The crystallization of both native P. putida transcriptional regulator PtxS and its complex with its DNA recognition sequence using the counter-diffusion method are reported.
Keywords: PtxS–DNA, glucose, counter-diffusion, microfocus
Abstract
Pseudomonas putida PtxS is a member of the LacI protein family of transcriptional regulators involved in glucose metabolism. All genes involved in this pathway are clustered into two operons, kgu and gad. PtxS controls the expression of the kgu and gad operons as well as its own transcription. The PtxS operator is a perfect palindrome, 5′-TGAAACCGGTTTCA-3′, which is present in all three promoters. Crystallization of native PtxS failed, and PtxS–DNA crystals were finally produced by the counter-diffusion technique. A portion of the capillary used for crystal growth was attached to the end of a SPINE standard cap and directly flash-cooled in liquid nitrogen for diffraction tests. A full data set was collected with a beam size of 10 × 10 µm. The crystal belonged to the trigonal space group P3, with unit-cell parameters a = b = 213.71, c = 71.57 Å. Only unhandled crystals grown in capillaries of 0.1 mm inner diameter diffracted X-rays to 1.92 Å resolution.
1. Introduction
Glucose is a metabolite used as a source of carbon by the genus Pseudomonas through a series of pathways that take place in the periplasm and the cytoplasm. In the periplasm, glucose is oxidized by both glucose dehydrogenase and gluconate dehydrogenase to generate 2-ketogluconate. This resulting product is then transported to the cytoplasm and transformed into 6-phosphogluconate, which enters the Entner–Doudoroff pathway (Entner & Doudoroff, 1952 ▶). All of the genes involved in this process are grouped into two different operons termed kgu and gad. Bioinformatics studies have shown that all of the key enzymes for the synthesis of 6-phosphogluconate from glucose via glucose-6-phosphate or via gluconate are present in several members of the genus such as P. fluorescens Pf-5, P. syringae DC3000 and P. aeruginosa PAO1. BLAST analysis revealed a high degree of sequence conservation for all gene products ranging from 70 to 95% similarity (Del Castillo et al., 2007 ▶).
In P. putida, glucose metabolism is regulated by PtxS, a member of the LacI protein family of transcriptional regulators which controls the expression of the kgu and gad operons as well as its own transcription. Interestingly, in the human pathogen P. aeruginosa the PtxS homologue was found to additionally regulate the expression of the primary virulence factor exotoxin A (Colmer & Hamood, 1998 ▶). Since the nonpathogenic saprophyte P. putida is devoid of an exotoxin A gene, the primary biological function of PtxS in this species thus appears to be restricted to the regulation of carbohydrate metabolism (Daddaoua et al., 2010 ▶).
PtxS is predicted to contain two domains: a helix–turn–helix motif-containing DNA-binding domain located at the N-terminus and a C-terminal domain that binds the effector 2-ketogluconate. Differential scanning calorimetry assays revealed that PtxS unfolds via two events centred at 321.2 and 330.7 K. In the presence of 2-ketogluconate the unfolding of the effector-binding domain occurs at a higher temperature, providing further evidence for 2-ketogluconate–PtxS interactions (Daddaoua et al., 2010 ▶). Purified PtxS is a dimer in solution that binds to the target promoters with affinities in the range 1–3 µM. Footprint analyses revealed that PtxS binds to a palindrome with the sequence 5′-TGAAACCGGTTTCA-3′ that is present in each of the target promoters (Daddaoua et al., 2010 ▶). This palindrome overlaps with the RNA polymerase binding site and the regulatory action of PtxS consists of competition with the RNA polymerase for the promoter.
In this work, we summarize the efforts carried out to shed light on the structural basis of PtxS-mediated transcriptional regulation. For this purpose, we produced PtxS crystals in complex with a recognition DNA fragment comprising its operator site. Initial three-dimensional crystals were grown by the counter-diffusion technique in capillaries of 0.1 mm inner diameter. These crystals were found to be very fragile after extraction from the capillaries for incubation in several cryoprotectant solutions. Only unhandled crystals that remained in the capillary used for growth diffracted to a maximum resolution of 1.92 Å. For this purpose, a capillary portion was mounted directly on a SPINE cap and exposed to X-rays on the microfocous beamline ID23-2 at the European Synchrotron Radiation Facility (ESRF; Flot et al., 2010 ▶).
2. Materials and methods
2.1. Overexpression and purification
Plasmid pET24b_PtxS was used for the overexpression of P. putida PtxS (339 amino acids, molecular weight 36 789 Da; accession No. Q88HH7) fused to a C-terminal polyhistidine tag. The construction of this plasmid as well as the expression and purification of the protein have been described previously (Daddaoua et al., 2010 ▶). Briefly, Escherichia coli BL21b cells containing pET24b_PtxS were grown at 303 K until the culture reached an OD600 of 0.6. Subsequently, the temperature was set to 291 K and the cells were grown for an additional 1 h before the addition of 1 mM IPTG. Growth was then continued at 291 K overnight. The cells were harvested by centrifugation and subsequently resuspended in buffer A [50 mM Tris–HCl pH 7.9, 500 mM NaCl, 1 mM DTT, 10 mM imidazole, 10%(v/v) glycerol]. Cell disruption was performed using a French press. After a centrifugation step, the supernatant was loaded onto a HisTrap HP column (GE Healthcare) and eluted with an imidazole gradient to buffer B [50 mM Tris–HCl pH 7.9, 500 mM NaCl, 1 mM DTT, 500 mM imidazole, 10%(v/v) glycerol]. Protein-containing fractions were pooled, concentrated and dialyzed against buffer C [50 mM HEPES pH 7.9, 100 mM NaCl, 1 mM DTT, 10%(v/v) glycerol]. The protein was concentrated to 70 mg ml−1 using 0.5 ml concentration units (Amicon Ultra-0.5, Merck Millipore) prior to incubation with an equimolar concentration of the DNA.
2.2. Crystallization and data collection
Initial crystallization conditions for purified native PtxS were determined by the counter-diffusion technique (García-Ruiz, 2003 ▶) using capillaries of 0.1 mm inner diameter with the CSK-24 crystallization screening kit (Triana S&T, Granada, Spain). Initial conditions (2 M potassium/sodium phosphate, 0.1 M Na HEPES pH 7.5 and 1.5 M ammonium sulfate, 0.1 M sodium citrate pH 5.60) were further optimized by the vapour-diffusion method using a hanging-drop configuration. Plate-shaped crystals were obtained by mixing 1 µl protein solution (30 mg ml−1) and 1 µl reservoir solution consisting of 20%(v/v) PEG 4000, 0.25 M ammonium sulfate, 100 mM sodium acetate pH 4.8. The crystals were cryoprotected in mother liquor supplemented with either 20%(w/v) PEG 400 or 20%(v/v) glycerol and flash-cooled in liquid nitrogen.
For formation of the PtxS–DNA complex, an aliquot of purified native PtxS at 70 mg ml−1 was incubated with an equimolar concentration of duplex DNA (cognate promoter DNA) with sequence 5′-TGAAACCGGTTTCA-3′ previously generated by the annealing of two synthesized complementary oligonucleotides. The DNA/protein mixture was then used to perform crystallization experiments using the counter-diffusion method and the CSK-24 kit (Triana S&T, Granada, Spain). Three-dimensional PtxS–DNA crystals with hexagonal faces were grown in several conditions. Post-refinement of initial hits allowed improved crystals to be obtained using 20%(v/v) PEG 400, 0.1 M HEPES pH 7.5, 0.1 M calcium chloride.
X-ray diffraction data sets were collected using synchrotron radiation on beamline ID23-2 at the ESRF. Data sets were collected from PtxS–DNA crystals using a MAR 225 detector (oscillation range 0.5°, crystal-to-detector distance of 196.3 mm). The diffraction data were processed using XDS (Kabsch, 2010 ▶) and scaled using SCALA (Evans, 2006 ▶) from the CCP4 suite (Winn et al., 2011 ▶) to a resolution of 1.92 Å.
3. Results and discussion
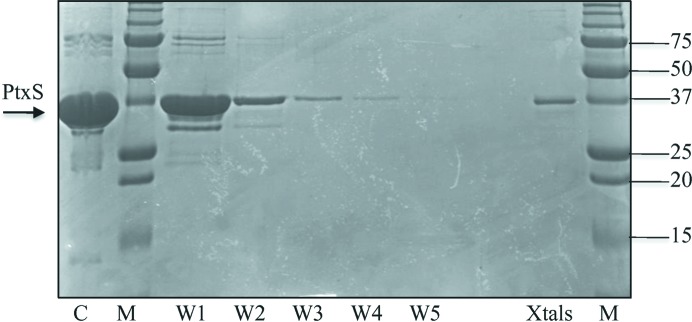
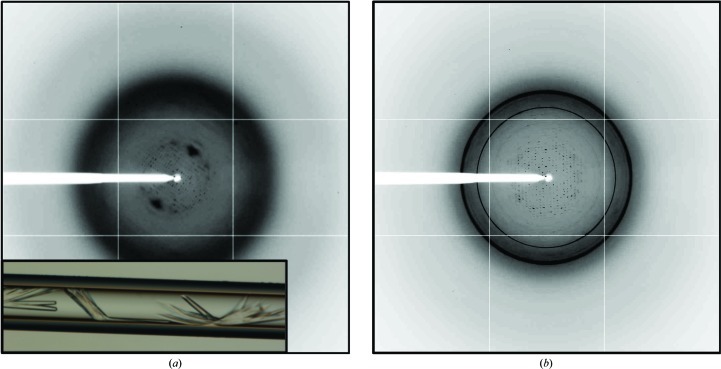
Screening of crystallization conditions for native PtxS was initially carried out using the counter-diffusion technique in capillaries of 0.1 mm inner diameter. Rod-shaped crystals were obtained in two separate conditions: 2 M potassium/sodium phosphate, 0.1 M Na HEPES pH 7.5 and 1.5 M ammonium sulfate, 0.1 M sodium citrate pH 5.6. Crystal analysis by SDS–PAGE confirmed the presence of full-length PtxS (Fig. 1 ▶). However, X-ray analysis on beamline ID14-4 at the ESRF produced poor diffraction data that could not be improved by the use of cryoprotectant solutions (Fig. 2 ▶ a). All attempts to improve the crystallization conditions, including variation of protein concentration, high-resolution pH screening and the use of capillaries with a larger inner diameter, did not improve either the crystal quality or size. Alternatively, the same crystallization conditions were used to grow similar crystals by the hanging-drop vapour-diffusion technique. Crystals grown by the hanging-drop method were screened for cryoprotection using glycerol, MPD, sorbitol and PEG 400 at different concentrations and equilibration times. The resulting crystals were cryoprotected and tested for diffraction on beamline ID14-4 at the ESRF, revealing that only the use of PEG 400 at high concentration [20%(w/v)] did not destroy the sample during crystal soaking. Unfortunately, the crystals only diffracted to a maximum resolution of 6 Å, frequently showing the presence of ice rings and clear deformation of the spot shape (Fig. 2 ▶ b).
Figure 1.
SDS–PAGE gel of PtxS crystals. Purified PtxS used in crystallization trials is shown in lane C. Lanes W1, W2, W3, W4 and W5 contain samples obtained after washing the crystals with reservoir solution. The final sample containing PtxS crystals can be observed in lane Xtals. Lanes M contain protein marker (Bio-Rad; labelled in kDa).
Figure 2.
Initial native PtxS crystals and their diffraction pattern. (a) Native PtxS crystals initially obtained by the counter-diffusion technique in 0.1 mm capillaries are shown together with their diffraction pattern after cryoprotection with mother liquor supplemented with 20%(v/v) glycerol. (b) Diffraction image of refined PtxS crystals grown by the vapour-diffusion method in the hanging-drop configuration. The image was obtained following cryoprotection with mother liquor supplemented with 20%(w/v) PEG 400.
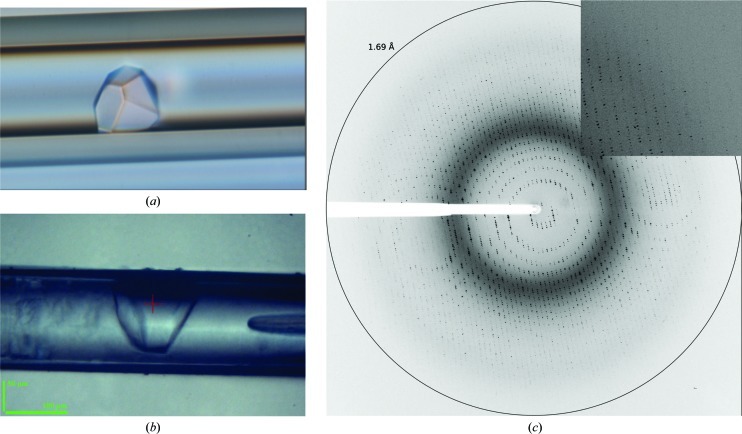
Our efforts then focused on promoting crystal improvement through better protein stabilization. To this end, native PtxS was incubated with a duplex DNA fragment comprising its operator site at a 1:1 protein:DNA molar ratio prior to crystallization experiments. As described above, initial screening was carried out by the counter-diffusion technique in capillaries of 0.1 mm inner diameter. PtxS–DNA crystals with different shapes grew in several conditions and were larger than those obtained without DNA (Fig. 3 ▶ a). The refined crystallization solution consisted of 20%(v/v) PEG 400, 0.1 M HEPES pH 7.5, 0.1 M calcium chloride. Crystals were subsequently reproduced using both the hanging-drop vapour-diffusion and the counter-diffusion methods. Diffraction tests of crystals grown inside 0.2 mm capillaries were performed in-house at room temperature. The crystals diffracted to a maximum resolution of 3 Å, but decayed rapidly. In order to obtain higher resolution data, further tests were performed on the ID23-2 microfocus beamline at the ESRF. Initially, cryoprotected crystals extracted from both hanging drops and capillaries were tested for diffraction, but only low-resolution diffraction data could be recorded. Analysis of the diffraction images indicated an abrupt loss of diffraction power with an increase in mosaicity. To avoid crystal extraction from the capillary and the stress provoked by the cryoprotection protocol, we attempted the direct diffraction of crystals in 0.1 mm capillaries (Martínez-Rodríguez et al., 2008 ▶). Capillary portions of approximately 1 cm, ideally containing a single crystal, were attached to the end of a SPINE standard cap and flash-cooled in liquid nitrogen. The 20%(w/v) PEG 400 present in the crystallization solution was sufficient to prevent the formation of ice crystals (Figs. 3 ▶ b and 3 ▶ c).
Figure 3.
Capillary counter-diffusion PtxS–DNA crystal and diffraction pattern. (a) Initial counter-diffusion-grown crystals of the PtxS–DNA complex. (b) Portion of a capillary containing a PtxS–DNA crystal cryocooled and automatically mounted and centred on the ID23-2 beamline. (c) Corresponding diffraction pattern of an unextracted crystal obtained by counter-diffusion.
The automatic mounting system available at ID23-2 at the ESRF was able to handle the standard pin with the portion of capillary attached to it. Initially, the auto-centring routine was used to detect the capillary (Lavault et al., 2006 ▶), but further fine-positioning of the crystal was performed manually. As a result, the quality of the diffraction pattern of PtxS–DNA crystals that were not extracted from the capillary and that were not brought into contact with a cryoprotectant was highly improved (Figs. 3 ▶ b and 3 ▶ c) compared with analogous experiments using handled crystals. A full data set was collected from a single crystal that diffracted X-rays to a maximum resolution of 1.92 Å (Table 1 ▶).
Table 1. Summary of X-ray data and statistics for the PtxS–DNA complex crystal.
Values in parentheses are for the highest resolution shell.
| Beamline | ID23-2, ESRF |
| Wavelength (Å) | 0.8726 |
| Beam size (µm) | 10 × 10 |
| Space group | P3 |
| Unit-cell parameters (Å) | a = b = 213.71, c = 71.57 |
| Resolution range (Å) | 56.61–1.92 (1.99–1.92) |
| Observed reflections | 654069 (61526) |
| Independent reflections | 275363 (26994) |
| Data completeness (%) | 98.6 (98.5) |
| R merge † (%) | 7.9 (43.5) |
| Average I/σ(I) | 10.3 (2.7) |
| Multiplicity | 2.4 (2.3) |
| Molecules per asymmetric unit | 3 |
| Matthews coefficient (Å3 Da−1) | 2.35 |
| Solvent content (%) | 47.55 |
R
merge = 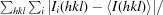
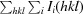 , where Ii(hkl) is the ith measurement of reflection hkl and 〈I(hkl)〉 is the weighted mean of all measurements.
, where Ii(hkl) is the ith measurement of reflection hkl and 〈I(hkl)〉 is the weighted mean of all measurements.
4. Concluding remarks
Our experiments have shown that PtxS crystals (native or in complex with DNA) are highly sensitivite to cryo-buffer treatment, which impedes the collection of good-quality data. Only unhandled PtxS–DNA crystals grown in capillaries and flash-cooled in liquid nitrogen without supplementation with cryoprotectant yielded clear diffraction patterns to high resolution. A complete data set from a crystal grown in situ in a capillary that diffracted to 1.92 Å resolution was collected on the highly automated microfocus beamline ID23-2 at the ESRF. These results substantiate the usefulness of the capillary counter-diffusion technique as a powerful tool for high-throughput crystallization of sensitive samples.
Acknowledgments
The projects ‘Factoría de Cristalización’ and CSD2007-0005, Consolider-Ingenio and Pathogenomics ERANET BIO2008-04419-E (MICINN) and Proyecto de Excelencia RNM-5384 of the Junta de Andalucía provided financial support for this work. JAG acknowledges the financial support of project BIO2010-16800 of MICINN. EPM is supported by a ‘Ramón y Cajal’ research contract (MICINN). We are also grateful to the staff of beamlines ID14-4 and ID23-2 at ESRF, Grenoble, France for their help during data collection and to Dr Alfonso García Caballero for helpful and constructive reading of the manuscript.
References
- Castillo, T. del, Ramos, J. L., Rodríguez-Herva, J. J., Fuhrer, T., Sauer, U. & Duque, E. (2007). J. Bacteriol. 189, 5142–5152. [DOI] [PMC free article] [PubMed]
- Colmer, J. A. & Hamood, A. N. (1998). Mol. Gen. Genet. 258, 250–259. [DOI] [PubMed]
- Daddaoua, A., Krell, T., Alfonso, C., Morel, B. & Ramos, J.-L. (2010). J. Bacteriol. 192, 4357–4366. [DOI] [PMC free article] [PubMed]
- Entner, N. & Doudoroff, M. (1952). J. Biol. Chem. 196, 853–862. [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Flot, D., Mairs, T., Giraud, T., Guijarro, M., Lesourd, M., Rey, V., van Brussel, D., Morawe, C., Borel, C., Hignette, O., Chavanne, J., Nurizzo, D., McSweeney, S. & Mitchell, E. (2010). J. Synchrotron Rad. 17, 107–118. [DOI] [PMC free article] [PubMed]
- García-Ruiz, J. M. (2003). Methods Enzymol. 368, 130–154. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lavault, B., Ravelli, R. B. G. & Cipriani, F. (2006). Acta Cryst. D62, 1348–1357. [DOI] [PubMed]
- Martínez-Rodríguez, S., González-Ramírez, L. A., Clemente-Jiménez, J. M., Rodríguez-Vico, F., Las Heras-Vázquez, F. J., Gavira, J. A. & García-Ruiz, J. M. (2008). Acta Cryst. F64, 50–53. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.