The first structure of a glycerol-3-phosphate dehydrogenase from S. cerevisiae, GPD1, is reported at 2.45 Å resolution.
Keywords: glycerol-3-phosphate dehydrogenases, Saccharomyces cerevisiae, GPD1
Abstract
The interconversion of glycerol 3-phosphate and dihydroxyacetone phosphate by glycerol-3-phosphate dehydrogenases provides a link between carbohydrate and lipid metabolism and provides Saccharomyces cerevisiae with protection against osmotic and anoxic stress. The first structure of a glycerol-3-phosphate dehydrogenase from S. cerevisiae, GPD1, is reported at 2.45 Å resolution. The asymmetric unit contains two monomers, each of which is organized with N- and C-terminal domains. The N-terminal domain contains a classic Rossmann fold with the (β-α-β-α-β)2 motif typical of many NAD+-dependent enzymes, while the C-terminal domain is mainly α-helical. Structural and phylogenetic comparisons reveal four main structure types among the five families of glycerol-3-phosphate and glycerol-1-phosphate dehydrogenases and reveal that the Clostridium acetobutylican protein with PDB code 3ce9 is a glycerol-1-phosphate dehydrogenase.
1. Introduction
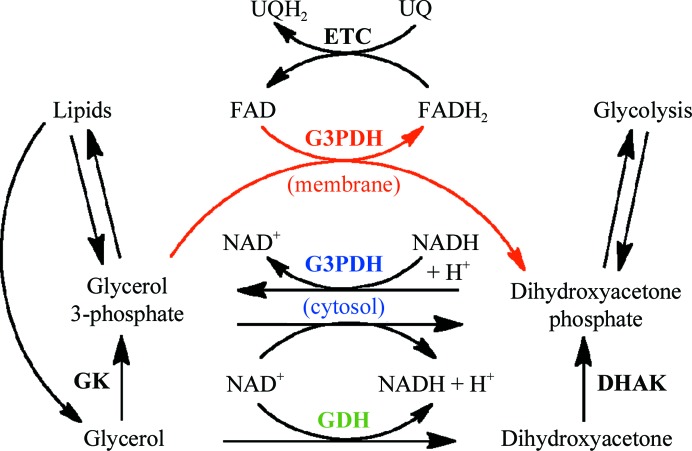
The interconversion of glycerol and dihydroxyacetone, either phosphorylated or unphosphorylated, is a key step in the integration of carbohydrate and lipid metabolism and in glycerol metabolism (Linn, 1976 ▶). The importance of this metabolic transition is evident from the evolution of at least five phylogenetically unrelated or distantly related families of enzymes that catalyze this reaction. The family with the widest metabolic impact includes the glycerol-3-phosphate dehydrogenases (G3PDHs), which are at the core of carbohydrate and lipid catabolism and anabolism. Glycerol 3-phosphate from the breakdown of phospholipids and triglycerides (via glycerol kinase) is converted into the glycolysis intermediate dihydroxyacetone phosphate, while the reverse reaction produces glycerol 3-phosphate, which is required for the synthesis of triglycerides and phospholipids (Fig. 1 ▶). In addition, the concerted action of cytosolic (NAD+-dependent) G3PDHs and membrane-bound (FAD-dependent) G3PDHs transfers reducing equivalents from cytosolic NADH into the electron-transport chain of both bacteria and mitochondria (Fig. 1 ▶; Larsson et al., 1998 ▶; Shen et al., 2003 ▶). The yeast cytosolic G3PDHs GPD1 and GPD2 can functionally complement one another but are differentially regulated, with GPD1 responding to osmotic stress for elevated glycerol production and GPD2 responding to anoxic conditions, presumably to convert excess NADH to NAD+ (Ansell et al., 1997 ▶). In addition, the reduction of dihydroxyacetone phosphate by G3PDH prevents its conversion into the potentially toxic methylglyoxalate (Richard, 1984 ▶; Lo et al., 1994 ▶; Martins et al., 2001 ▶).
Figure 1.
Schematic outline of the roles of G3PDH and GDH in metabolism. The cytosolic G3PDH (blue) interconverts glycerol 3-phosphate and dihydroxyacetone phosphate depending on the catabolic/anabolic state of the cell. The membrane-bound G3PDH (red) preferentially oxidizes glycerol 3-phosphate and transfers the electrons to ubiquinone in the respiratory electron-transport chain for ATP production. GDHs (green) preferentially oxidize glycerol to dihydroxyacetone. Both glycerol and dihydroxyacetone can be phosphorylated at the expense of ATP by glycerol kinase (GK) and dihydroxyacetone kinase (DHAK), respectively.
Glycerolipids are most commonly based on glycerol 3-phosphate, but some species, particularly among the archaebacteria, utilize the enantiomeric glycerol 1-phosphate. Consequently, G3PDHs are supplemented in some organisms with NAD(P)+-dependent glycerol-1-phosphate dehydrogenases (G1PDHs; Koga et al., 1998 ▶; Nishihara & Koga, 1995 ▶), which surprisingly show lower sequence similarity to G3PDHs than to bacterial and eukaryotic glycerol dehydrogenases (GDHs). The GDHs are primarily responsible for the oxidation of glycerol to dihydroxyacetone, which must be phosphorylated before entering the glycolytic pathway, making them less versatile metabolically than the G3PDHs (Linn, 1976 ▶). Examples of this enzyme can be found among the broad-substrate-range polyol dehydrogenases (Ruzheinikov et al., 2001 ▶) and the aldo-keto reductases (Richter et al., 2010 ▶).
Structures are available for at least one member of each of these families of G(P)DHs except the G1PDHs. For example, the G3PDHs from Archaeoglobis fulgidis (PDB entry 1txg; Sakasegawa et al., 2004 ▶), Thermotoga maritima (PDB entry 1z82; Lesley et al., 2002 ▶), Coxiella burnetii (PDB entry 3k96; Center for Structural Genomics of Infectious Diseases, unpublished work), Plasmodium falciparum (PDB entry 1yj8; Structural Genomics of Pathogenic Protozoa Consortium, unpublished work), Leishmania mexicana (PDB entry 1n1e; Choe et al., 2003 ▶) and Homo sapiens (PDB entry 1x0v; Ou et al., 2006 ▶) are NAD+-dependent and cytosolic, while the enzymes from Escherichia coli (PDB entry 2qcu; Yeh et al., 2008 ▶) and Bacillus halodurans (PDB entry 3da1; Northeast Structural Genomics Consortium, unpublished work) are FAD-dependent and membrane-associated. The NAD+-dependent GDHs from B. stearothermophilus (PDB entry 1jpu; Ruzheinikov et al., 2001 ▶), Sinorhizobium meliloti (PDB entry 3uhj; New York Structural Genomics Research Consortium, unpublished work), Clostridium acetobutylicum (PDB entry 3ce9; Joint Center for Structural Genomics, unpublished work), Schizosaccharomyces pombe (PDB entry 1ta9; A. M. Mulichak, unpublished work) and T. maritima (PDB entry 1kq3; Lesley et al., 2002 ▶) are members of the diverse group of polyol dehydrogenases, while the NADP+-dependent GDH from Gluconobacter oxydans (PDB entry 3n2t; Richter et al., 2010 ▶) is a member of the aldo-keto reductase family. To date, the structure of a G1PDH has not been reported.
In light of the physiological information pertaining to the G3PDHs from Saccharomyces cerevisiae, the absence of a structure of a representative enzyme from this species is notable. To fill this void, we report here the structure of GPD1 from S. cerevisiae refined to 2.45 Å resolution and compare the structures and phylogenetic groupings of the GPDH and GDH families, which reveals that a G1PDH structure has been reported but incorrectly annotated.
2. Materials and methods
2.1. Expression and purification of GPD1
The clone for GPD1, YDL022W (Gelperin et al., 2005 ▶), was purchased from Open Biosystems. The open reading frame was amplified using the primers 5′-AGCAGAATTCCATATGTCTGCTGCTGC (forward) and 5′-AAGAAGGATCCTTAATCTTCATGTAG (reverse) and transferred into pKS+ for sequence confirmation and then into pET28b using the restriction enzymes BamHI and NdeI to generate pET28b-GPD1. Expression in E. coli BL21 was induced with 0.1 mM IPTG and the cells were incubated at 310 K for 16 h. The cells were lysed using a French press and the crude extract was fractionated with ammonium sulfate followed by ion-exchange chromatography on DEAE cellulose. The purified protein was judged to be >95% pure by SDS–PAGE, with a specific activity of 0.03 µmol dihydroxyacetone phosphate reduced per minute per milligram of protein at room temperature, which is slightly lower than the specific activity of the enzyme from chicken liver (White & Kaplan, 1969 ▶).
2.2. Crystallization and refinement of GPD1
GPD1 was crystallized by the hanging-drop vapour-diffusion method using a reservoir solution consisting of 12% polyethylene glycol 8000, 0.1 M Tris–HCl pH 8.5, 0.3 M MgCl2. The first crystals appeared on the second day and continued growing to dimensions of 150 × 30 µm. Crystals flash-cooled using 20% glycerol as a cryoprotectant were used for data collection to 2.45 Å resolution on the ID29 beamline at the ESRF, Grenoble (Table 1 ▶). Data were processed with the XDS package (Kabsch, 2010 ▶) and a molecular-replacement solution was found with Phaser (McCoy et al., 2007 ▶) using the N-terminal and C-terminal domains of a monomer of the human enzyme (PDB entry 1x0v; sequence identity of 45.6%) as the search model. The protein structure was refined using REFMAC (Murshudov et al., 2011 ▶) and manually modelled with the molecular-graphics program Coot (Emsley & Cowtan, 2004 ▶). A final refinement cycle with BUSTER (Bricogne et al., 2011 ▶) produced R and R free values of 20.0% and 20.8%, respectively (Table 1 ▶). The coordinates have been deposited in the PDB as entry 4fgw.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data-collection statistics | |
| Space group | P43 |
| Unit-cell parameters | |
| a (Å) | 64.43 |
| b (Å) | 64.43 |
| c (Å) | 198.05 |
| α = β = γ (°) | 90.0 |
| Resolution (Å) | 29.42–2.45 (2.51–2.45) |
| Unique reflections | 29349 (2152) |
| Completeness (%) | 99.8 (99.9) |
| R merge | 0.075 (0.510) |
| 〈I/σ(I)〉 | 16.7 (3.0) |
| Multiplicity | 4.2 (4.2) |
| Model-refinement statistics | |
| No. of reflections | 29349 |
| R cryst (%) | 20.0 |
| R free (%) | 20.8 |
| Non-H atoms | 5132 |
| Water molecules | 68 |
| Average B factor (Å2) | 65.8 |
| Coordinate error† (Å) | 0.47 |
| R.m.s.d. bonds (Å) | 0.008 |
| R.m.s.d. angles (°) | 1.19 |
Based on maximum likelihood.
2.3. Phylogenetic analysis
The sequences listed in Table 2 ▶ were aligned using ClustalW and phylogenetic inferences were determined using programs from the PHYLIP 3.69 package (Felsenstein, 1989 ▶), including SEQBOOT, PROTDIST, FITCH and CONSENSE. The quality of the branching patterns was assessed by bootstrap resampling of the data sets using 100 replications.
Table 2. Glycerol (phosphate) dehydrogenase sequences used for phylogenetic analysis in Fig. 4 ▶ .
| No. | Strain | GenBank No. | PDB code |
|---|---|---|---|
| 1 | Archaeoglobus fulgidus | 1txg | |
| 2 | Saccharomyces cerevisiae | CAA80827.1 | 4fgw |
| 3 | Saccharomyces cerevisiae | CAA62526.1 | |
| 4 | Schizosaccharomyces pombe | CAA91239.1 | |
| 5 | Schizosaccharomyces pombe | CAA39630.1 | |
| 6 | Homo sapiens | 1x0v | |
| 7 | Plasmodium falciparum | 1yj8 | |
| 8 | Bacillus halodurans C-125 | NP_242506.1 | |
| 9 | Coxiella burnetii | 3k96 | |
| 10 | Thermotoga maritima | 1z82 | |
| 11 | Leishmania mexicana | 1n1g | |
| 12 | Shigella sp. D9 | ZP_08394385.1 | |
| 13 | Escherichia coli IAI1 | YP_002388887.1 | 2qcu |
| 14 | Klebsiella oxytoca KCTC 1686 | YP_005016918.1 | |
| 15 | Bacillus megaterium DSM 319 | YP_003595771.1 | |
| 16 | Planococcus donghaensis | ZP_08095479.1 | |
| 17 | Bacillus subtilis subsp. subtilis RO-NN-1 | YP_005556002.1 | |
| 18 | Bacillus halodurans | 3da1 | |
| 19 | Homo sapiens | AAB60403.1 | |
| 20 | Saccharomyces cerevisiae | CAA86123.1 | |
| 21 | Pseudomonas aeruginosa PA7 | YP_001349596.1 | |
| 22 | Gluconobacter morbifer G707 | ZP_09092611.1 | |
| 23 | Gluconobacter oxydans | 3n2t | |
| 24 | Commensalibacter intestini A911 | ZP_09012598.1 | |
| 25 | Acetobacter tropicalis NBRC 101654 | ZP_08643811.1 | |
| 26 | Streptococcus mutans UA159 | AAN58240.1 | |
| 27 | Bacillus stearothermophilus | 1jpu | |
| 28 | Thermotoga maritima MSB8 | EHA59067.1 | |
| 29 | Enterococcus faecalis TX1467 | EGG53141.1 | |
| 30 | Sinorhizobium meliloti | 3uhj | |
| 31 | Escherichi coli MS 107-1 | ZP_07099561.1 | |
| 32 | Schizosaccharomyces pombe | O13702.1 | 1ta9 |
| 33 | Methanosaeta concilii GP6 | YP_004383617.1 | |
| 34 | Methanococcoides burtonii DSM 6242 | ABE51967.1 | |
| 35 | Thermoproteus uzoniensis 768-20 | AEA13476.1 | |
| 36 | Acetobacterium woodii DSM 1030 | AFA47572.1 | |
| 37 | Eubacterium limosum KIST612 | ADO36353.1 | |
| 38 | Thermoanaerobacterium xylanolyticum LX-11 | YP_004470282.1 | |
| 39 | Mahella australiensis 50-1 BON | YP_004462215.1 | |
| 40 | Paenibacillus polymyxa M1 | CCC84600.1 | |
| 41 | Clostridium acetobutylicum | 3ce9 | |
| 42 | Anaplasma centrale str. Israel | YP_003328243.1 |
3. Results and discussion
3.1. Cloning, expression and crystallization of GPD1
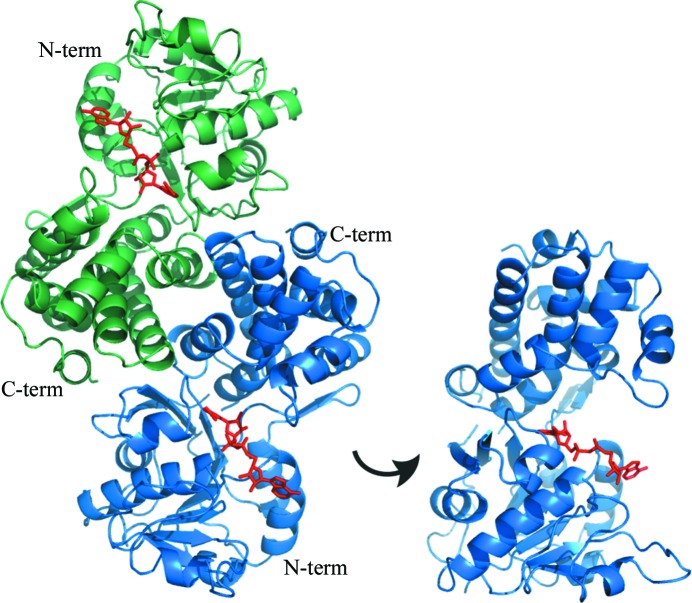
A clone of S. cerevisiae YDL022W encoding GPD1 was purchased from Open Biosystems and transferred into pKS+ and pET28b for sequence verification and expression, respectively. The purified protein was crystallized and its structure was determined at 2.45 Å resolution (Table 1 ▶). The homodimeric structure of the enzyme is reflected by the presence of two subunits in the asymmetric unit related by an exact noncrystallographic twofold axis (Fig. 2 ▶). The electron-density maps defined a nearly continuous backbone from residue 35 to the C-terminal residue 385 in both subunits. However, in one of the subunits electron density was poor for the protein region from Glu74 to Gln91, which was likely to be a result of conformational variability as suggested by the relatively high temperature-factor values, which nevertheless did not affect the overall quality of the refined model (Table 1 ▶).
Figure 2.
Structure of S. cerevisiae GPD1. Left: the dimer found in the asymmetric unit and the biologically active form of the enzyme. The A subunit is shown in blue and the B subunit is shown in green. The N-terminal domain (N-term) and C-terminal domain (C-term) are labelled. NAD+ molecules are coloured red and their locations are deduced from the binding site in the human enzyme (PDB entry 1x0v). Right: the A subunit rotated by 90° to provide a better view of the substrate-binding cleft.
The structure determined corresponds to the apoenzyme and is very similar to those of other cytosolic G3PDHs with the subunits organized in two domains. The N-terminal domain contains a classic Rossmann fold (β-α-β-α-β)2 (Rao & Rossmann, 1973 ▶) with parallel β-strands common to NAD-binding proteins and is extended by an (α-β-α-β) motif but with the two additional β-strands oriented antiparallel to the first six. The C-terminal domain consisting of residues 236–385 is largely α-helical. The crevasse formed in the waist of the subunit between the two domains harbours the NAD+-binding site along the Rossmann fold (Fig. 2 ▶). The human and yeast enzymes differ primarily in two extended loops in the human enzyme that protrude into the binding pocket, making it somewhat smaller.
3.2. Comparison with other glycerol dehydrogenase structures
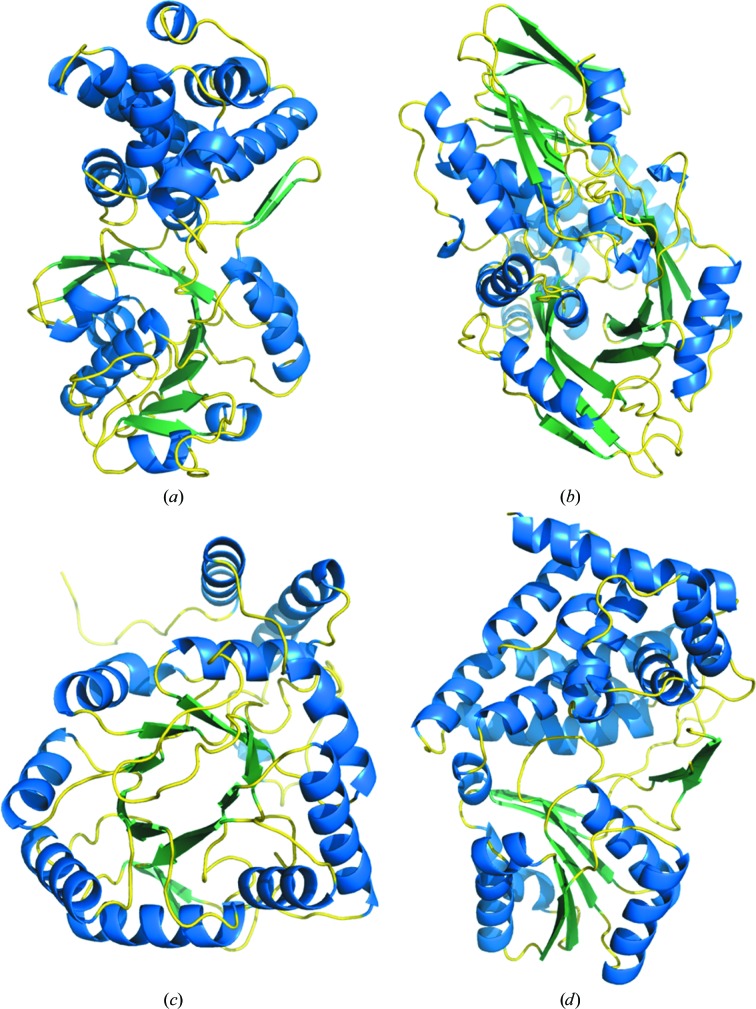
GPD1 superimposes well onto the structures of the six cytosolic enzymes (PDB entries 1n1g, 1txg, 1x0v, 1yj8, 1z82 and 3k96) with root-mean-square deviations of 1.8–2.7 Å for 290–330 Cα atoms. However, attempts to superimpose GPD1 onto either of the two membrane-bound G3PDHs or any of the six GDHs revealed three distinct groupings of substantially different structures that precluded superimposition (Fig. 3 ▶). The membrane-bound G3PDHs from E. coli (PDB entry 2qcu) and B. halodurans (PDB entry 3da1) contain three (β-α-β) motifs in the N-terminal 170 residues that are not organized in a classic Rossmann fold and the remainder of the protein is rich in β-sheets, mainly with an antiparallel organization (Fig. 3 ▶ b). The GDH from G. oxydans (PDB entry 3n2t) shares the repeating (α-β)8 motif common to the aldo-keto reductase family that is organized with the β-strands forming a central barrel surrounded by the helical segments (Fig. 3 ▶ c). Lastly, the GDHs from B. stearothermophilus (PDB entry 1jpu), Sinorhizobium meliloti (PDB entry 3uhj), Schizosaccharomyces pombe (PDB entry 1ta9), Thermotoga maritima (PDB entry 1kq3) and Clostridium acetobutylicum (PDB entry 3ce9; but see below) contain a modified Rossmann fold with the first (β-α-β-α-β) motif (residues 11–67) followed by an (α-β-α-β) motif (residues 70–118) (Fig. 3 ▶ d).
Figure 3.
Comparison of monomer structures of representatives of each of the five families of glycerol (phosphate) dehydrogenases. (a) S. cerevisiae GPD1 (PDB entry 4fgw) represents the cytosolic G3PDHs. (b) E. coli GlpD (PDB entry 2qcu) represents the membrane-bound G3PDHs. (c) G. oxydans AKR11B4 (PDB entry 3n2t) represents GDHs resembling aldo-keto reductases. (d) B. stearothermophilus GDH (PDB entry 1jpu) represents GDHs from the polyol dehydrogenase family and also the G1PDHs. The classic Rossmann fold is positioned at the bottom in (a). The high β-sheet content in GlpD is evident at the top and bottom of (b). Modified Rossmann folds are evident in the core of AKR11B4 in (c) and at the bottom of GDH in (d).
3.3. PDB entry 3ce9 is a glycerol-1-phosphate dehydrogenase structure
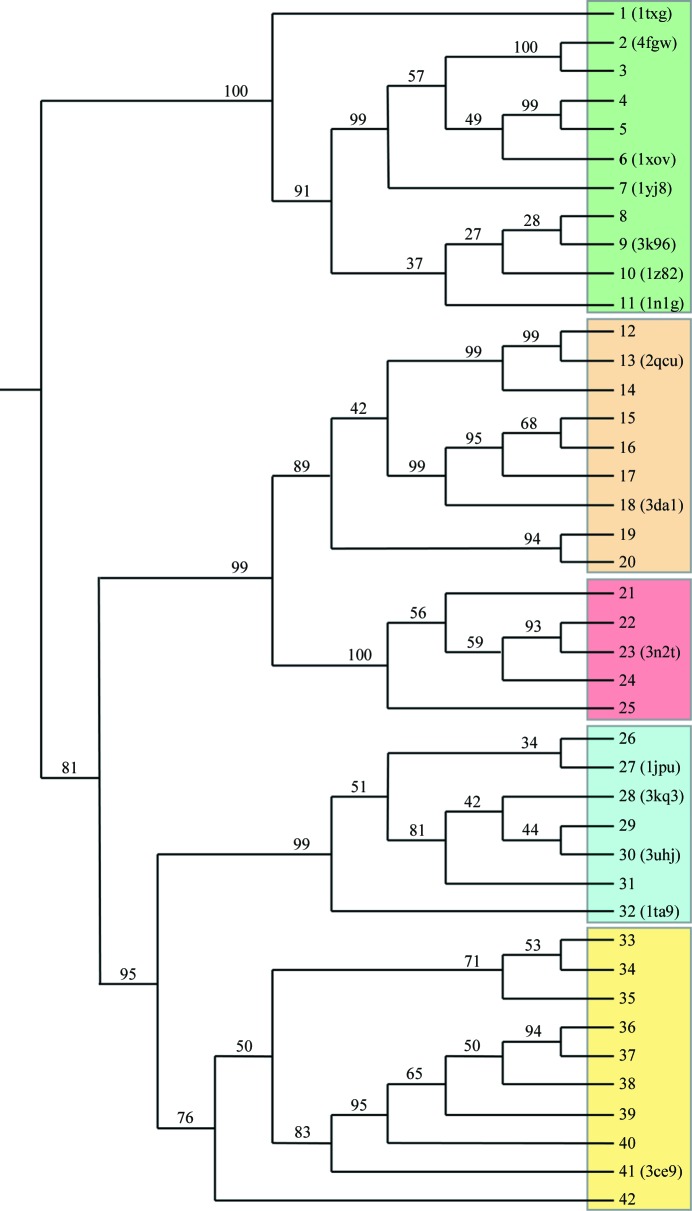
In an attempt to gain insight into how the family of G1PDHs may be related to these different structural groups, representative sequences from each family were aligned and subjected to phylogenetic analysis (Fig. 4 ▶). The resulting tree contains the expected five groups or clades: the cytosolic G3PDHs (clade A in Fig. 4 ▶), the membrane-bound G3PDHs (clade B), the aldo-keto reductase-like GDHs (clade C), the polyol dehydrogenase-like GDHs (clade D) and the G1PDHs (clade E). The surprise in this analysis is the location of the supposed GDH of C. acetobutylicum (protein 41 in Fig. 4 ▶) among the G1PDH-family sequences, suggesting that the protein with PDB code 3ce9 should be re-annotated as a G1PDH. Serendipitously, this outcome fills the hole created by the lack of a G1PDH structure; there is in fact at least one representative structure from each of the five clades of glycerol (phosphate) dehydrogenases. The close relationship between the polyol dehydrogenase-like GDHs and the G1PDHs on the same branch of the tree in Fig. 4 ▶ is further substantiated by the structure 3ce9 being very similar to the structure 1jpu in Fig. 3 ▶(d). C. acetobutylicum G1PDH shares 21% sequence similarity with B. stearothermophilus GDH and the root-mean-square deviation (r.m.s.d.) on 295 Cα atoms after superimposition is 2.6 Å. This can be compared with the greater similarity among the four GDHs, with 45–50% sequence similarity and r.m.s.d. values of 1.0–1.2 Å on 330–350 Cα atoms for superimposition of 1jpu on 3kq3, 1ta9 and 3uhj.
Figure 4.
Phylogenetic analysis of the glycerol (phosphate) dehydrogenase family. The tree was constructed using FITCH from the 42 protein sequences listed in Table 2 ▶ aligned using ClustalW. The percentage of replicates in the bootstrap resampling that contain the branch shown are indicated and PDB codes are included for those proteins for which structures have been reported. Group A contains the cytosolic G3PDHs (structures in Fig. 3 ▶ a), group B contains the membrane-bound G3PDHs (Fig. 3 ▶ b), group C contains the aldo-keto reductase-like proteins (Fig. 3 ▶ c), group D contains the polyol dehydrogenase-like GDHs (Fig. 3 ▶ d) and group E contains the G1PDHs. It should be noted that protein 41, which was annotated in PDB entry 3ce9 as being a GDH and has a structure very similar to that of B. stearothermophilus GDH (PDB entry 1jpu) in Fig. 3 ▶(d), clearly groups with the G1PDHs.
4. Conclusions
The structure of S. cerevisiae GPD1 has been refined to 2.45 Å resolution. Each monomer is organized with distinct N-terminal and C-terminal domains, with the N-terminal domain containing a classic Rossmann-fold (β-α-β-α-β)2 motif. The β-sheet element is extended by an additional (α-β-α-β) motif, albeit with the added two β-strands oriented antiparallel to the strands in the main fold. The structure of GPD1 is very similar to those of other cytosolic G3PDHs, but differs substantially from the eight other reported G3PDH and GDH structures. Phylogenetic analysis of glycerol (phosphate) dehydrogenase proteins confirmed the existence of five families or clades and revealed that one protein previously annotated as a GDH with its structure reported as PDB entry 3ce9 was actually a G1PDH. Thus, there is one representative structure for each of the five families and the close relationship between GDHs and G1PDHs is clearly evident from the structures of the polyol dehydrogenase-like GDHs being very similar to the G1PDH structure.
Supplementary Material
PDB reference: GPD1, 4fgw
Acknowledgments
This work was supported by a Discovery Grant 9600 from the Natural Sciences and Engineering Research Council of Canada, by the Canada Research Chair Program (to PCL) and by grant BFU2009-09268 from the Ministerio de Educación y Ciencia (MEC), Spain (to IF).
References
- Ansell, R., Granath, K., Hohmann, S., Thevelein, J. M. & Adler, L. (1997). EMBO J. 16, 2179–2187. [DOI] [PMC free article] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER Cambridge: Global Phasing Ltd.
- Choe, J., Guerra, D., Michels, P. A. M. & Hol, W. G. J. (2003). J. Mol. Biol. 329, 335–349. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Felsenstein, J. (1989). Cladistics, 5, 164–166.
- Gelperin, D. M., White, M. A., Wilkinson, M. L., Kon, Y., Kung, L. A., Wise, K. J., Lopez-Hoyo, N., Jiang, L., Piccirillo, S., Yu, H., Gerstein, M., Dumont, M. E., Phizicky, E. M., Snyder, M. & Grayhack, E. J. (2005). Genes Dev. 19, 2816–2826. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Koga, Y., Kyuragi, T., Nishihara, M. & Sone, N. (1998). J. Mol. Evol. 46, 54–63. [DOI] [PubMed]
- Larsson, C., Påhlman, I. L., Ansell, R., Rigoulet, M., Adler, L. & Gustafsson, L. (1998). Yeast, 14, 347–357. [DOI] [PubMed]
- Lesley, S. A. et al. (2002). Proc. Natl Acad. Sci. USA, 99, 11664–11669.
- Linn, E. C. C. (1976). Annu. Rev. Microbiol. 30, 535–578. [DOI] [PubMed]
- Lo, T. W. C., Westwood, M. E., McLellan, A. C., Selwood, T. & Thornalley, P. J. (1994). J. Biol. Chem. 269, 32299–32305. [PubMed]
- Martins, A. M. T. B. S., Cordeiro, C. A. A. & Ponces Freire, A. M. J. (2001). FEBS Lett. 499, 41–44. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nishihara, M. & Koga, Y. (1995). J. Biochem. 117, 933–935. [DOI] [PubMed]
- Ou, X., Ji, C., Han, X., Zhao, X., Li, X., Mao, Y., Wong, L.-L., Bartlam, M. & Rao, Z. (2006). J. Mol. Biol. 357, 858–869. [DOI] [PubMed]
- Rao, S. T. & Rossmann, M. G. (1973). J. Mol. Biol. 76, 241–256. [DOI] [PubMed]
- Richard, J. P. (1984). J. Am. Chem. Soc. 106, 4926–4936.
- Richter, N., Breicha, K., Hummel, W. & Niefind, K. (2010). J. Mol. Biol. 404, 353–362. [DOI] [PubMed]
- Ruzheinikov, S. N., Burke, J., Sedelnikova, S., Baker, P. J., Taylor, R., Bullough, P. A., Muir, N. M., Gore, M. G. & Rice, D. W. (2001). Structure, 9, 789–802. [DOI] [PubMed]
- Sakasegawa, S.-I., Hagemeier, C. H., Thauer, R. K., Essen, L.-O. & Shima, S. (2004). Protein Sci. 13, 3161–3171. [DOI] [PMC free article] [PubMed]
- Shen, W., Wei, Y., Dauk, M., Zheng, Z. & Zou, J. (2003). FEBS Lett. 536, 92–96. [DOI] [PubMed]
- White, H. B. & Kaplan, N. O. (1969). J. Biol. Chem. 244, 6031–6039. [PubMed]
- Yeh, J. I., Chinte, U. & Du, S. (2008). Proc. Natl Acad. Sci. USA, 105, 3280–3285. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: GPD1, 4fgw