Abstract
In the title compound, C21H26ClN2O4S.Cl, also known as tianeptine hydrochloride, the seven-membered ring adopts a boat conformation. The dihedral angle between the mean planes of the benzene rings is 44.44 (7)°. There is an intramolecular hydrogen bond formed via S= O⋯H—N. In the crystal, molecules are connected via pairs of N—H.·O, N—H⋯Cl and O—H⋯Cl hydrogen bonds, forming inversion dimers, which are consolidated by C—H⋯O interactions. The dimers are linked by C—H⋯O and C—H⋯Cl interactions, forming a two-dimensional network lying parallel to (011).
Related literature
For general information about tianeptine and its preparation, see: Guzman et al. (2010 ▶). For related structures, see: Orola et al. (2012 ▶).
Experimental
Crystal data
C21H26ClN2O4S+·Cl−
M r = 473.40
Triclinic,
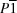
a = 9.5439 (2) Å
b = 10.0910 (2) Å
c = 13.1802 (3) Å
α = 104.000 (1)°
β = 101.538 (1)°
γ = 105.018 (1)°
V = 1139.04 (4) Å3
Z = 2
Mo Kα radiation
μ = 0.41 mm−1
T = 190 K
0.24 × 0.20 × 0.14 mm
Data collection
Nonius KappaCCD diffractometer
7552 measured reflections
4985 independent reflections
4015 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.100
S = 1.03
4985 reflections
273 parameters
H-atom parameters constrained
Δρmax = 0.76 e Å−3
Δρmin = −0.34 e Å−3
Data collection: COLLECT (Hooft, 1998 ▶); cell refinement: HKL DENZO (Otwinowski & Minor, 1997 ▶); data reduction: SCALEPACK (Otwinowski & Minor, 1997 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: SHELXL97 and publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global. DOI: 10.1107/S1600536812042432/pv2586sup1.cif
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H3A⋯Cl2i | 0.90 | 2.31 | 3.154 (2) | 157 |
| N2—H3B⋯O3ii | 0.90 | 2.32 | 2.821 (2) | 115 |
| C16—H11B⋯O3ii | 0.97 | 2.56 | 3.201 (2) | 124 |
| O4—H6⋯Cl2iii | 0.82 | 2.22 | 3.043 (2) | 176 |
| C4—H3⋯Cl2iv | 0.93 | 2.82 | 3.651 (2) | 150 |
| C18—H6A⋯O4v | 0.97 | 2.56 | 3.467 (2) | 157 |
| C7—H7⋯Cl2 | 0.98 | 2.59 | 3.534 (2) | 162 |
| N2—H3B⋯O2 | 0.90 | 2.02 | 2.802 (2) | 144 |
Symmetry codes: (i) 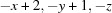 ; (ii)
; (ii) 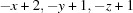 ; (iii)
; (iii) 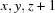 ; (iv)
; (iv) 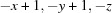 ; (v)
; (v) 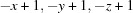 .
.
Acknowledgments
This work was supported by the European Regional Development Fund (No. 2011/0014/2DP/2.1.1.1.0/10/APIA/VIAA/092).
supplementary crystallographic information
Comment
Tianeptine salts are of wide interest since they are crystalline. Although the synthesis of the title compound, tianeptine hydrochloride, has been described (Guzman et al., 2010) we describe in this article an improved method of its synthesis and its crystal structure.
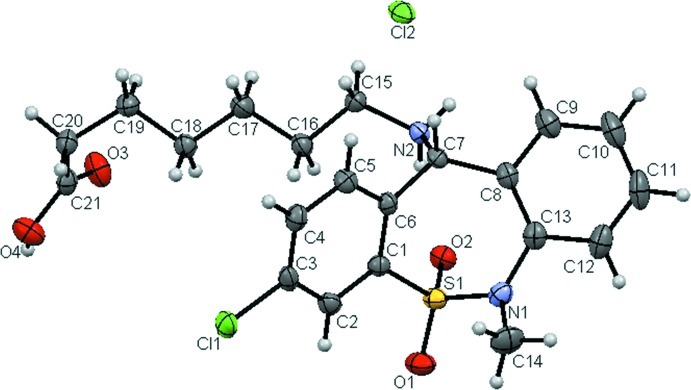
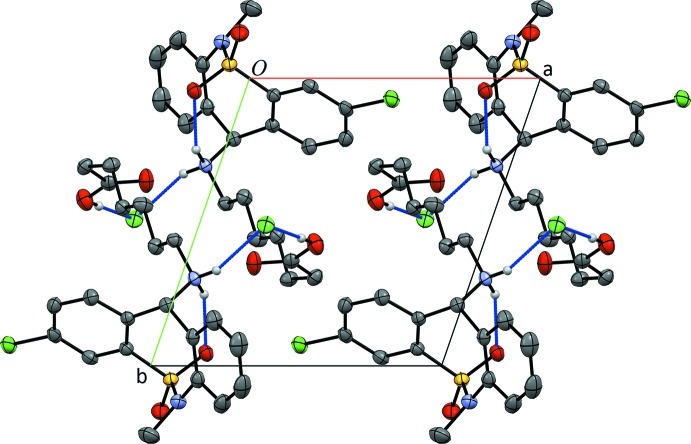
In the title compound (Fig. 1), the seven-membered ring adopts a boat conformation with the values of torsion angles: C1—S1—N1—C13 = -78.4 (2) and C1—C6—C7—C8 = -56.3 (3)°. The dihedral angle between the mean planes of the two benzene rings (C1–C6 and C8–C13) is 44.44 (7)°. There is an intramolecular hydrogen bond in the title molecule which is formed via S1═O2···H3B—N2 that stabilizes the molecular structure. In the crystal, the molecules are connected via hydrogen bonds between carboxyl and amine groups and chloride anion, O4—H6···Cl2, N2—H3A···Cl2 and N2—H3B···O3. The crystal structure is further consolidated by intermolecular interactions, C18—H6A···O4, C4—H3···Cl2, C16—H11B···O3 and C7—H7···Cl2 (Table 1 and Fig. 2). The supramolecular structure of tianeptine hydrochloride consists of parallel oriented tricyclic fragment and parallel oriented carbon atom chains (heptanoic acid). Carbon atom chains are linked with hydrogen bonds via chloride anion, amine and carboxylgroup. The torsion angle C8—C7—N2—C15 is -168.4 (2)° so that the carbon atom chain C15—C20 is almost parallel to the benzene ring C8—C13.
The crystal structures of tianeptine polymorphs have been reported recently (Orola et al., 2012). The title structure is more similar with polymorph A structure in which tianeptine molecules are linked via hydrogen bonds between amine and carboxyl groups. The tianeptine molecules in the structure of tianeptine polymorph B are in a zwiterrion form.
Experimental
Tianeptine sodium salt (0.5 g;1.09 mmol) was dissolved in 20 ml deionized water in a Erlenmeyer flask and added ~3 mmol of hydrochloric acid. Mixture were stirred for 6 h. After 6 h suspension was filtered and washed with cold water. The product was dried and recrystallized from water by slow evaporation at room temperature.
Refinement
All hydrogen atoms were positioned geometrically with C—H distances ranging from 0.93 to 0.97 Å and refined as riding on their parent atoms with Uiso(H) = 1.5 Ueq(C) for methyl groups and Uiso(H) = 1.2 Ueq(C) for others.
Figures
Fig. 1.
The molecular structure of the title compound showing 50% probability ellipsoids and hydrogen atoms are shown as small spheres of arbitrary radii.
Fig. 2.
Packing diagram of the title compound viewed along the c axis. Blue lines indicate hydrogen bonds.
Crystal data
| C21H26ClN2O4S+·Cl− | Z = 2 |
| Mr = 473.40 | F(000) = 496 |
| Triclinic, P1 | Dx = 1.380 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.5439 (2) Å | Cell parameters from 6759 reflections |
| b = 10.0910 (2) Å | θ = 1.0–27.1° |
| c = 13.1802 (3) Å | µ = 0.41 mm−1 |
| α = 104.4000 (12)° | T = 190 K |
| β = 101.538 (1)° | Plate, colourless |
| γ = 105.0180 (11)° | 0.24 × 0.20 × 0.14 mm |
| V = 1139.04 (4) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 4015 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.021 |
| Graphite monochromator | θmax = 27.1°, θmin = 3.1° |
| CCD scans | h = −11→12 |
| 7552 measured reflections | k = −12→12 |
| 4985 independent reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.100 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0363P)2 + 0.641P] where P = (Fo2 + 2Fc2)/3 |
| 4985 reflections | (Δ/σ)max = 0.018 |
| 273 parameters | Δρmax = 0.76 e Å−3 |
| 0 restraints | Δρmin = −0.34 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.48761 (6) | 0.92769 (6) | 0.32682 (5) | 0.03721 (15) | |
| S1 | 1.08561 (5) | 1.04778 (5) | 0.36222 (4) | 0.02568 (12) | |
| Cl2 | 0.77093 (5) | 0.48993 (5) | −0.09234 (4) | 0.03147 (13) | |
| O2 | 1.17517 (14) | 0.95570 (14) | 0.37600 (11) | 0.0274 (3) | |
| N2 | 1.04759 (17) | 0.69630 (16) | 0.20172 (13) | 0.0218 (3) | |
| H3A | 1.1139 | 0.6682 | 0.1689 | 0.026* | |
| H3B | 1.0994 | 0.7516 | 0.2708 | 0.026* | |
| O4 | 0.57739 (17) | 0.42297 (18) | 0.67606 (12) | 0.0375 (4) | |
| H6 | 0.6331 | 0.4443 | 0.7378 | 0.056* | |
| O1 | 1.08744 (17) | 1.15746 (16) | 0.45495 (12) | 0.0371 (4) | |
| O3 | 0.76590 (16) | 0.35639 (18) | 0.62834 (12) | 0.0382 (4) | |
| N1 | 1.13800 (18) | 1.12491 (17) | 0.27471 (14) | 0.0281 (4) | |
| C5 | 0.7141 (2) | 0.7622 (2) | 0.13441 (16) | 0.0264 (4) | |
| H2 | 0.6903 | 0.6926 | 0.0667 | 0.032* | |
| C4 | 0.5978 (2) | 0.7900 (2) | 0.17507 (17) | 0.0288 (4) | |
| H3 | 0.4974 | 0.7398 | 0.1352 | 0.035* | |
| C6 | 0.8655 (2) | 0.83537 (19) | 0.19193 (15) | 0.0212 (4) | |
| C2 | 0.7818 (2) | 0.9710 (2) | 0.33545 (16) | 0.0265 (4) | |
| H5 | 0.8047 | 1.0416 | 0.4025 | 0.032* | |
| C18 | 0.7174 (2) | 0.4501 (2) | 0.40383 (16) | 0.0266 (4) | |
| H6A | 0.6521 | 0.5092 | 0.4011 | 0.032* | |
| H6B | 0.7988 | 0.4970 | 0.4713 | 0.032* | |
| C7 | 0.9826 (2) | 0.7887 (2) | 0.14329 (15) | 0.0216 (4) | |
| H7 | 0.9253 | 0.7226 | 0.0698 | 0.026* | |
| C3 | 0.6328 (2) | 0.8933 (2) | 0.27553 (17) | 0.0272 (4) | |
| C21 | 0.6408 (2) | 0.3647 (2) | 0.60345 (16) | 0.0261 (4) | |
| C20 | 0.5396 (2) | 0.3075 (2) | 0.48869 (16) | 0.0282 (4) | |
| H10A | 0.4775 | 0.3685 | 0.4796 | 0.034* | |
| H10B | 0.4726 | 0.2109 | 0.4761 | 0.034* | |
| C16 | 0.8769 (2) | 0.5855 (2) | 0.30509 (16) | 0.0268 (4) | |
| H11A | 0.8158 | 0.6486 | 0.3035 | 0.032* | |
| H11B | 0.9622 | 0.6312 | 0.3702 | 0.032* | |
| C1 | 0.8963 (2) | 0.9404 (2) | 0.29264 (15) | 0.0228 (4) | |
| C15 | 0.9334 (2) | 0.5636 (2) | 0.20475 (16) | 0.0249 (4) | |
| H13A | 0.9785 | 0.4873 | 0.2007 | 0.030* | |
| H13B | 0.8475 | 0.5310 | 0.1405 | 0.030* | |
| C13 | 1.1778 (2) | 1.0475 (2) | 0.18368 (16) | 0.0273 (4) | |
| C19 | 0.6274 (2) | 0.3021 (2) | 0.40408 (16) | 0.0291 (4) | |
| H15A | 0.6963 | 0.2486 | 0.4177 | 0.035* | |
| H15B | 0.5570 | 0.2497 | 0.3322 | 0.035* | |
| C8 | 1.1112 (2) | 0.8998 (2) | 0.12618 (15) | 0.0244 (4) | |
| C17 | 0.7835 (3) | 0.4409 (2) | 0.30818 (18) | 0.0338 (5) | |
| H17A | 0.7014 | 0.3949 | 0.2411 | 0.041* | |
| H17B | 0.8465 | 0.3796 | 0.3104 | 0.041* | |
| C9 | 1.1641 (2) | 0.8421 (3) | 0.03912 (17) | 0.0338 (5) | |
| H18 | 1.1217 | 0.7438 | 0.0004 | 0.041* | |
| C14 | 1.0767 (3) | 1.2418 (2) | 0.2609 (2) | 0.0444 (6) | |
| H19A | 1.1297 | 1.2932 | 0.2203 | 0.067* | |
| H19B | 1.0895 | 1.3074 | 0.3313 | 0.067* | |
| H19C | 0.9710 | 1.2007 | 0.2221 | 0.067* | |
| C12 | 1.2910 (2) | 1.1316 (3) | 0.1514 (2) | 0.0381 (5) | |
| H20 | 1.3341 | 1.2301 | 0.1891 | 0.046* | |
| C10 | 1.2774 (3) | 0.9263 (3) | 0.0089 (2) | 0.0437 (6) | |
| H21 | 1.3110 | 0.8851 | −0.0489 | 0.052* | |
| C11 | 1.3399 (3) | 1.0725 (3) | 0.0656 (2) | 0.0437 (6) | |
| H22 | 1.4151 | 1.1306 | 0.0455 | 0.052* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0319 (3) | 0.0313 (3) | 0.0597 (4) | 0.0146 (2) | 0.0278 (3) | 0.0173 (3) |
| S1 | 0.0248 (2) | 0.0244 (2) | 0.0245 (2) | 0.00797 (19) | 0.00501 (19) | 0.0035 (2) |
| Cl2 | 0.0313 (3) | 0.0342 (3) | 0.0234 (2) | 0.0100 (2) | 0.0077 (2) | 0.0005 (2) |
| O2 | 0.0264 (7) | 0.0312 (7) | 0.0254 (7) | 0.0130 (6) | 0.0048 (6) | 0.0086 (6) |
| N2 | 0.0221 (8) | 0.0241 (8) | 0.0221 (8) | 0.0099 (6) | 0.0091 (6) | 0.0075 (7) |
| O4 | 0.0396 (9) | 0.0494 (9) | 0.0282 (8) | 0.0243 (8) | 0.0087 (7) | 0.0107 (7) |
| O1 | 0.0359 (8) | 0.0334 (8) | 0.0324 (8) | 0.0106 (7) | 0.0060 (6) | −0.0025 (7) |
| O3 | 0.0293 (8) | 0.0611 (10) | 0.0343 (8) | 0.0210 (7) | 0.0112 (6) | 0.0242 (8) |
| N1 | 0.0265 (8) | 0.0238 (8) | 0.0337 (9) | 0.0080 (7) | 0.0070 (7) | 0.0102 (7) |
| C5 | 0.0272 (10) | 0.0288 (10) | 0.0235 (10) | 0.0096 (8) | 0.0062 (8) | 0.0091 (8) |
| C4 | 0.0223 (10) | 0.0304 (11) | 0.0353 (11) | 0.0078 (8) | 0.0088 (8) | 0.0132 (9) |
| C6 | 0.0222 (9) | 0.0227 (9) | 0.0225 (9) | 0.0088 (7) | 0.0083 (7) | 0.0108 (8) |
| C2 | 0.0321 (10) | 0.0236 (10) | 0.0290 (10) | 0.0129 (8) | 0.0132 (8) | 0.0096 (8) |
| C18 | 0.0287 (10) | 0.0273 (10) | 0.0288 (10) | 0.0114 (8) | 0.0123 (8) | 0.0119 (9) |
| C7 | 0.0234 (9) | 0.0242 (9) | 0.0180 (9) | 0.0085 (7) | 0.0065 (7) | 0.0068 (8) |
| C3 | 0.0277 (10) | 0.0262 (10) | 0.0398 (11) | 0.0140 (8) | 0.0192 (9) | 0.0179 (9) |
| C21 | 0.0273 (10) | 0.0261 (10) | 0.0300 (10) | 0.0086 (8) | 0.0106 (8) | 0.0160 (9) |
| C20 | 0.0254 (10) | 0.0293 (10) | 0.0291 (10) | 0.0047 (8) | 0.0082 (8) | 0.0119 (9) |
| C16 | 0.0299 (10) | 0.0284 (10) | 0.0264 (10) | 0.0109 (8) | 0.0135 (8) | 0.0103 (9) |
| C1 | 0.0230 (9) | 0.0219 (9) | 0.0249 (9) | 0.0086 (7) | 0.0072 (7) | 0.0086 (8) |
| C15 | 0.0274 (10) | 0.0221 (9) | 0.0271 (10) | 0.0081 (8) | 0.0112 (8) | 0.0083 (8) |
| C13 | 0.0233 (10) | 0.0323 (11) | 0.0311 (10) | 0.0111 (8) | 0.0076 (8) | 0.0163 (9) |
| C19 | 0.0324 (11) | 0.0262 (10) | 0.0272 (10) | 0.0064 (8) | 0.0107 (8) | 0.0074 (9) |
| C8 | 0.0227 (9) | 0.0314 (10) | 0.0235 (9) | 0.0114 (8) | 0.0072 (8) | 0.0133 (8) |
| C17 | 0.0452 (12) | 0.0280 (11) | 0.0340 (11) | 0.0132 (9) | 0.0209 (10) | 0.0105 (9) |
| C9 | 0.0341 (11) | 0.0434 (12) | 0.0309 (11) | 0.0161 (10) | 0.0141 (9) | 0.0162 (10) |
| C14 | 0.0553 (15) | 0.0302 (12) | 0.0531 (15) | 0.0198 (11) | 0.0141 (12) | 0.0171 (11) |
| C12 | 0.0286 (11) | 0.0400 (13) | 0.0507 (14) | 0.0077 (9) | 0.0107 (10) | 0.0270 (12) |
| C10 | 0.0401 (13) | 0.0665 (17) | 0.0386 (13) | 0.0212 (12) | 0.0233 (11) | 0.0272 (13) |
| C11 | 0.0323 (12) | 0.0622 (17) | 0.0528 (15) | 0.0146 (11) | 0.0216 (11) | 0.0391 (14) |
Geometric parameters (Å, º)
| Cl1—C3 | 1.7332 (19) | C7—H7 | 0.9800 |
| S1—O1 | 1.4240 (15) | C21—C20 | 1.499 (3) |
| S1—O2 | 1.4354 (14) | C20—C19 | 1.522 (3) |
| S1—N1 | 1.6315 (17) | C20—H10A | 0.9700 |
| S1—C1 | 1.7629 (19) | C20—H10B | 0.9700 |
| N2—C15 | 1.507 (2) | C16—C15 | 1.513 (3) |
| N2—C7 | 1.521 (2) | C16—C17 | 1.515 (3) |
| N2—H3A | 0.9000 | C16—H11A | 0.9700 |
| N2—H3B | 0.9000 | C16—H11B | 0.9700 |
| O4—C21 | 1.328 (2) | C15—H13A | 0.9700 |
| O4—H6 | 0.8200 | C15—H13B | 0.9700 |
| O3—C21 | 1.204 (2) | C13—C8 | 1.397 (3) |
| N1—C13 | 1.436 (3) | C13—C12 | 1.400 (3) |
| N1—C14 | 1.480 (3) | C19—H15A | 0.9700 |
| C5—C4 | 1.387 (3) | C19—H15B | 0.9700 |
| C5—C6 | 1.392 (3) | C8—C9 | 1.401 (3) |
| C5—H2 | 0.9300 | C17—H17A | 0.9700 |
| C4—C3 | 1.381 (3) | C17—H17B | 0.9700 |
| C4—H3 | 0.9300 | C9—C10 | 1.384 (3) |
| C6—C1 | 1.398 (3) | C9—H18 | 0.9300 |
| C6—C7 | 1.513 (2) | C14—H19A | 0.9600 |
| C2—C3 | 1.388 (3) | C14—H19B | 0.9600 |
| C2—C1 | 1.393 (3) | C14—H19C | 0.9600 |
| C2—H5 | 0.9300 | C12—C11 | 1.369 (3) |
| C18—C17 | 1.512 (3) | C12—H20 | 0.9300 |
| C18—C19 | 1.518 (3) | C10—C11 | 1.381 (4) |
| C18—H6A | 0.9700 | C10—H21 | 0.9300 |
| C18—H6B | 0.9700 | C11—H22 | 0.9300 |
| C7—C8 | 1.529 (2) | ||
| O1—S1—O2 | 119.59 (9) | C15—C16—C17 | 109.79 (16) |
| O1—S1—N1 | 108.15 (9) | C15—C16—H11A | 109.7 |
| O2—S1—N1 | 106.88 (8) | C17—C16—H11A | 109.7 |
| O1—S1—C1 | 108.63 (9) | C15—C16—H11B | 109.7 |
| O2—S1—C1 | 109.38 (8) | C17—C16—H11B | 109.7 |
| N1—S1—C1 | 102.91 (8) | H11A—C16—H11B | 108.2 |
| C15—N2—C7 | 115.42 (14) | C2—C1—C6 | 122.19 (17) |
| C15—N2—H3A | 108.4 | C2—C1—S1 | 118.86 (15) |
| C7—N2—H3A | 108.4 | C6—C1—S1 | 118.74 (14) |
| C15—N2—H3B | 108.4 | N2—C15—C16 | 114.61 (16) |
| C7—N2—H3B | 108.4 | N2—C15—H13A | 108.6 |
| H3A—N2—H3B | 107.5 | C16—C15—H13A | 108.6 |
| C21—O4—H6 | 109.5 | N2—C15—H13B | 108.6 |
| C13—N1—C14 | 117.16 (17) | C16—C15—H13B | 108.6 |
| C13—N1—S1 | 120.96 (13) | H13A—C15—H13B | 107.6 |
| C14—N1—S1 | 115.98 (15) | C8—C13—C12 | 119.4 (2) |
| C4—C5—C6 | 121.92 (19) | C8—C13—N1 | 125.42 (17) |
| C4—C5—H2 | 119.0 | C12—C13—N1 | 115.16 (19) |
| C6—C5—H2 | 119.0 | C18—C19—C20 | 113.90 (17) |
| C3—C4—C5 | 119.18 (18) | C18—C19—H15A | 108.8 |
| C3—C4—H3 | 120.4 | C20—C19—H15A | 108.8 |
| C5—C4—H3 | 120.4 | C18—C19—H15B | 108.8 |
| C5—C6—C1 | 117.17 (17) | C20—C19—H15B | 108.8 |
| C5—C6—C7 | 117.27 (17) | H15A—C19—H15B | 107.7 |
| C1—C6—C7 | 125.46 (16) | C13—C8—C9 | 117.75 (18) |
| C3—C2—C1 | 118.31 (18) | C13—C8—C7 | 128.70 (17) |
| C3—C2—H5 | 120.8 | C9—C8—C7 | 113.54 (18) |
| C1—C2—H5 | 120.8 | C18—C17—C16 | 114.47 (17) |
| C17—C18—C19 | 112.27 (17) | C18—C17—H17A | 108.6 |
| C17—C18—H6A | 109.2 | C16—C17—H17A | 108.6 |
| C19—C18—H6A | 109.2 | C18—C17—H17B | 108.6 |
| C17—C18—H6B | 109.2 | C16—C17—H17B | 108.6 |
| C19—C18—H6B | 109.2 | H17A—C17—H17B | 107.6 |
| H6A—C18—H6B | 107.9 | C10—C9—C8 | 122.2 (2) |
| C6—C7—N2 | 111.23 (14) | C10—C9—H18 | 118.9 |
| C6—C7—C8 | 120.45 (16) | C8—C9—H18 | 118.9 |
| N2—C7—C8 | 109.32 (14) | N1—C14—H19A | 109.5 |
| C6—C7—H7 | 104.8 | N1—C14—H19B | 109.5 |
| N2—C7—H7 | 104.8 | H19A—C14—H19B | 109.5 |
| C8—C7—H7 | 104.8 | N1—C14—H19C | 109.5 |
| C4—C3—C2 | 121.21 (18) | H19A—C14—H19C | 109.5 |
| C4—C3—Cl1 | 119.27 (15) | H19B—C14—H19C | 109.5 |
| C2—C3—Cl1 | 119.51 (16) | C11—C12—C13 | 121.6 (2) |
| O3—C21—O4 | 122.96 (19) | C11—C12—H20 | 119.2 |
| O3—C21—C20 | 123.80 (19) | C13—C12—H20 | 119.2 |
| O4—C21—C20 | 113.22 (17) | C11—C10—C9 | 119.1 (2) |
| C21—C20—C19 | 112.61 (16) | C11—C10—H21 | 120.4 |
| C21—C20—H10A | 109.1 | C9—C10—H21 | 120.4 |
| C19—C20—H10A | 109.1 | C12—C11—C10 | 119.9 (2) |
| C21—C20—H10B | 109.1 | C12—C11—H22 | 120.0 |
| C19—C20—H10B | 109.1 | C10—C11—H22 | 120.0 |
| H10A—C20—H10B | 107.8 | ||
| O1—S1—N1—C13 | 166.82 (14) | N1—S1—C1—C2 | −116.33 (15) |
| O2—S1—N1—C13 | 36.83 (17) | O1—S1—C1—C6 | 173.00 (14) |
| C1—S1—N1—C13 | −78.34 (16) | O2—S1—C1—C6 | −54.86 (16) |
| O1—S1—N1—C14 | −41.01 (17) | N1—S1—C1—C6 | 58.50 (16) |
| O2—S1—N1—C14 | −170.99 (15) | C7—N2—C15—C16 | −92.52 (19) |
| C1—S1—N1—C14 | 73.83 (17) | C17—C16—C15—N2 | −170.85 (16) |
| C6—C5—C4—C3 | 0.0 (3) | C14—N1—C13—C8 | −117.4 (2) |
| C4—C5—C6—C1 | −0.8 (3) | S1—N1—C13—C8 | 34.5 (3) |
| C4—C5—C6—C7 | 175.82 (17) | C14—N1—C13—C12 | 61.3 (2) |
| C5—C6—C7—N2 | −102.80 (18) | S1—N1—C13—C12 | −146.80 (15) |
| C1—C6—C7—N2 | 73.5 (2) | C17—C18—C19—C20 | −171.27 (17) |
| C5—C6—C7—C8 | 127.40 (18) | C21—C20—C19—C18 | −68.0 (2) |
| C1—C6—C7—C8 | −56.3 (2) | C12—C13—C8—C9 | 1.3 (3) |
| C15—N2—C7—C6 | 56.1 (2) | N1—C13—C8—C9 | 179.94 (17) |
| C15—N2—C7—C8 | −168.43 (15) | C12—C13—C8—C7 | −177.35 (18) |
| C5—C4—C3—C2 | 1.0 (3) | N1—C13—C8—C7 | 1.3 (3) |
| C5—C4—C3—Cl1 | −179.69 (14) | C6—C7—C8—C13 | 27.4 (3) |
| C1—C2—C3—C4 | −1.2 (3) | N2—C7—C8—C13 | −103.2 (2) |
| C1—C2—C3—Cl1 | 179.56 (13) | C6—C7—C8—C9 | −151.28 (17) |
| O3—C21—C20—C19 | −26.4 (3) | N2—C7—C8—C9 | 78.08 (19) |
| O4—C21—C20—C19 | 155.14 (17) | C19—C18—C17—C16 | −178.95 (17) |
| C3—C2—C1—C6 | 0.3 (3) | C15—C16—C17—C18 | −178.47 (17) |
| C3—C2—C1—S1 | 174.95 (13) | C13—C8—C9—C10 | −0.6 (3) |
| C5—C6—C1—C2 | 0.7 (3) | C7—C8—C9—C10 | 178.25 (19) |
| C7—C6—C1—C2 | −175.65 (17) | C8—C13—C12—C11 | −0.9 (3) |
| C5—C6—C1—S1 | −174.00 (13) | N1—C13—C12—C11 | −179.73 (19) |
| C7—C6—C1—S1 | 9.7 (2) | C8—C9—C10—C11 | −0.5 (3) |
| O1—S1—C1—C2 | −1.84 (17) | C13—C12—C11—C10 | −0.2 (3) |
| O2—S1—C1—C2 | 130.31 (15) | C9—C10—C11—C12 | 0.9 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H3A···Cl2i | 0.90 | 2.31 | 3.154 (2) | 157 |
| N2—H3B···O3ii | 0.90 | 2.32 | 2.821 (2) | 115 |
| C16—H11B···O3ii | 0.97 | 2.56 | 3.201 (2) | 124 |
| O4—H6···Cl2iii | 0.82 | 2.22 | 3.043 (2) | 176 |
| C4—H3···Cl2iv | 0.93 | 2.82 | 3.651 (2) | 150 |
| C18—H6A···O4v | 0.97 | 2.56 | 3.467 (2) | 157 |
| C7—H7···Cl2 | 0.98 | 2.59 | 3.534 (2) | 162 |
| N2—H3B···S1 | 0.90 | 2.98 | 3.533 (2) | 121 |
| N2—H3B···O2 | 0.90 | 2.02 | 2.802 (2) | 144 |
| C2—H5···O1 | 0.93 | 2.52 | 2.894 (2) | 104 |
| C14—H19B···O1 | 0.96 | 2.48 | 2.881 (3) | 105 |
Symmetry codes: (i) −x+2, −y+1, −z; (ii) −x+2, −y+1, −z+1; (iii) x, y, z+1; (iv) −x+1, −y+1, −z; (v) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2586).
References
- Guzman, H., Popov, A., Rammeloo, T. J. L., Remenar, J., Saoud, J. B. & Tawa, M. (2010). US Patent No. 20,100,112,051 A1 20100506.
- Hooft, R. (1998). COLLECT Nonius B V, Delft, The Netherlands.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Orola, L., Veidis, M. V., Sarcevica, I., Actins, A., Belyakov, S. & Platonenko, A. (2012). Int. J. Pharm. 432, 50–56. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global. DOI: 10.1107/S1600536812042432/pv2586sup1.cif
Additional supplementary materials: crystallographic information; 3D view; checkCIF report