The solution structure of the cold-shock-like protein from R. rickettsii, the causative agent of Rocky Mountain spotted fever, is reported.
Keywords: NMR, cold-shock domains, OB folds, Rocky Mountain spotted fever, Rickettsia rickettsii
Abstract
Rocky Mountain spotted fever is caused by Rickettsia rickettsii infection. R. rickettsii can be transmitted to mammals, including humans, through the bite of an infected hard-bodied tick of the family Ixodidae. Since the R. rickettsii genome contains only one cold-shock-like protein and given the essential nature of cold-shock proteins in other bacteria, the structure of the cold-shock-like protein from R. rickettsii was investigated. With the exception of a short α-helix found between β-strands 3 and 4, the solution structure of the R. rickettsii cold-shock-like protein has the typical Greek-key five-stranded β-barrel structure found in most cold-shock domains. Additionally, the R. rickettsii cold-shock-like protein, with a ΔG of unfolding of 18.4 kJ mol−1, has a similar stability when compared with other bacterial cold-shock proteins.
1. Introduction
In 1906, Howard Ricketts discovered the bacterium that causes Rocky Mountain spotted fever and this bacterium was ultimately named Rickettsia rickettsii after him (Gross & Schäfer, 2011 ▶; Ricketts, 1906a ▶,b ▶). R. rickettsii is an intracellular pathogen that is transmitted to mammals, including humans, through the bite of an infected tick (Gross & Schäfer, 2011 ▶; Dumler & Walker, 2005 ▶). Even with treatment, 5–10% of humans with an R. rickettsii infection will die of Rocky Mountain spotted fever (Dumler & Walker, 2005 ▶). Ticks can become infected either through bacterial transfer from an infected tick to its eggs or by transfer from a tick biting an infected mammal (McDade & Newhouse, 1986 ▶; Azad & Beard, 1998 ▶; Burgdorfer & Varma, 1967 ▶). It is interesting that environmental stimuli, such as the changes in temperature that the bacterium might experience while residing in a tick or when transferred to a mammalian host, cause limited change in relative mRNA transcription levels in R. rickettsii (Ellison et al., 2009 ▶). In particular, the relative mRNA transcription levels of the R. rickettsii cold-shock-like protein (Rr-Csp) are not significantly changed at numerous temperatures (Ellison et al., 2009 ▶). It may be that, as with other cold-shock proteins, protein levels of Rr-Csp are determined based on translational control versus transcriptional control (Giuliodori et al., 2004 ▶, 2010 ▶; Horn et al., 2007 ▶).
Traditionally, cold-shock proteins are thought to function as RNA chaperones during cold shock through melting and binding to mRNA, thereby reducing or preventing the formation of mRNA secondary structure (Horn et al., 2007 ▶; Chaikam & Karlson, 2010 ▶). This allows continued translation during cold adaptation (Horn et al., 2007 ▶; Chaikam & Karlson, 2010 ▶). Cold-shock proteins have a cold-shock domain structure, also termed an oligosaccharide/oligonucleotide (OB) binding fold, which consist of a five-stranded antiparallel β-barrel that binds to single-stranded nucleic acids (Horn et al., 2007 ▶; Chaikam & Karlson, 2010 ▶). This domain architecture is also found in eukaryotes, including plants and animals (Horn et al., 2007 ▶; Chaikam & Karlson, 2010 ▶). Other conserved structural features of cold-shock proteins include two nonspecific RNA-binding sequence motifs, RNP1 and RNP2 (ribonucleoprotein motifs 1 and 2, respectively), which are involved in binding to single-stranded RNA or DNA (Horn et al., 2007 ▶; Chaikam & Karlson, 2010 ▶). Given that Rr-Csp is the only cold-shock-like protein in the R. rickettsii genome (Ellison et al., 2009 ▶) and given the essential nature of bacterial cold-shock proteins for bacterial survival, the solution structure of the R. rickettsii cold-shock-like protein and its ΔG of unfolding are presented and compared with those of other bacterial cold-shock proteins.
2. Materials and methods
Undergraduate students taking a biochemistry laboratory course and/or a physical chemistry laboratory course completed the majority of these experiments during a single academic semester.
2.1. Protein expression and purification
A codon-optimized gene coding for a SUMO-Rr-Csp fusion was obtained from GenScript (Piscataway, New Jersey, USA). The UniProt code for Rr-Csp is A8GT84_RICRS. Escherichia coli BL21 (DE3) cells containing the pET28a-His6-SUMO-Rr-Csp construct were grown in Luria broth or [U-15N/13C] M9 minimal medium to an OD600 of ∼0.6 before protein expression was induced for 4–5 h using 1 mM isopropyl β-d-1-thiogalactopyranoside. Cell pellets were collected by centrifugation and stored at 253 K until processing. Cells were resuspended in buffer A (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole pH 8.0), lysed by sonication and clarified using centrifugation (15 000g for 15 min). The supernatant was loaded onto ∼2 ml His60 nickel resin for 30 min and the column was washed with 30 ml buffer A. The His6-SUMO-Rr-Csp fusion protein was eluted using a buffer consisting of 50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole pH 8.0. Fractions containing His6-SUMO-Rr-Csp based on SDS–PAGE were pooled with 400 µg SUMO/ubiquitin-like protease 1 (Ulp-1) and dialyzed against 20 mM sodium phosphate, 50 mM NaCl pH 8.0 for 2 d at 277 K. After dialysis and digestion with Ulp-1, the dialysate was loaded onto a His60 nickel resin column and the flowthrough and buffer A wash were collected. The flowthrough and wash were exchanged into buffer consisting of 20 mM sodium phosphate, 50 mM NaCl pH 6.0 and concentrated to ∼600 µl using ultrafiltration. The molecular weight of purified Rr-Csp was confirmed using MALDI-TOF mass spectrometry (measured m/z 7771.1, expected m/z 7770.7). Size-exclusion chromatography was performed in 200 mM sodium phosphate pH 7.5 using a Zorbax GF-450 column at a flow rate of 0.5 ml min−1.
2.2. NMR spectroscopy and structure determination
Data were acquired at the Medical College of Wisconsin’s NMR facility on a Bruker 500 MHz spectrometer equipped with a triple-resonance cryoprobe and were processed using NMRPipe (Delaglio et al., 1995 ▶). A complete list of the collected NMR spectra can be found in the Supplementary Material1. The NMR sample consisted of 1.5 mM Rr-Csp in 20 mM sodium phosphate pH 6.0 with 50 mM NaCl, 10% D2O and 0.2% NaN3. All NMR spectra were collected at a sample temperature of 298 K. Backbone chemical shift assignments were generated by GARANT (Bartels et al., 1996 ▶). Manual checking of the backbone chemical shift assignments indicated that they were correct. Side chains were assigned manually and overall assignments were 98% complete. 1Hα, 13Cα, 13Cβ, 13C′ and 15N chemical shifts and TALOS+ were used to generate backbone dihedral angle constraints (Shen et al., 2009 ▶). Distance restraints were generated from three-dimensional 15N-edited NOESY–HSQC, 13C-edited NOESY–HSQC and 13C(aromatic)-edited NOESY–HSQC spectra (τmix = 80 ms). The NOEASSIGN module of the torsion-angle dynamics program CYANA 3.0 was used to assign the NOESY spectra, determine initial distance restraints and calculate initial structures (Herrmann et al., 2002 ▶). CYANA 3.0 was used for subsequent manual refinement (Herrmann et al., 2002 ▶). However, the initial ensembles calculated using the NOEASSIGN module had high precision and almost no constraint violations (target-function values of 0.01 or 0.00 Å2) and required little manual refinement. The X-PLOR program was used for further refinement of the protein structure in explicit water solvent by adding physical force-field terms to the experimental constraints (Linge et al., 2003 ▶; Brünger, 1992 ▶; Brunger, 2007 ▶; Schwieters et al., 2003 ▶). Table 1 ▶ lists the statistics from the PSVS suite (Bhattacharya et al., 2007 ▶), PROCHECK-NMR (Laskowski et al., 1996 ▶) and WHAT_CHECK (Hooft et al., 1996 ▶) for validation of the final 20 conformers, which were the 20 lowest energy conformers of the 100 calculated. Heteronuclear NOE spectra were obtained using the Bruker hsqcnoef3gpsi pulse program.
Table 1. Statistics for 20 Rr-Csp conformers (PDB entry 2lss; BMRB entry 18442).
| Completeness of resonance assignments† (%) | 98 |
| Constraints | |
| Nonredundant distance constraints | |
| Total | 1813 |
| Intraresidue (i = j) | 1123 |
| Sequential [(i j) = 1] | 298 |
| Medium [1 (i j) 5] | 86 |
| Long | 306 |
| Dihedral angle constraints ( and ) | 120 |
| Constraints per residue | |
| Average No. of constraints per residue | 27 |
| Constraint violations | |
| Average No. of distance-constraint violations per structure | |
| 0.10.2 | 17.05 |
| 0.20.5 | 1.45 |
| >0.5 | 0 |
| Average r.m.s. distance violation per constraint () | 0.02 |
| Maximum distance violation () | 0.36 |
| Average No. of dihedral angle violations per structure | |
| 110 | 3.75 |
| >10 | 0 |
| R.m.s. dihedral angle violation per constraint () | 0.33 |
| Maximum dihedral angle violation () | 3.4 |
| Average atomic r.m.s.d. to the mean structure () | |
| Residues 270 | |
| Backbone (C, C, N) | 0.49 0.09 |
| Heavy atoms | 0.93 0.09 |
| Deviations from idealized covalent geometry‡ | |
| Bond-length r.m.s.d. () | 0.017 |
| Torsion-angle violations r.m.s.d. () | 1.3 |
| LennardJones energy§ (kJmol1) | 1450 40 |
| Ramachandran statistics¶ (% of all residues) | |
| Most favored | 86.2 |
| Additionally allowed | 13.8 |
| Generously allowed | 0 |
| Disallowed | 0 |
The missing chemical shifts are the H, H, Q, Q and Q of Met1, the H of Ala2 and the H of Phe31.
Final X-PLOR (Brnger, 1992 ▶) force constants (kcalmol1; 1cal = 4.186J) were 250 (bonds), 250 (angles), 300 (impropers), 100 (chirality), 100 (), 50 (NOE constraints) and 200 (torsion-angle constraints). Idealized covalent geometry is from Engh Huber (1991 ▶).
Nonbonded energy was calculated in X-PLOR-NIH (Schwieters et al., 2003 ▶).
Values are from PROCHECK-NMR (Laskowski et al., 1996 ▶).
2.3. ΔG of unfolding
A two-state equilibrium between native and denatured Rr-Csp was assumed, similar to other Csp studies (Motono et al., 2008 ▶; Kumar et al., 2001 ▶; Perl et al., 2000 ▶). Guanidine-denaturation curves were used to determine the ΔG of unfolding of 5 µM Rr-Csp in 100 mM sodium phosphate at pH 7.0 as described by Shirley (1995 ▶). ΔG was determined at various guanidine hydrochloride concentrations using the equation
where F N is the intrinsic tryptophan fluorescence intensity at 350 nm of the native protein, F D is the fluorescence of the denatured protein and F [GuHCl] is the fluorescence at a given guanidine concentration. Fluorescence intensities were measured using an F2500 Hitachi fluorescence spectrophotometer with an excitation wavelength of 285 nm and an emission scan from 300 to 500 nm. The ΔG of denaturation under native conditions was extrapolated using a plot of ΔG [GuHCl] versus guanidine hydrochloride concentration (Shirley, 1995 ▶).
3. Results and discussion
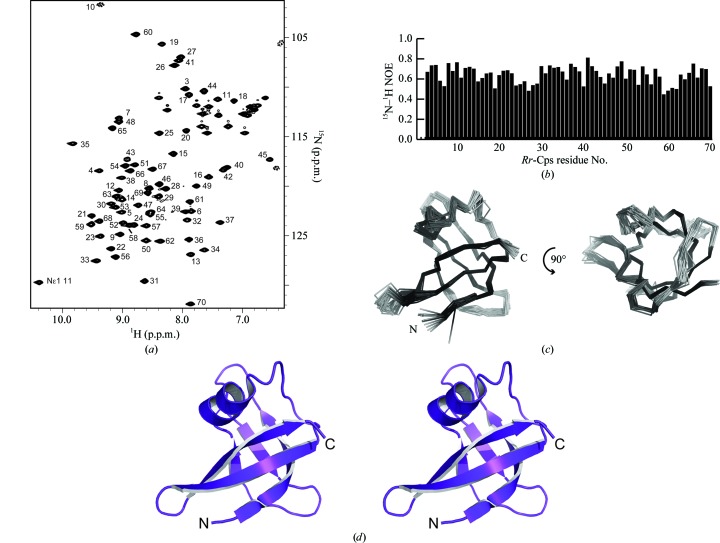
Rr-Csp, which consists of 70 residues, was purified as a His6-SUMO fusion using immobilized metal-affinity chromatography. Incubation with His6-Ulp-1 and subsequent immobilized metal-affinity chromatography was used to separate the His6-SUMO fusion from Rr-Csp (Supplementary Fig. S1 ▶ 1). After concentration and buffer exchange, the resulting two-dimensional 15N–1H HSQC spectrum of Rr-Csp showed a homogenous spectrum with distinct peaks distributed throughout the spectrum, indicating that the protein was folded (Fig. 1 ▶ a). In size-exclusion chromatography Rr-Csp eluted at a higher retention time than SUMO (small ubiquitin-like modifier), a monomeric protein of 11 kDa, suggesting that Rr-Csp is monomeric like nearly all of its bacterial homologs (Horn et al., 2007 ▶). Heteronuclear NOE values indicated that the entire Rr-Csp protein was structured (Fig. 1 ▶ b). Standard NMR techniques were used to solve the structure of Rr-Csp (Markley et al., 2003 ▶), and the Rr-Csp ensemble of structures was deposited in the Protein Data Bank (Bernstein et al., 1977 ▶) as entry 2lss. Chemical shift assignments and structural restraints were deposited in the Biological Magnetic Resonance Bank (Ulrich et al., 2008 ▶; BMRB entry 18442). Two orientations of the ensemble are shown in Fig. 1 ▶(c) and a stereo image of the Rr-Csp structure is shown in Fig. 1 ▶(d). As expected, Rr-Csp shows the conserved five-stranded β-barrel fold characteristic of cold-shock proteins, with the exception of a short α-helix between strands 3 and 4. This helix was first identified by TALOS+ (Shen et al., 2009 ▶) and was confirmed by NOE patterns consistent with an α-helix in the 15N NOESY.
Figure 1.
Solution structure of the R. rickettsii cold-shock-like protein (Rr-Csp). (a) 15N–1H HSQC spectrum acquired from 1.5 mM [U-15N/13C] Rr-Csp at 298 K using a 500 MHz Bruker spectrometer. (b) 15N–1H heteronuclear NOEs plotted for each Rr-Csp residue. (c) Ensemble of 20 Rr-Csp structures shown as two views, with the second being a 90° rotation along the x axis with respect to the first. (d) A stereoview of the lowest energy structure from the Rr-Csp ensemble shown as a ribbon diagram.
Although the NMR spectra indicated single peaks for each NMR-active atom, they also indicated that an unidentified molecule copurified with Rr-Csp. This was most apparent in the 13C HCCH–TOCSY, which showed signals with carbon and proton chemical shifts that most closely matched shifts common to nucleic acids. Most notable were two C atoms with chemical shifts at 89.8 p.p.m. and 87.2 p.p.m., shifts that are consistent with those observed for the C1′ and C4′ atoms of nucleotides (Supplementary Fig. S21). No C atoms from proteins resonate in this region. Given that Rr-Csp is a cold-shock domain, also known as an oligosaccharide/oligonucleotide (OB) binding fold, it is possible that this signal comes from a nucleotide that copurified with Rr-Csp. Like other cold-shock proteins, Rr-Csp contains an RNP1-like motif (amino acids 17–23) and an RNP2-like motif (amino acids 30–34). The RNP1 and RNP2 motifs in other cold-shock proteins form a single-stranded nucleic acid-binding site (Horn et al., 2007 ▶; Chaikam & Karlson, 2010 ▶; Max et al., 2006 ▶). However, we were not able to identify NOEs between this copurifying molecule and Rr-Csp. With the exception of observing that the chemical shifts of the molecule are consistent with those of a nucleotide, identifying the molecule is beyond the scope of this structure report.
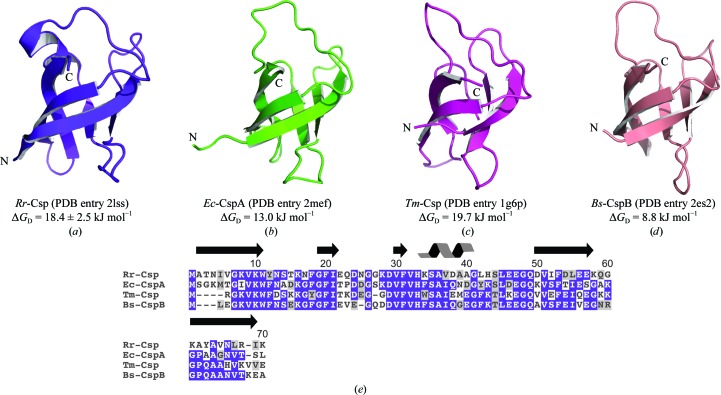
In addition to being studied for their nucleic acid-binding properties and their roles in bacterial cold adaption, bacterial cold-shock proteins from mesophiles, thermophiles and hyperthermophiles have been investigated for factors that contribute to the stability and thermostability of a protein (Motono et al., 2008 ▶; Kumar et al., 2001 ▶; Perl et al., 2000 ▶). As shown in Fig. 2 ▶, Rr-Csp is homologous in both sequence and structure to other bacterial cold-shock proteins from E. coli, Thermotoga maritima and Bacillus subtilis for which the values of ΔG of denaturation (ΔG D) at room temperature are known (Kumar et al., 2001 ▶). To allow a comparison, the ΔG D at room temperature for Rr-Csp was determined (Supplementary Fig. S31; Shirley, 1995 ▶). At 18.4 ± 2.5 kJ mol−1, the ΔG D value for Rr-Csp is higher than the values reported for other cold-shock proteins from mesophiles, such as 13.0 kJ mol−1 for E. coli CspA (Ec-CspA) and 8.8 kJ mol−1 for B. subtilis CspB (Bs-CspB) (Kumar et al., 2001 ▶). The ΔG D of Rr-Csp more closely matches that of 19.7 kJ mol−1 (Kumar et al., 2001 ▶) for the Csp from the hyperthermophile T. maritima.
Figure 2.
Rr-Csp and structural homologs with known ΔG D values. Ribbon diagrams and ΔG D values at room temperature for (a) Rr-Csp (PDB entry 2lss), (b) E. coli CspA (Ec-CspA; PDB entry 3mef; Feng et al., 1998 ▶), (c) T. maritima Csp (Tm-Csp; PDB entry 1g6p; Kremer et al., 2001 ▶) and (d) B. subtilis CspB (Bs-CspB; PDB entry 2es2; Max et al., 2006 ▶). The ΔG D values for E. coli CspA, T. maritima Csp and B. subtilis CspB are from Kumar et al. (2001 ▶). (e) Multiple sequence alignment of Rr-Csp with E. coli CspA, T. maritima Csp and B. subtilis CspB. The backgrounds of identical residues are blue and those of conserved residues are gray. The residue numbering corresponds to that of Rr-Csp.
A potential hypothesis that might explain the ΔG D of Rr-Csp being similar to the ΔG D of a Csp from a hyperthermophile is that salt bridges may stabilize the Rr-Csp structure. Salt bridges have been suggested to contribute to the increased stability of proteins from thermophiles and hyperthermophiles (Kumar, Tsai & Nussinov, 2000 ▶; Kumar, Ma et al., 2000 ▶; Kumar, Tsai, Ma et al., 2000 ▶; Costantini et al., 2008 ▶; Kumar et al., 2001 ▶). Tm-Csp, the Csp from T. maritima, contains an arginine at position 2 that is conserved in and is known to increase the stability of Csps from thermophiles and hyperthermophiles (Perl et al., 2000 ▶; Kremer et al., 2001 ▶). Kremer and coworkers suggest that this arginine is a part of an ion cluster, a cluster of acidic and basic amino acids, that contributes to the increased stability of Tm-Csp (Kremer et al., 2001 ▶). Two salt bridges identified in the lowest energy conformer of the solution structure of Tm-Csp support this claim. Rr-Csp does not contain the conserved arginine found in the Csps of thermophiles and hyperthermophiles (Fig. 2 ▶ e). However, each structure in the Rr-Csp ensemble contains an average of four salt bridges. Analysis of the structures of Bs-CspB and Ec-CspA, which are Csps from mesophiles, revealed one salt bridge in Bs-CspB and none in Ec-CspA. Salt bridges were identified by the program ESBRI (Costantini et al., 2008 ▶) using a cutoff distance of 4 Å between charged residues. One salt bridge in Rr-Csp was also suggested by the seven close contacts that were identified during validation of the Rr-Csp ensemble. Each close contact is in a different structure of the ensemble and consists of either an Hζ1 or an Hζ3 atom of Lys10 at a distance of 1.55–1.60 Å from either the O∊1 or the O∊2 atom of Glu22. This potential salt bridge is not likely to be present in Ec-CspA, which contains an identical lysine but has a threonine instead of the glutamate found in Rr-Csp. Bs-CspB does contain the corresponding lysine and glutamate. However, the side-chain amino group of the lysine and the carboxyl group of the glutamate in Bs-CspB are 5.4 Å apart and were not identified as a salt bridge by ESBRI (Costantini et al., 2008 ▶).
Future experiments are needed to test the hypothesis that potential salt bridges in Rr-Csp are responsible for the increased stability. However, this structure, which is the first solution structure from the R. rickettsii genome to be deposited in the PDB, provides an example of how structural biology techniques can be successfully learned and applied in an undergraduate laboratory course.
Supplementary Material
PDB reference: cold-shock-like protein, 2lss
Supporting information file. DOI: 10.1107/S174430911203881X/kw5052sup1.pdf
Acknowledgments
We would like to thank the UW-Whitewater Chemistry Department and the Medical College of Wisconsin’s NMR facility for reagents, instrument time and support. We thank Dr Steve Anderson for his critical review of the manuscript. CTV was partially supported by National Institutes of Health grant 1-R15CA159202-01.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: KW5052).
References
- Azad, A. F. & Beard, C. B. (1998). Emerg. Infect. Dis. 4, 179–186. [DOI] [PMC free article] [PubMed]
- Bartels, C., Billeter, M., Güntert, P. & Wüthrich, K. (1996). J. Biomol. NMR, 7, 207–213. [DOI] [PubMed]
- Bernstein, F. C., Koetzle, T. F., Williams, G. J., Meyer, E. F. Jr, Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol. 112, 535–542. [DOI] [PubMed]
- Bhattacharya, A., Tejero, R. & Montelione, G. T. (2007). Proteins, 66, 778–795. [DOI] [PubMed]
- Brünger, A. T. (1992). X-PLOR Version 3.1. A System for X-ray Crystallography and NMR New Haven: Yale University Press.
- Brunger, A. T. (2007). Nature Protoc. 2, 2728–2733. [DOI] [PubMed]
- Burgdorfer, W. & Varma, M. G. (1967). Annu. Rev. Entomol. 12, 347–376. [DOI] [PubMed]
- Chaikam, V. & Karlson, D. T. (2010). BMB. Rep. 43, 1–8. [DOI] [PubMed]
- Costantini, S., Colonna, G. & Facchiano, A. M. (2008). Bioinformation, 3, 137–138. [DOI] [PMC free article] [PubMed]
- Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995). J. Biomol. NMR, 6, 277–293. [DOI] [PubMed]
- Dumler, J. S. & Walker, D. H. (2005). N. Engl. J. Med. 353, 551–553. [DOI] [PubMed]
- Ellison, D. W., Clark, T. R., Sturdevant, D. E., Virtaneva, K. & Hackstadt, T. (2009). PLoS One, 4, e5612. [DOI] [PMC free article] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Feng, W., Tejero, R., Zimmerman, D. E., Inouye, M. & Montelione, G. T. (1998). Biochemistry, 37, 10881–10896. [DOI] [PubMed]
- Giuliodori, A. M., Brandi, A., Gualerzi, C. O. & Pon, C. L. (2004). RNA, 10, 265–276. [DOI] [PMC free article] [PubMed]
- Giuliodori, A. M., Di Pietro, F., Marzi, S., Masquida, B., Wagner, R., Romby, P., Gualerzi, C. O. & Pon, C. L. (2010). Mol. Cell, 37, 21–33. [DOI] [PubMed]
- Gross, D. & Schäfer, G. (2011). Microbes Infect. 13, 10–13. [DOI] [PubMed]
- Herrmann, T., Güntert, P. & Wüthrich, K. (2002). J. Mol. Biol. 319, 209–227. [DOI] [PubMed]
- Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. (1996). Nature (London), 381, 272. [DOI] [PubMed]
- Horn, G., Hofweber, R., Kremer, W. & Kalbitzer, H. R. (2007). Cell. Mol. Life Sci. 64, 1457–1470. [DOI] [PMC free article] [PubMed]
- Kremer, W., Schuler, B., Harrieder, S., Geyer, M., Gronwald, W., Welker, C., Jaenicke, R. & Kalbitzer, H. R. (2001). Eur. J. Biochem. 268, 2527–2539. [DOI] [PubMed]
- Kumar, S., Ma, B., Tsai, C.-J. & Nussinov, R. (2000). Proteins, 38, 368–383. [DOI] [PubMed]
- Kumar, S., Tsai, C.-J., Ma, B. & Nussinov, R. (2000). J. Biomol. Struct. Dyn. 17, 79–85. [DOI] [PubMed]
- Kumar, S., Tsai, C.-J. & Nussinov, R. (2000). Protein Eng. 13, 179–191. [DOI] [PubMed]
- Kumar, S., Tsai, C.-J. & Nussinov, R. (2001). Biochemistry, 40, 14152–14165. [DOI] [PubMed]
- Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996). J. Biomol. NMR, 8, 477–486. [DOI] [PubMed]
- Linge, J. P., Williams, M. A., Spronk, C. A., Bonvin, A. M. & Nilges, M. (2003). Proteins, 50, 496–506. [DOI] [PubMed]
- Markley, J. L., Ulrich, E. L., Westler, W. M. & Volkman, B. F. (2003). Methods Biochem. Anal. 44, 89–113. [PubMed]
- Max, K. E., Zeeb, M., Bienert, R., Balbach, J. & Heinemann, U. (2006). J. Mol. Biol. 360, 702–714. [DOI] [PubMed]
- McDade, J. E. & Newhouse, V. F. (1986). Annu. Rev. Microbiol. 40, 287–309. [DOI] [PubMed]
- Motono, C., Gromiha, M. M. & Kumar, S. (2008). Proteins, 71, 655–669. [DOI] [PubMed]
- Perl, D., Mueller, U., Heinemann, U. & Schmid, F. X. (2000). Nature Struct. Biol. 7, 380–383. [DOI] [PubMed]
- Ricketts, H. T. (1906a). JAMA, 47, 33–36.
- Ricketts, H. T. (1906b). JAMA, 47, 1067–1069.
- Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Clore, G. M. (2003). J. Magn. Reson. 160, 65–73. [DOI] [PubMed]
- Shen, Y., Delaglio, F., Cornilescu, G. & Bax, A. (2009). J. Biomol. NMR, 44, 213–223. [DOI] [PMC free article] [PubMed]
- Shirley, B. A. (1995). Methods Mol. Biol. 40, 177–190. [DOI] [PubMed]
- Ulrich, E. L. et al. (2008). Nucleic Acids Res. 36, D402–D408. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: cold-shock-like protein, 2lss
Supporting information file. DOI: 10.1107/S174430911203881X/kw5052sup1.pdf