Abstract
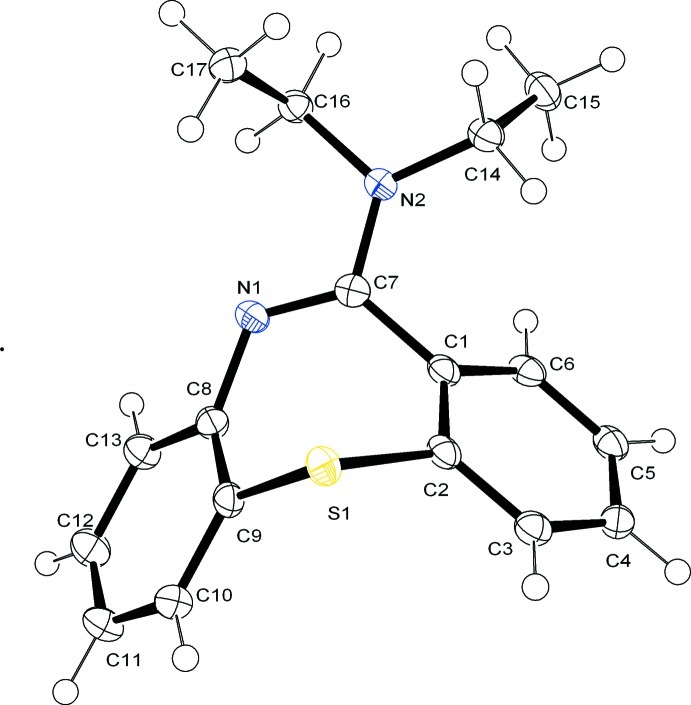
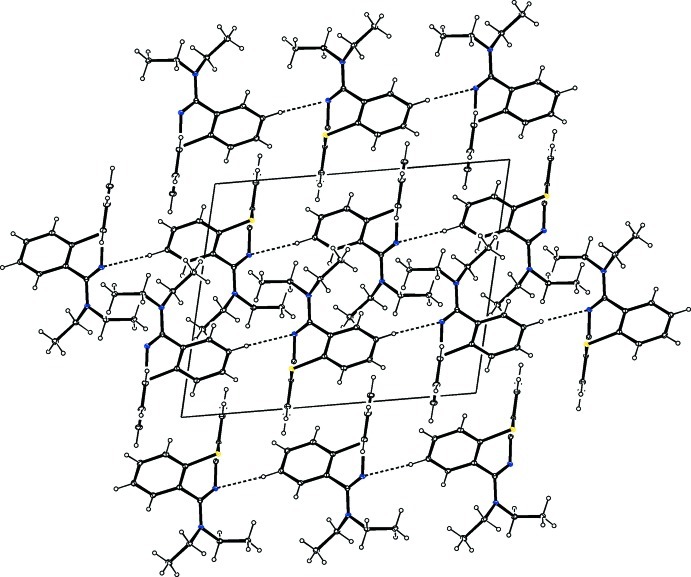
In the title compound, C17H18N2S, the thiazepine ring adopts a boat conformation and the dihedral angle between the benzene rings is 75.92 (5)°, resulting in a butterfly-like conformation. In the crystal, molecules are connected via weak Caromatic—H⋯N contacts involving the imine N atom as acceptor and through a quite short C—H⋯π interaction. The resulting molecular chains propagate along the c-axis direction.
Related literature
For ‘privileged structures’, that is ‘structures able to provide high affinity ligands for more than one type of receptor’, see: Evans et al. (1988 ▶); Patchett & Nargund (2000 ▶); Fedi et al. (2008 ▶). For the clinical use of dibenzothiazepine derivatives, see: Ganesh et al. (2011 ▶); Pettersson et al. (2009 ▶); Riedel et al. (2007 ▶); Warawa et al. (2001 ▶). For structure–property relationships in (6,7,6)-tricyclic ring systems, see: Ravikumar & Sridhar (2005 ▶); Altamura et al. (2008 ▶, 2009 ▶, 2011 ▶). For geometrical data and descriptors, see: Duax et al. (1976 ▶); Bertolasi et al. (1982 ▶); Allen et al. (1987 ▶).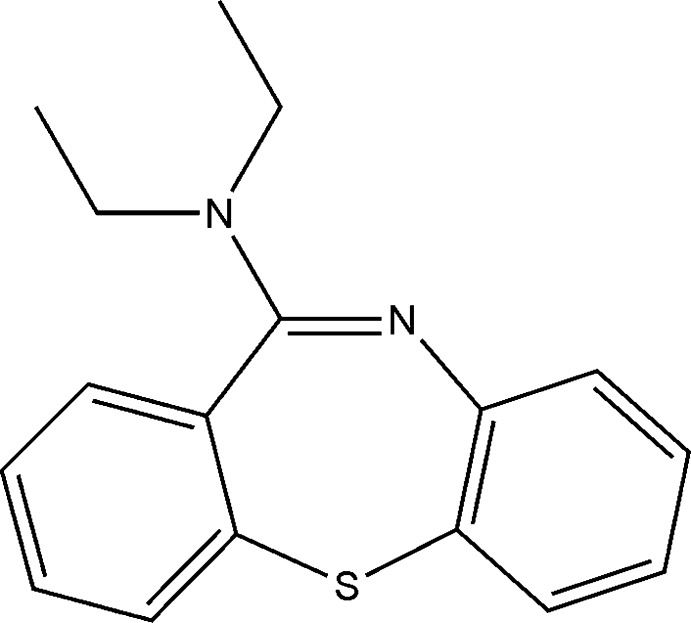
Experimental
Crystal data
C17H18N2S
M r = 282.40
Monoclinic,
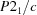
a = 12.0137 (2) Å
b = 8.2257 (1) Å
c = 15.0513 (2) Å
β = 102.952 (1)°
V = 1449.54 (4) Å3
Z = 4
Cu Kα radiation
μ = 1.89 mm−1
T = 150 K
0.20 × 0.18 × 0.03 mm
Data collection
Oxford Diffraction XcaliburPX diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2006 ▶) T min = 0.722, T max = 0.945
6173 measured reflections
2364 independent reflections
1837 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.032
wR(F 2) = 0.086
S = 1.05
2364 reflections
181 parameters
H-atom parameters constrained
Δρmax = 0.23 e Å−3
Δρmin = −0.26 e Å−3
Data collection: CrysAlis CCD (Oxford Diffraction, 2006 ▶); cell refinement: CrysAlis CCD; data reduction: CrysAlis RED (Oxford Diffraction, 2006 ▶); program(s) used to solve structure: SIR92 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: PARST (Nardelli, 1995 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536812042328/nc2296sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812042328/nc2296Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C8–C12 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5⋯N1i | 0.95 | 2.70 | 3.576 (2) | 154 |
| C4—H4⋯Cg i | 0.95 | 2.81 | 3.5759 (18) | 139 |
Symmetry code: (i) 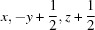 .
.
Acknowledgments
The authors acknowledge the CRIST (Centro di Cristallografia Strutturale, University of Firenze) where the data collection was performed.
supplementary crystallographic information
Comment
Many antidepressant drugs have a tricyclic structure with two aromatic rings fused to a central seven membered ring, on which one or more heteroatoms can be present. In this context, dibenzothiazepines are a class of heterocyclic scaffolds containing nitrogen and sulfur which belong to the class of the so-called "privileged structures", i.e. structures "able to provide high affinity ligands for more than one type of receptor" (Evans et al. 1988; Patchett & Nargund 2000; Fedi et al. 2008). In fact the dibenzothiazepine skeleton has a broad spectrum of medical applications; its derivatives are used to treat schizophrenia and also find applications as neuroleptics, antidepressants, antihistaminic, just to name a few (Ganesh et al. 2011; Pettersson et al. 2009; Riedel et al. 2007; Warawa et al. 2001). Within our programme research concerning the structural elucidation of tricyclic molecules in order to gain further insights into structure–property relationships (Altamura et al., 2008; Altamura et al., 2009; Altamura et al., 2011) we investigated the crystal and molecular structure of the title compound. The overall shape of the tricyclic skeleton is controlled both by the conformation of the central seven-membered ring and the relative arrangement of the aromatic rings bound to it. The central thiazepine ring adopts the usual boat conformation, with C1, C2, C8 and C9 as the basal plane, the S atom as the bow and the N1—C7 bond as the stern. The deviation from a pure boat conformation is quite small as can be seen from the asymmetry index ΔCs which is 3.97°; the bow angle is 51.04 (8)° and the stern angle is 41.89 (8)°(Duax et al. 1976; Bertolasi et al. 1982; Ravikumar & Sridhar, 2005). Finally, the dihedral angle between the benzene rings is 104.08 (5)°. As a consequence the dibenzothiazepine tricyclic skeleton assumes an overall butterfly-like shape (Fig. 1). The N2—C7 bond is shorter than an usual N—C single bond [1.368 (2) Å compared to 1.416 Å (Allen et al. 1987)] and the sum of the bond angles about N2 is 358°; consequently, N2 has a partial sp2 character. Molecular chains, which propagate along the c axis, are formed through intermolecular interactions (Fig. 2): a weak C—Haromatic···N contact which involves the imine nitrogen atom as acceptor and a quite short C—H···π interaction (Table 1).
Experimental
The synthesis of the title compound started from the commercially available tricyclic lactam (dibenzo[b,f][1,4]thiazepin-11(10H)-one) (0.89 g, 3.91 mmol), that was dissolved into 10 ml of phosphorus oxychloride and refluxed for two hours under nitrogen atmosphere. After removal of excess POCl3 under vacuum, the corresponding iminochloride (11-chlorodibenzo[b,f][1,4]thiazepine) was obtained as a yellow oil, that was dissolved in 20 ml of anhydrous toluene. An excess of diethylamine (20 ml, 191 mmol) was added and the mixture refluxed until complete conversion (monitored by HPLC; about 6 h were needed for complete conversion). After removal of the solvent in vacuo and purification by flash chromatography (eluent: gradient CH2Cl2/cyclohexane from 50:50 to 100% CH2Cl2), dibenzo[b,f][1,4]thiazepin-11-yl-diethyl-amine was obtained as a yellow oil (0.66 g, 60% yield), which became a white solid on standing at room temperature for several days. Crystals suitable for single-crystal X-ray diffraction analysis were obtained by slow evaporation of a water/DMSO solution of dibenzo[b,f][1,4]thiazepin-11-yl-diethyl-amine.
Refinement
All the H atoms were positioned with idealized geometry using a riding model and refined with Uiso(H) 1.2 times Ueq(C) (1.5 for methyl H atoms).
Figures
Fig. 1.
Crystal structure of the title compound with labelling and displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
Crystal structure of the title compound with view along the b-axis. Intermolecular H-bonding interactions are shown as dashed lines.
Crystal data
| C17H18N2S | F(000) = 600 |
| Mr = 282.40 | Dx = 1.294 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.5418 Å |
| a = 12.0137 (2) Å | Cell parameters from 3716 reflections |
| b = 8.2257 (1) Å | θ = 3.8–64.6° |
| c = 15.0513 (2) Å | µ = 1.89 mm−1 |
| β = 102.952 (1)° | T = 150 K |
| V = 1449.54 (4) Å3 | Platelet, colourless |
| Z = 4 | 0.20 × 0.18 × 0.03 mm |
Data collection
| Oxford Diffraction XcaliburPX diffractometer | 2364 independent reflections |
| Radiation source: Enhance (Cu) X-ray Source | 1837 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.019 |
| Detector resolution: 8.1241 pixels mm-1 | θmax = 64.7°, θmin = 3.8° |
| ω scans | h = −13→11 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2006) | k = −9→9 |
| Tmin = 0.722, Tmax = 0.945 | l = −16→17 |
| 6173 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.032 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.086 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0564P)2] where P = (Fo2 + 2Fc2)/3 |
| 2364 reflections | (Δ/σ)max < 0.001 |
| 181 parameters | Δρmax = 0.23 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.83045 (3) | 0.13292 (5) | 0.85602 (3) | 0.01958 (15) | |
| N1 | 0.67962 (11) | 0.44375 (17) | 0.84814 (9) | 0.0179 (3) | |
| N2 | 0.51660 (11) | 0.33602 (17) | 0.87760 (9) | 0.0181 (3) | |
| C1 | 0.69759 (13) | 0.2400 (2) | 0.97172 (11) | 0.0177 (4) | |
| C2 | 0.78691 (14) | 0.1378 (2) | 0.96160 (11) | 0.0175 (4) | |
| C3 | 0.84541 (14) | 0.0447 (2) | 1.03477 (11) | 0.0198 (4) | |
| H3 | 0.9047 | −0.0264 | 1.0268 | 0.024* | |
| C4 | 0.81731 (14) | 0.0556 (2) | 1.11916 (11) | 0.0211 (4) | |
| H4 | 0.8564 | −0.0089 | 1.1687 | 0.025* | |
| C5 | 0.73182 (14) | 0.1613 (2) | 1.13056 (11) | 0.0211 (4) | |
| H5 | 0.7137 | 0.1715 | 1.1886 | 0.025* | |
| C6 | 0.67280 (13) | 0.2522 (2) | 1.05782 (11) | 0.0197 (4) | |
| H6 | 0.6143 | 0.3241 | 1.0666 | 0.024* | |
| C7 | 0.63322 (13) | 0.3418 (2) | 0.89419 (11) | 0.0169 (4) | |
| C8 | 0.79782 (14) | 0.4649 (2) | 0.85930 (11) | 0.0188 (4) | |
| C9 | 0.87701 (14) | 0.3391 (2) | 0.85923 (11) | 0.0188 (4) | |
| C10 | 0.99058 (14) | 0.3727 (2) | 0.85997 (11) | 0.0237 (4) | |
| H10 | 1.0430 | 0.2861 | 0.8604 | 0.028* | |
| C11 | 1.02769 (15) | 0.5322 (2) | 0.86002 (12) | 0.0290 (5) | |
| H11 | 1.1051 | 0.5551 | 0.8596 | 0.035* | |
| C12 | 0.95124 (15) | 0.6578 (2) | 0.86075 (12) | 0.0280 (5) | |
| H12 | 0.9767 | 0.7673 | 0.8618 | 0.034* | |
| C13 | 0.83815 (14) | 0.6251 (2) | 0.86002 (11) | 0.0218 (4) | |
| H13 | 0.7867 | 0.7128 | 0.8600 | 0.026* | |
| C14 | 0.45010 (14) | 0.2064 (2) | 0.90866 (11) | 0.0200 (4) | |
| H14A | 0.5032 | 0.1237 | 0.9421 | 0.024* | |
| H14B | 0.4014 | 0.1529 | 0.8548 | 0.024* | |
| C15 | 0.37461 (15) | 0.2692 (2) | 0.97035 (12) | 0.0252 (4) | |
| H15A | 0.3324 | 0.1782 | 0.9891 | 0.038* | |
| H15B | 0.3206 | 0.3493 | 0.9371 | 0.038* | |
| H15C | 0.4224 | 0.3203 | 1.0245 | 0.038* | |
| C16 | 0.45022 (14) | 0.4423 (2) | 0.80635 (11) | 0.0188 (4) | |
| H16A | 0.4865 | 0.5510 | 0.8112 | 0.023* | |
| H16B | 0.3723 | 0.4555 | 0.8170 | 0.023* | |
| C17 | 0.44140 (14) | 0.3772 (2) | 0.71044 (11) | 0.0209 (4) | |
| H17A | 0.3965 | 0.4527 | 0.6661 | 0.031* | |
| H17B | 0.4039 | 0.2707 | 0.7046 | 0.031* | |
| H17C | 0.5181 | 0.3662 | 0.6988 | 0.031* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0205 (2) | 0.0207 (3) | 0.0190 (2) | −0.00074 (18) | 0.00747 (17) | −0.00055 (18) |
| N1 | 0.0153 (7) | 0.0199 (8) | 0.0191 (7) | −0.0019 (6) | 0.0049 (6) | −0.0011 (6) |
| N2 | 0.0153 (7) | 0.0197 (8) | 0.0196 (8) | −0.0014 (6) | 0.0042 (6) | 0.0016 (6) |
| C1 | 0.0148 (8) | 0.0195 (9) | 0.0187 (9) | −0.0059 (7) | 0.0034 (7) | −0.0009 (7) |
| C2 | 0.0163 (9) | 0.0188 (9) | 0.0177 (9) | −0.0058 (7) | 0.0041 (7) | −0.0019 (7) |
| C3 | 0.0170 (9) | 0.0189 (10) | 0.0232 (9) | −0.0023 (7) | 0.0041 (7) | −0.0009 (8) |
| C4 | 0.0190 (10) | 0.0234 (10) | 0.0190 (9) | −0.0051 (8) | 0.0003 (7) | 0.0029 (8) |
| C5 | 0.0192 (9) | 0.0286 (10) | 0.0156 (9) | −0.0079 (8) | 0.0043 (7) | −0.0029 (7) |
| C6 | 0.0154 (9) | 0.0233 (10) | 0.0202 (9) | −0.0046 (8) | 0.0035 (7) | −0.0041 (7) |
| C7 | 0.0172 (9) | 0.0175 (9) | 0.0162 (8) | 0.0000 (7) | 0.0043 (7) | −0.0041 (7) |
| C8 | 0.0177 (9) | 0.0232 (10) | 0.0150 (8) | −0.0038 (7) | 0.0023 (7) | −0.0002 (7) |
| C9 | 0.0190 (9) | 0.0230 (10) | 0.0144 (8) | −0.0041 (7) | 0.0037 (7) | 0.0013 (7) |
| C10 | 0.0171 (9) | 0.0309 (11) | 0.0230 (9) | 0.0009 (8) | 0.0044 (7) | 0.0037 (8) |
| C11 | 0.0164 (9) | 0.0389 (12) | 0.0312 (11) | −0.0071 (9) | 0.0044 (8) | 0.0044 (9) |
| C12 | 0.0235 (10) | 0.0276 (11) | 0.0316 (11) | −0.0108 (8) | 0.0035 (8) | 0.0012 (9) |
| C13 | 0.0201 (9) | 0.0207 (10) | 0.0247 (10) | −0.0018 (8) | 0.0055 (8) | −0.0008 (8) |
| C14 | 0.0177 (9) | 0.0208 (10) | 0.0211 (9) | −0.0025 (8) | 0.0039 (7) | −0.0011 (8) |
| C15 | 0.0229 (10) | 0.0295 (11) | 0.0250 (9) | −0.0061 (8) | 0.0090 (8) | 0.0001 (8) |
| C16 | 0.0159 (9) | 0.0172 (9) | 0.0233 (9) | 0.0016 (7) | 0.0046 (7) | 0.0018 (7) |
| C17 | 0.0182 (9) | 0.0224 (10) | 0.0214 (9) | 0.0024 (8) | 0.0031 (7) | 0.0020 (7) |
Geometric parameters (Å, º)
| S1—C2 | 1.7814 (16) | C9—C10 | 1.390 (2) |
| S1—C9 | 1.7831 (18) | C10—C11 | 1.385 (3) |
| N1—C7 | 1.291 (2) | C10—H10 | 0.9500 |
| N1—C8 | 1.403 (2) | C11—C12 | 1.384 (3) |
| N2—C7 | 1.368 (2) | C11—H11 | 0.9500 |
| N2—C14 | 1.470 (2) | C12—C13 | 1.383 (2) |
| N2—C16 | 1.472 (2) | C12—H12 | 0.9500 |
| C1—C6 | 1.397 (2) | C13—H13 | 0.9500 |
| C1—C2 | 1.398 (2) | C14—C15 | 1.526 (2) |
| C1—C7 | 1.502 (2) | C14—H14A | 0.9900 |
| C2—C3 | 1.395 (2) | C14—H14B | 0.9900 |
| C3—C4 | 1.388 (2) | C15—H15A | 0.9800 |
| C3—H3 | 0.9500 | C15—H15B | 0.9800 |
| C4—C5 | 1.385 (2) | C15—H15C | 0.9800 |
| C4—H4 | 0.9500 | C16—C17 | 1.521 (2) |
| C5—C6 | 1.383 (2) | C16—H16A | 0.9900 |
| C5—H5 | 0.9500 | C16—H16B | 0.9900 |
| C6—H6 | 0.9500 | C17—H17A | 0.9800 |
| C8—C13 | 1.403 (2) | C17—H17B | 0.9800 |
| C8—C9 | 1.406 (2) | C17—H17C | 0.9800 |
| C2—S1—C9 | 96.16 (8) | C9—C10—H10 | 119.9 |
| C7—N1—C8 | 124.28 (14) | C12—C11—C10 | 119.53 (17) |
| C7—N2—C14 | 125.08 (14) | C12—C11—H11 | 120.2 |
| C7—N2—C16 | 118.54 (14) | C10—C11—H11 | 120.2 |
| C14—N2—C16 | 114.71 (13) | C13—C12—C11 | 120.46 (17) |
| C6—C1—C2 | 118.19 (15) | C13—C12—H12 | 119.8 |
| C6—C1—C7 | 120.08 (15) | C11—C12—H12 | 119.8 |
| C2—C1—C7 | 121.64 (14) | C12—C13—C8 | 121.32 (17) |
| C3—C2—C1 | 120.48 (15) | C12—C13—H13 | 119.3 |
| C3—C2—S1 | 119.67 (13) | C8—C13—H13 | 119.3 |
| C1—C2—S1 | 119.79 (13) | N2—C14—C15 | 112.82 (14) |
| C4—C3—C2 | 120.26 (16) | N2—C14—H14A | 109.0 |
| C4—C3—H3 | 119.9 | C15—C14—H14A | 109.0 |
| C2—C3—H3 | 119.9 | N2—C14—H14B | 109.0 |
| C5—C4—C3 | 119.52 (16) | C15—C14—H14B | 109.0 |
| C5—C4—H4 | 120.2 | H14A—C14—H14B | 107.8 |
| C3—C4—H4 | 120.2 | C14—C15—H15A | 109.5 |
| C6—C5—C4 | 120.27 (15) | C14—C15—H15B | 109.5 |
| C6—C5—H5 | 119.9 | H15A—C15—H15B | 109.5 |
| C4—C5—H5 | 119.9 | C14—C15—H15C | 109.5 |
| C5—C6—C1 | 121.19 (16) | H15A—C15—H15C | 109.5 |
| C5—C6—H6 | 119.4 | H15B—C15—H15C | 109.5 |
| C1—C6—H6 | 119.4 | N2—C16—C17 | 113.16 (14) |
| N1—C7—N2 | 118.19 (15) | N2—C16—H16A | 108.9 |
| N1—C7—C1 | 124.74 (14) | C17—C16—H16A | 108.9 |
| N2—C7—C1 | 116.77 (14) | N2—C16—H16B | 108.9 |
| N1—C8—C13 | 117.10 (16) | C17—C16—H16B | 108.9 |
| N1—C8—C9 | 125.17 (16) | H16A—C16—H16B | 107.8 |
| C13—C8—C9 | 117.30 (15) | C16—C17—H17A | 109.5 |
| C10—C9—C8 | 121.12 (16) | C16—C17—H17B | 109.5 |
| C10—C9—S1 | 119.41 (14) | H17A—C17—H17B | 109.5 |
| C8—C9—S1 | 119.46 (13) | C16—C17—H17C | 109.5 |
| C11—C10—C9 | 120.26 (17) | H17A—C17—H17C | 109.5 |
| C11—C10—H10 | 119.9 | H17B—C17—H17C | 109.5 |
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C8–C12 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···N1i | 0.95 | 2.70 | 3.576 (2) | 154 |
| C4—H4···Cgi | 0.95 | 2.81 | 3.5759 (18) | 139 |
Symmetry code: (i) x, −y+1/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NC2296).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Altamura, M., Dapporto, P., Guidi, A., Harmat, N., Jerry, L., Libralesso, E., Paoli, P. & Rossi, P. (2008). New J. Chem. 32, 1617–1627.
- Altamura, M., Fedi, V., Giannotti, D., Paoli, P. & Rossi, P. (2009). New J. Chem. 33, 2219–2231.
- Altamura, M., Guidi, A., Jerry, L., Paoli, P. & Rossi, P. (2011). CrystEngComm, 13, 2310–2317.
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst. 32, 115–119.
- Bertolasi, V., Ferretti, V., Gilli, G. & Borea, P. A. (1982). Cryst. Struct. Commun. 11, 1481–1486.
- Duax, W. L., Weeks, C. M. & Rohrer, D. C. (1976). Topics in Stereochemistry, edited by E. L. Eliel and N. Allinger. New York: John Wiley & Sons.
- Evans, B. E., Rittle, K. E., Bock, M. G., DiPardo, R. M., Freidinger, R. M., Whittle, W. L., Lundell, G. F., Veber, D. F., Anderson, P. S., Chang, R. S. L., Lotti, V. J., Cerino, D. J., Chen, T. B., Kling, P. J., Kunkel, K. A., Springer, J. P. & Hirshfield, J. (1988). J. Med. Chem. 31, 2235–2246.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Fedi, V., Guidi, A. & Altamura, M. (2008). Mini Rev. Med. Chem. 8, 1464–1484. [DOI] [PubMed]
- Ganesh, D. M., Yogesh, M. K., Ashok, K., Dharmendra, S., Kisan, M. K. & Suresh, B. M. (2011). Indian J. Chem. 50B, 1196–, 1201.
- Nardelli, M. (1995). J. Appl. Cryst. 28, 659.
- Oxford Diffraction (2006). CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd, Abingdon, England.
- Patchett, A. A. & Nargund, R. P. (2000). Annu. Rep. Med. Chem. 35, 289–298.
- Pettersson, H., Bulow, A., Ek, F., Jensen, J., Ottesen, L. K., Fejzic, A., Ma, J.-N., Del Tredici, A. L., Currier, E. A., Gardell, L. R., Tabatabaei, A., Craig, D., McFarland, K., Ott, T. R., Piu, F., Burstein, E. S. & Olsson, R. (2009). J. Med. Chem. 52, 1975–1982. [DOI] [PubMed]
- Ravikumar, K. & Sridhar, B. (2005). Acta Cryst. E61, o3245–o3248. [DOI] [PubMed]
- Riedel, M., Mueller, N., Strassnig, M., Spellman, I., Severus, E. & Moeller, H.-J. (2007). Neuropsyc. Dis. Treat. 3, 219–235. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Warawa, E. J., Migler, B. M., Ohnmacht, C. J., Needles, A. L., Gatos, G. C., Mclaren, F. M., Nelson, C. L. & Kirkland, K. M. (2001). J. Med. Chem. 44, 372–389. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, New_Global_Publ_Block. DOI: 10.1107/S1600536812042328/nc2296sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812042328/nc2296Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report