentation methods against the three datasets with known manual segmentations. Since those same datasets were also used in the optimization of the prior atlas probability maps (see 2.5, we employed a leave-one-out scheme for all parts of the validation. Thus, for each validation dataset the atlas probability maps were optimized only over the other 2 datasets. A clear improvement in the inferior frontal, temporal and posterior occipital lobe is visible. These regions are at a lower stage of myelination at 1 year of age. We further favorably compared the performance of our proposed IGM-EM versus a conventional EM method and FSL’s FIRST method (Fig. 4) using the Tanimoto volumetric overlap error against the manual segmentation.
Fig. 3.
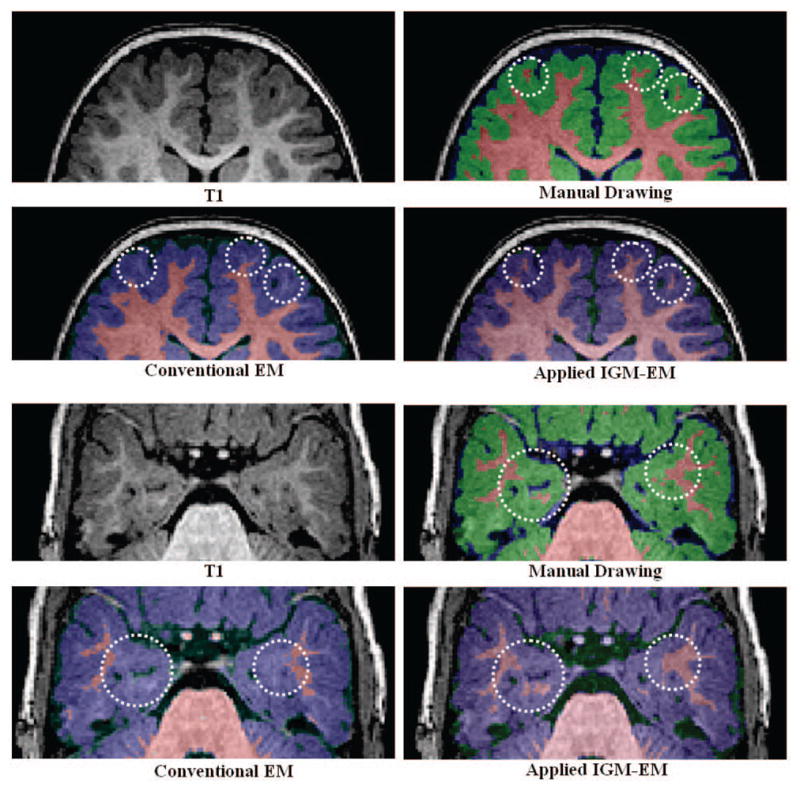
Visual comparison of segmentation. The WM was under-estimated in prefrontal and inferior temporal lobe (white dot circles) using the conventional EM algorithm as compared to the IGM-EM results and the expert ground truth.
Fig. 4.
Tanimoto Error results to the ground truth. The results of IGM-EM show the lowest error in WM and GM.
4. CONCLUSION
The quantitative analyses of postnatal development using brain tissue volume and cortical thickness measurements, which are main components of most traditional anatomical MRI studies, are based on accurate brain tissue segmentation. Here, we propose a correction and tissue segmentation methodology that allows a standard brain tissue segmentation method to handle low myelination areas in 1-year old brain MRIs. The method is based mainly on a trained longitudinal model of MR intensity change from 1-year to 2-year old. We furthermore presented a EM-based optimization of adapting existing non-optimal prior probability maps to fit know expert rater segmentations.
Acknowledgments
The IBIS (Infant Brain Imaging Study) Network is an NIH funded Autism Center of Excellence (HDO55741) and consists of a consortium of 7 Universities in the U.S. and Canada. Clinical Sites: University of North Carolina: J. Piven (IBIS Network PI), H.C. Hazlett, C. Chappell; University of Washington: S. Dager, A. Estes; Washington University: K. Botteron, R. McKinstry, J. Contstantino, L. Flake; Childrens Hospital of Philadelphia: R. Schultz, S. Paterson; University of Alberta: L. Zwaigenbaum. Data Coordinating Center: Montreal Neurological Institute: A. Evans, L. Collins, B. Pike, R. Aleong, S. Das. Image Processing Core: University of Utah: G. Gerig; University of North Carolina: M. Styner. Statistical Analysis Core: University of North Carolina: H. Gu. Genetics Analysis Core: University of North Carolina: P. Sullivan, F. Wright.
Footnotes
References
- 1.Fu JC, Chen CC, Chai JW, Wong STC, Li IC. Image segmentation by EM-based adaptive pulse coupled neural networks in brain magnetic resonance imaging. Computerized Medical Imaging and Graphics. 2010;34(4):308–320. doi: 10.1016/j.compmedimag.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Dyrby TB, Rostrup E, Baaré WFC, van Straaten ECW, Barkhof F, Vrenken H, Ropele S, Schmidt R, Erkinjuntti T, Wahlund LO, et al. Segmentation of age-related white matter changes in a clinical multi-center study. NeuroImage. 2008;41(2):335–345. doi: 10.1016/j.neuroimage.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Lin GC, Wang WJ, Wang CM, Sun SY. Automated classification of multi-spectral MR images using Linear Discriminant Analysis. Computerized Medical Imaging and Graphics. 2010;34(4):251–268. doi: 10.1016/j.compmedimag.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Shi F, Yap PT, Fan Y, Gilmore JH, Lin W, Shen D. Construction of multi-region-multi-reference atlases for neonatal brain MRI segmentation. Neuroimage. 2010;51(2):684–693. doi: 10.1016/j.neuroimage.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. Medical Imaging, IEEE Transactions on. 2002;17(1):87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 6.Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins DL, Peters TM, Evans AC. Automated 3D nonlinear deformation procedure for determination of gross morphometric variability in human brain,” in. Proceedings of SPIE. 1994;2359:180. [Google Scholar]
- 8.Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL. Unbiased average age-appropriate atlases for pediatric studies. NeuroImage. 2010 doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding XQ, Kucinski T, Wittkugel O, Goebell E, Grzyska U, Görg M, Kohlschütter A, Zeumer H. Normal brain maturation characterized with age-related T2 relaxation times: an attempt to develop a quantitative imaging measure for clinical use. Investigative radiology. 2004;39(12):740. doi: 10.1097/00004424-200412000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. Journal of Neuroscience. 2004;24(38):8223. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilmes JA. A gentle tutorial of the EM algorithm and its application to parameter estimation for Gaussian mixture and hidden Markov models. International Computer Science Institute. 1998;4 [Google Scholar]