The FliH–FliI complex from the bacterial flagellar type III export apparatus has been expressed, purified and crystallized, and the crystals have been characterized by X-ray diffraction.
Keywords: FliH–FliI complex, type III export apparatus, bacterial flagellum
Abstract
The bacterial flagellar proteins are translocated into the central channel of the flagellum by a specific protein-export apparatus for self-assembly at the distal growing end. FliH and FliI are soluble components of the export apparatus and form an FliH2–FliI heterotrimer in the cytoplasm. FliI is an ATPase and the FliH2–FliI complex delivers export substrates from the cytoplasm to an export gate made up of six integral membrane proteins of the export apparatus. In this study, an FliHC fragment consisting of residues 99–235 was co-purified with FliI and the FliHC2–FliI complex was crystallized. Crystals were obtained using the hanging-drop vapour-diffusion technique with PEG 400 as a precipitant. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 133.7, b = 147.3, c = 164.2 Å, and diffracted to 3.0 Å resolution.
1. Introduction
Bacteria move in liquid environments by rotating a long helical filamentous organelle called the flagellum, which is composed of a basal body, a hook and a filament. For construction of the flagellum, most flagellar proteins are synthesized in the cytoplasm, translocated into the central channel of the growing flagellum by the flagellar type III protein-export apparatus and then travel through the channel to the growing end. The export apparatus consists of six integral membrane proteins, FlhA, FlhB, FliO, FliP, FliQ and FliR, and three soluble proteins, FliH, FliI and FliJ. These proteins are highly homologous to those of the type III secretion system of pathogenic bacteria, which directly injects virulence factors into eukaryotic host cells (Macnab, 2004 ▶; Minamino & Namba, 2004 ▶; Minamino et al., 2008 ▶).
FliI is a Walker-type ATPase (Fan & Macnab, 1996 ▶) and forms a homohexameric ring to fully exert its ATPase activity (Claret et al., 2003 ▶; Minamino et al., 2006 ▶). The entire crystal structure of FliI highly resembles the α and β subunits of F1-ATPase (Imada et al., 2007 ▶). The formation of the hexamer ring and the ATPase activity are promoted by FliJ, which shows a remarkable structural similarity to the coiled-coil part of the F1 γ subunit. FliJ binds to the central hole of the FliI ring to form an FliI6–FliJ complex in a similar manner to the F1 α3β3γ complex (Ibuki et al., 2011 ▶).
FliI forms a heterotrimeric complex, with an FliH dimer working as the ATPase regulator (Auvray et al., 2002 ▶; González-Pedrajo et al., 2002 ▶; Minamino & Macnab, 2000 ▶; Minamino et al., 2001 ▶). The FliH homodimer binds to the extreme N-terminal region of FliI and prevents FliI from forming the homohexameric ring, thereby suppressing the ATPase activity in the cytoplasm (Minamino & Macnab, 2000 ▶; Okabe et al., 2009 ▶). FliT, a flagellar export chaperone that is specific for the filament-capping protein FliD, also binds to the N-terminal region of FliI through its core (FliT94) and competes with FliH in FliI binding during the export process of FliD (Minamino et al., 2012 ▶).
The FliH2–FliI complex, along with FliJ, recruits export substrates from the cytoplasm to their docking platform in the export gate made up of six integral membrane proteins of the export apparatus and induces the initial entry of the substrates into the gate (Minamino et al., 2003 ▶; Minamino & Namba, 2008 ▶; Thomas et al., 2004 ▶). Unfolding and translocation of the substrates through the gate is powered by the proton motive force (PMF) across the cytoplasmic membrane, while the chemical energy derived from the hydrolysis of ATP by FliI is used for the release of FliH and FliI from the gate and substrate translocation (Minamino & Namba, 2008 ▶; Kazetani et al., 2009 ▶). The docking platform is made up of the cytoplasmic domains of FlhA and FlhB, and a specific interaction between FliJ and FlhA brought about by FliH and FliI is required for efficient PMF-driven protein export (Minamino et al., 2011 ▶).
FliH from Salmonella enterica serovar Typhimurium (UniProt ID P15934) consists of 235 amino-acid residues and has an elongated shape (Minamino & Macnab, 2000 ▶). The extreme N-terminal region of FliH is responsible not only for the interaction with the flagellar C-ring protein FliN but also for stable association of the FliI hexamer with the docking platform (Minamino et al., 2009 ▶). A central region between residues 101 and 140 is required for homodimer formation and contains a sequence with a significant probability of an α-helical coiled-coil structure (González-Pedrajo et al., 2002 ▶). The C-terminal region of FliH provides a binding site for FliI (González-Pedrajo et al., 2002 ▶; Minamino et al., 2002 ▶). Interestingly, the N- and C-terminal regions of FliH are homologous to the β and δ subunits of FoF1-ATP synthase, respectively (Pallen et al., 2006 ▶). Since the β and δ subunits form the peripheral stalk that connects F1 to Fo, these similarities suggest that FliH may anchor the FliI6–FliJ ring complex to the export gate. However, the action of FliH during flagellar protein export remains obscure. In order to understand the role of FliH in the protein-export process, the atomic structure of FliH is essential. Here, we copurified an FliHC fragment consisting of residues 99–235 with FliI and crystallized the FliHC2–FliI heterotrimeric complex. We also report the preliminary X-ray crystallographic study of the complex.
2. Materials and methods
2.1. Protein expression and purification
An NdeI–BamHI fragment encoding the fliI gene from pMM1701 (Minamino & Macnab, 2000 ▶) was subcloned into the NdeI–BglII site of pETDuet-1 (Novagen) to create the plasmid pYU001. An NdeI–BamHI fragment encoding the fliHC2 gene from pBGhHC2 (González-Pedrajo et al., 2002 ▶) was inserted into the NdeI–BamHI site of pET15b (Novagen) to create the plasmid pYU002. An NcoI–BamHI fragment encoding N-terminally His-tagged FliHC2 (His-FliHC2) was subcloned into the NcoI–BamHI site of pYU001 to give the plasmid pYU003. A 10 ml overnight culture of Escherichia coli BL21 (DE3) cells transformed with pYU003, which encodes both His-FliHC2 and FliI, was inoculated into 1 l LB medium (10 g Bacto tryptone, 5 g yeast extract, 10 g NaCl per litre) containing 100 mg ml−1 ampicillin. Cells were grown at 310 K until the culture density reached an OD600 of 0.6. The expression of His-FliHC2 and FliI was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.1 mM and the culture was continued overnight at 291 K. The cells were harvested by centrifugation (6400g, 10 min, 277 K) and stored at 193 K. The cells were thawed, suspended in binding buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl) with a tablet of Complete protease-inhibitor cocktail (Boehringer Mannheim) and sonicated (Astrason model XL2020 sonicator, Misonix Inc.). The cell lysate was centrifuged (14 000g, 40 min, 277 K) to remove cell debris. The supernatant was loaded onto a HisTrap HP column (GE Healthcare) equilibrated with binding buffer. After washing the column with 15 ml binding buffer containing 50 mM imidazole, proteins were eluted using a linear gradient of 50–500 mM imidazole and fractions containing the His-FliHC2–FliI complex were collected. To remove the His tag from the N-terminus of FliHC2, 25 units of thrombin (Amersham Biosciences) were added to the eluted fractions containing the His-FliHC2–FliI complex. The protease cleavage produced FliHC2–FliI with an extra three residues, glycine, serine and histidine, attached to the N-terminus as a remnant of the His tag. The reaction mixture was dialyzed overnight against binding buffer at room temperature and was loaded onto a HisTrap HP column. After washing the column with 15 ml binding buffer, proteins were eluted in 20 ml binding buffer containing 50 mM imidazole. The fractions containing the FliHC2–FliI complex were applied onto a HiLoad Superdex 75 (26/60) column (GE Healthcare) equilibrated with 25 mM HEPES–NaOH buffer pH 8.0 containing 150 mM NaCl and 1 mM DTT. The purity of the product was analyzed by SDS–PAGE and MALDI–TOF mass spectrometry (Voyager DE/PRO). The purified protein sample was dialyzed overnight against 10 mM HEPES–NaOH pH 8.0, 10 mM NaCl and concentrated to 7.0 mg ml−1 for further use.
2.2. Crystallization
Crystallization screening of the FliHC2–FliI complex was performed at 293 K by the sitting-drop vapour-diffusion technique using the following screening kits: Wizard I and II, Cryo I and II (Emerald BioStructures), Crystal Screen and Crystal Screen 2 (Hampton Research). Each drop was prepared by mixing 0.5 µl protein solution (7.0 mg ml−1) containing 1 mM ADP and 1 mM MgCl2 with 0.5 µl reservoir solution and was equilibrated against 70 µl reservoir solution. Within a week, small crystals were observed in drops containing 14–18% PEG 400 at pH 7.5. We optimized the conditions by varying the precipitant concentration, pH and additives using the hanging-drop method. Finally, crystals suitable for X-ray analysis were obtained from drops prepared by mixing 2 µl protein solution (7 mg ml−1) containing 2 mM ADP with 2 µl reservoir solution consisting of 0.1 M HEPES–NaOH pH 7.2, 5%(v/v) PEG 400, 0.1 M magnesium acetate at 285 K using the microseeding technique. The crystals appeared within one week.
2.3. Data collection and processing
All X-ray diffraction data were collected on beamline BL41XU at SPring-8, Harima, Japan. The crystals were soaked in a solution consisting of 90%(v/v) reservoir solution and 10%(v/v) MPD for a few seconds, cooled in liquid nitrogen and mounted in a 40 K helium-gas flow (Rigaku). The diffraction data were recorded on an ADSC Quantum 315 CCD detector (Area Detector Systems Corporation). The diffraction data were indexed, integrated and scaled using the programs MOSFLM (Leslie, 1992 ▶) and SCALA from the CCP4 program suite (Winn et al., 2011 ▶). The statistics of data collection are summarized in Table 1 ▶.
Table 1. Summary of the data statistics for FliHC2–FliI.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 133.7, b = 147.3, c = 164.2 |
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 60.0–3.0 (3.16–3.00) |
| Observations | 298335 (39579) |
| Unique reflections | 65054 (9429) |
| Completeness (%) | 99.4 (99.6) |
| Multiplicity | 4.6 (4.2) |
| 〈I/σ(I)〉 | 5.6 (2.9) |
| R merge (%) | 8.1 (39.7) |
3. Results and discussion
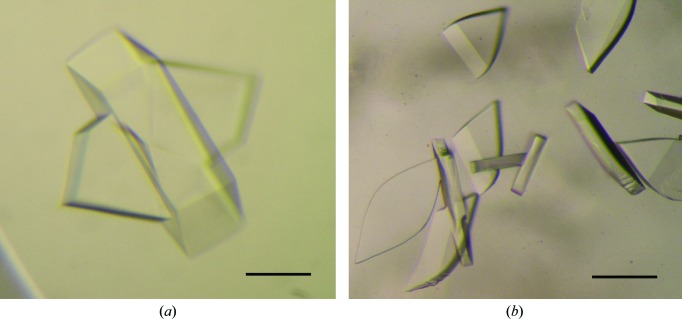
Initially, we prepared the FliHC2–FliI complex by mixing individually purified FliHC and His-FliI. After proteolytic removal of the His tag from FliI, the complex was further purified by size-exclusion chromatography and used for crystallization. Crystals were successfully grown from a solution consisting of 0.1 M glycine–NaOH pH 9.0–9.8, 6–14%(w/v) PEG 8000 at 285 K (Fig. 1 ▶ a). However, these crystals only diffracted to 5 Å resolution and no better crystals were obtained from this sample. We then constructed a pETDuet-1-based plasmid co-expressing both FliHC and FliI and copurified the complex. Crystals from this sample had a tendency to grow as clusters. However, they diffracted to ∼3.5 Å resolution. We then applied a microseeding technique to prepare single crystals and finally obtained leaf-shaped crystals with typical dimensions of 0.1 × 0.05 × 0.2 mm (Fig. 1 ▶ b). The concentration of PEG 400 in the optimized condition was not sufficiently high for cryoprotection. We used MPD as a cryoprotectant, as soaking the crystals in a high-concentration PEG 400 solution caused serious damage.
Figure 1.
Crystals of FliHC2–FliI. (a) Crystals obtained using 11% PEG 8000, 0.1 M glycine–NaOH pH 9.4. (b) Crystals obtained using 5% PEG 400, 0.1 M magnesium acetate, 0.1 M HEPES–NaOH pH 7.2 with the microseeding technique. The scale bar is 0.1 mm in length.
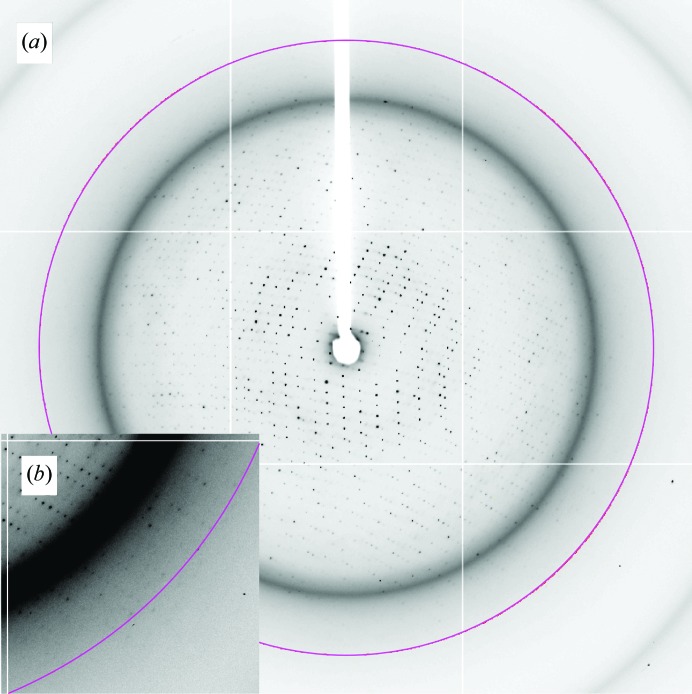
The crystals diffracted to 3.0 Å resolution (Fig. 2 ▶). To avoid radiation damage, we shifted the irradiation position of the crystal frame by frame during measurement. Moreover, cryocooling of the crystal to around 40 K was essential for full data collection.
Figure 2.
Diffraction image of an FliHC2–FliI crystal grown from the optimized condition. (a) A typical X-ray pattern of an FliHC2–FliI crystal collected on beamline BL41XU at SPring-8. The circle indicates a resolution of 3 Å. (b) Close-up view of the diffraction image.
The crystal belonged to the orthorhombic space group P212121, with unit-cell parameters a = 133.7, b = 147.3, c = 164.2 Å. The Matthews coefficient (V M; Matthews, 1968 ▶) suggests the presence of three, four or five FliHC2–FliI complexes in the asymmetric unit, with solvent contents of 64.3, 52.4 or 53.0%, respectively. The self-rotation function calculation showed no significant peak, implying that local twofold axes are parallel to the crystallographic axes or that the molecules in the asymmetric unit are related by local translational symmetry. Structural analysis by molecular replacement using FliI as a search model is now in progress.
Acknowledgments
We thank N. Shimizu and K. Hasegawa at SPring-8 for technical help in the use of beamline BL41XU. This work was supported in part by Grants-in-Aid for Scientific Research to KI (23115008), to TM (22570161) and to KN (21227006) and the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Science and Culture of Japan.
References
- Auvray, F., Ozin, A. J., Claret, L. & Hughes, C. (2002). J. Mol. Biol. 318, 941–950. [DOI] [PMC free article] [PubMed]
- Claret, L., Calder, S. R., Higgins, M. & Hughes, C. (2003). Mol. Microbiol. 48, 1349–1355. [DOI] [PMC free article] [PubMed]
- Fan, F. & Macnab, R. M. (1996). J. Biol. Chem. 271, 31981–31988. [DOI] [PubMed]
- González-Pedrajo, B., Fraser, G. M., Minamino, T. & Macnab, R. M. (2002). Mol. Microbiol. 45, 967–982. [DOI] [PubMed]
- Ibuki, T., Imada, K., Minamino, T., Kato, T., Miyata, T. & Namba, K. (2011). Nature Struct. Mol. Biol. 18, 277–282. [DOI] [PubMed]
- Imada, K., Minamino, T., Tahara, A. & Namba, K. (2007). Proc. Natl Acad. Sci. USA, 104, 131–137. [DOI] [PMC free article] [PubMed]
- Kazetani, K., Minamino, T., Miyata, T., Kato, T. & Namba, K. (2009). Biochem. Biophys. Res. Commun. 388, 323–327. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Macnab, R. M. (2004). Biochim. Biophys. Acta, 1694, 207–217. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Minamino, T., González-Pedrajo, B., Kihara, M., Namba, K. & Macnab, R. M. (2003). J. Bacteriol. 185, 3983–3988. [DOI] [PMC free article] [PubMed]
- Minamino, T., González-Pedrajo, B., Oosawa, K., Namba, K. & Macnab, R. M. (2002). J. Mol. Biol. 322, 281–290. [DOI] [PubMed]
- Minamino, T., Imada, K. & Namba, K. (2008). Mol. Biosyst. 4, 1105–1115. [DOI] [PubMed]
- Minamino, T., Kazetani, K., Tahara, A., Suzuki, H., Furukawa, Y., Kihara, M. & Namba, K. (2006). J. Mol. Biol. 360, 510–519. [DOI] [PubMed]
- Minamino, T., Kinoshita, M., Imada, K. & Namba, K. (2012). Mol. Microbiol. 83, 168–178. [DOI] [PubMed]
- Minamino, T. & Macnab, R. M. (2000). Mol. Microbiol. 37, 1494–1503. [DOI] [PubMed]
- Minamino, T., Morimoto, Y. V., Hara, N. & Namba, K. (2011). Nature Commun. 2, 475. [DOI] [PMC free article] [PubMed]
- Minamino, T. & Namba, K. (2004). J. Mol. Microbiol. Biotechnol. 7, 5–17. [DOI] [PubMed]
- Minamino, T. & Namba, K. (2008). Nature (London), 451, 485–488. [DOI] [PubMed]
- Minamino, T., Tame, J. R. H., Namba, K. & Macnab, R. M. (2001). J. Mol. Biol. 312, 1027–1036. [DOI] [PubMed]
- Minamino, T., Yoshimura, S. D., Morimoto, Y. V., González-Pedrajo, B., Kami-Ike, N. & Namba, K. (2009). Mol. Microbiol. 74, 1471–1483. [DOI] [PubMed]
- Okabe, M., Minamino, T., Imada, K., Namba, K. & Kihara, M. (2009). FEBS Lett. 583, 743–748. [DOI] [PubMed]
- Pallen, M. J., Bailey, C. M. & Beatson, S. A. (2006). Protein Sci. 15, 935–941. [DOI] [PMC free article] [PubMed]
- Thomas, J., Stafford, G. P. & Hughes, C. (2004). Proc. Natl Acad. Sci. USA, 101, 3945–3950. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.