Recombinant dihydroorotase domain of human CAD was expressed in E. coli, purified and crystallized. An X-ray diffraction data set was collected to 1.75 Å resolution.
Keywords: human CAD, dihydroorotase domain, pyrimidine biosynthesis
Abstract
CAD is a 243 kDa eukaryotic multifunctional polypeptide that catalyzes the first three reactions of de novo pyrimidine biosynthesis: glutamine-dependent carbamyl phosphate synthetase, aspartate transcarbamylase and dihydroorotase (DHO). In prokaryotes, these activities are associated with monofunctional proteins, for which crystal structures are available. However, there is no detailed structural information on the full-length CAD protein or any of its functional domains apart from that it associates to form a homohexamer of ∼1.5 MDa. Here, the expression, purification and crystallization of the DHO domain of human CAD are reported. The DHO domain forms homodimers in solution. Crystallization experiments yielded small crystals that were suitable for X-ray diffraction studies. A diffraction data set was collected to 1.75 Å resolution using synchrotron radiation at the SLS, Villigen, Switzerland. The crystals belonged to the orthorhombic space group C2221, with unit-cell parameters a = 82.1, b = 159.3, c = 61.5 Å. The Matthews coefficient calculation suggested the presence of one protein molecule per asymmetric unit, with a solvent content of 48%.
1. Introduction
Pyrimidine nucleotides are essential building blocks for nucleic acid synthesis, DNA repair and other cell functions (reviewed in Jones, 1980 ▶; Evans & Guy, 2004 ▶). In resting cells the need for pyrimidines is largely covered by the reutilization of preformed pyrimidines, whereas in proliferating cells, and particularly in tumour cells, de novo pyrimidine synthesis is essential to cover the high demand for DNA and RNA synthesis (Aoki & Weber, 1981 ▶; Fairbanks et al., 1995 ▶; Sigoillot et al., 2004 ▶). Consequently, the de novo biosynthetic pathway is a potential target for the development of antitumour drugs and also for the treatment of parasitic infections, since some pathogens (e.g. Plasmosdium species that cause malaria) are unable to recycle pyrimidines and depend on their de novo synthesis (Gero et al., 1984 ▶; Krungkrai et al., 1990 ▶; Christopherson et al., 2002 ▶).
In animals, pyrimidine biosynthesis is initiated and controlled by CAD, an ∼243 kDa multifunctional polypeptide that harbours the first three enzymatic activities of the pathway: glutamine-dependent carbamyl phosphate synthetase (GLN-CPS), aspartate transcarbamylase (ATC) and dihydroorotase (DHO) (Coleman et al., 1977 ▶; Kim et al., 1992 ▶; the acronym CAD is composed of the initials of the three different activities; Fig. 1 ▶ a). There is no direct structural information on CAD or on any of its domains apart from that it associates forming a homohexamer of ∼1.5 MDa (Lee et al., 1985 ▶) which, for reasons that are unclear, shuttles between the nucleus and the cytoplasm depending on the functional situation or the stage of the cell cycle (Sigoillot et al., 2005 ▶).
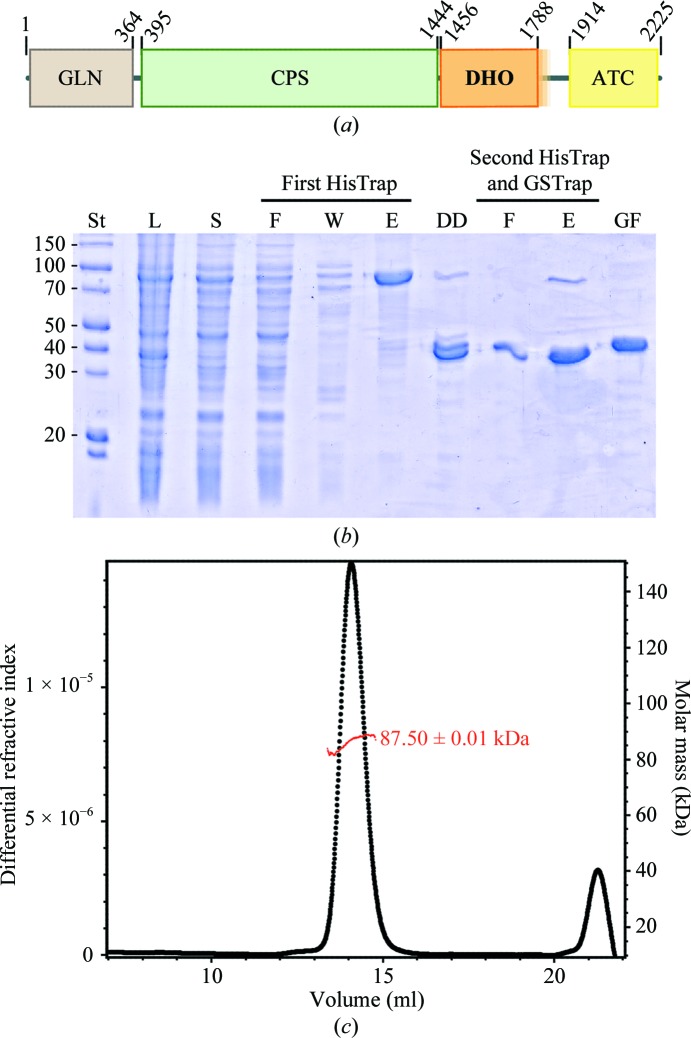
Figure 1.
Expression and purification of the DHO domain of human CAD. (a) Scheme of the domain organization of CAD. The DHO core domain (residues 1456–1788) is shown in orange and the uncertainty about the C-terminal boundary is indicated as a semitransparent extension into the linker region. (b) SDS–PAGE of human DHO purification. Lane St, molecular-mass standards (labelled in kDa); lane L, lysate; lane S, soluble fraction; lane F, flowthrough; lane W, column wash; lane E, elution; lane DD, dialyzed and digested with PreScission protease (note that the DHO domain without tag and the cleaved His6-MBP have similar masses); lane GF, final sample after gel filtration. (c) The molar mass of the protein was measured using gel filtration coupled with light scattering and indicates that the human DHO domain forms homodimers in solution.
Although the first three steps of pyrimidine biosynthesis are conserved among all species, the engineering of a single polypeptide carrying the three activities is unique to higher eukaryotes. In lower eukaryotes, such as Neurospora or yeast, the CPS and ATC activities are encoded within a single bifunctional CAD homologue that has an inactive DHO domain, and the DHO activity is encoded in a separate monofunctional protein (Lollier et al., 1995 ▶; Souciet et al., 1989 ▶). In contrast, in prokaryotes and plants CPS, ATC and DHO are separate monofunctional proteins that work independently, although they may associate into functional assemblies, as revealed by the DHO–ATC complex that is formed in the ancient hyperthermophilic bacterium Aquifex aeolicus (Ahuja et al., 2004 ▶; Zhang et al., 2009 ▶; Zrenner et al., 2006 ▶). A number of crystal structures of CPS, ATC and DHO from different bacteria or archaea have been determined. However, there is no structural information on any eukaryotic counterpart that could be applied to CAD.
DHO (EC 3.4.2.3) is a Zn2+ metalloenzyme that catalyzes the reversible conversion of carbamyl aspartate to dihydroorotate. Phylogenetic analyses (Fields et al., 1999 ▶) have classified the DHOs into two major groups. Type I are more ancient and larger (∼45 kDa); they are represented in all domains of life and include the DHO domain of CAD. Type II are smaller proteins (∼38 kDa) that are predominantly found in Gram-negative bacteria (e.g. Escherichia coli) and yeasts. The first reported structure of a DHO was that of the type II E. coli enzyme (Thoden et al., 2001 ▶), a dimer in which each subunit folds into an (α/β)8-barrel (or TIM-barrel) that is characteristic of the amidohydrolase superfamily of proteins (Holm & Sander, 1997 ▶). As in other members of this family, the active site of E. coli DHO presents four conserved histidines and one aspartate at specific positions in the β-barrel that coordinate two Zn2+ ions. Both metal ions are bridged by a carboxylated lysine and by a water molecule that is activated for nucleophilic attack (Porter et al., 2004 ▶; Lee et al., 2005 ▶, 2007 ▶).
A. aeolicus DHO provided the first structure of a type I DHO (Martin et al., 2005 ▶). This protein differed from E. coli DHO in having only one Zn2+ ion clearly visible in the active site and an aspartate replacing the carboxylated lysine. Interestingly, structural determination of A. aeolicus DHO in complex with ATC revealed a hollow dodecamer formed by two ATC trimers and six DHO monomers with rotational 32 symmetry (Zhang et al., 2009 ▶). It has been proposed that A. aeolicus DHO would closely resemble the DHO domain of CAD, since both proteins belong to the type I group and analysis of the isolated DHO of hamster CAD indicated the presence of only one Zn2+ per active site (Kelly et al., 1986 ▶) and did not detect the presence of a carboxylated lysine (Zhang et al., 2009 ▶); the A. aeolicus DHO–ATC complex thus might be a possible model for the core of the ∼1.5 MDa CAD complex.
Although crystallization and preliminary diffraction data have been reported for the DHO domain of hamster CAD (Maher et al., 2003 ▶), no structure was subsequently communicated. Here, we report the expression, purification and crystallization of the DHO domain of human CAD. Despite their small size, the protein crystals diffracted X-rays to high resolution.
2. Materials and methods
2.1. Cloning of human DHO
The cDNA of human CAD (UniProt P27708) was purchased from Open Biosystems (clone ID 5551082). Three different gene fragments covering the DHO domain plus additional C-terminal extensions into the DHO–ATC linker region were amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs). Constructs DHO-1, DHO-2 and DHO-3 started at residue 1456 and ended at residues 1788, 1800 and 1846, respectively. The forward primer (5′-AAGTTCTGTTTCAGGGCCCGatgacctcccaaaagcttgtg) and the reverse primers (DHO1, 5′-ATGGTCTAGAAAGCTTTAcaggaccacacggcggac; DHO2, 5′-ATGGTCTAGAAAGCTTTAcagaacctgcccatcgat; DHO3, 5′-ATGGTCTAGAAAGCTTTAatcaggaagccctgggatg) contained specific regions (shown in capitals) for cloning into the pOPIN-M expression vector (Oxford Protein Production Facility) using In-Fusion technology (Clontech). The resulting plasmids were verified by sequencing and transformed into E. coli Rosetta (DE3) pLysS cells (Novagen).
2.2. Protein expression and purification
Transformed bacterial cells were grown in autoinduction medium (Studier, 2005 ▶) supplemented with 100 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol in a shaking incubator for 6 h at 310 K followed by 21 h at 293 K. Cells were harvested by centrifugation, washed with PBS and stored at 193 K. The bacterial pellet was thawed and resuspended in buffer A (20 mM Tris–HCl pH 8, 0.5 M NaCl, 10 mM imidazole, 5% glycerol, 5 mM β-mercaptoethanol) with 2 mM phenylmethanesulfonyl fluoride (PMSF) and the cells were disrupted by sonication. The lysate was clarified by centrifugation in a Beckman Ti-45 rotor at 40 000 rev min−1 for 40 min. The supernatant was filtered through a 0.45 µm pore filter and applied onto a 5 ml Ni2+-loaded HisTrap Chelating FF column (GE Healthcare, USA). Following extensive washing of the column with buffer A containing 25 mM imidazole, the protein, which was tagged at the N-terminus with His6-tagged maltose-binding protein (His6-MBP), was eluted by increasing the imidazole concentration to 300 mM stepwise. Excess imidazole was removed and the tag was cleaved off by overnight dialysis against the same solution containing only 30 mM imidazole with the inclusion of GST-tagged PreScission protease (1/20th of the protein weight) within the dialysis bag. Two extra residues, a glycine and a proline, were left at the N-terminus of DHO (GP-1456MTSQ) after tag removal. The dialyzed and cleaved sample was loaded onto a second Ni2+-loaded HisTrap column connected to a 5 ml GSTrap FF column (GE Healthcare, USA) to retain the noncleaved protein, the His6-MBP tag and the protease. The cleaved DHO domain found in the flowthrough fractions was concentrated to ∼5 mg ml−1 using an Amicon Ultra system with a 10 kDa cutoff membrane and was further purified by size-exclusion chromatography on a Superdex 200 10/300 column (GE Healthcare, USA) equilibrated with GF buffer (20 mM Tris pH 8, 0.15 M NaCl, 20 µM ZnSO4, 0.2 mM TCEP). DHO eluted in a single peak that was pooled and concentrated as before to 5 mg ml−1. The sample was directly used for crystallization studies. In some instances the excess protein was supplemented with 20% glycerol, flash-frozen in liquid nitrogen before the gel-filtration step and stored at 193 K for several weeks, but this did not noticeably affect the crystallization properties of the protein. All purification steps were carried out at 277 K. The final sample purity was evaluated by SDS–PAGE with Coomassie staining. The protein concentration was determined from the absorbance at 280 nm using a theoretical ∊ of 38 960 M −1 cm−1.
2.3. Gel-filtration and multi-angle light-scattering (MALS) measurements
For molar-mass determination of the human DHO domain, 200 µl purified sample at 2 mg ml−1 was fractionated by gel filtration on a Superdex 200 10/300 column equilibrated in buffer GF using an ÄKTApurifier at a flow rate of 0.5 ml min−1. The eluted sample was characterized by in-line measurement of the refractive index and multi-angle light scattering using Optilab T-rEX and DAWN 8+ instruments, respectively (Wyatt). The data were analyzed using the ASTRA 6 software (Wyatt) to obtain the molar mass (Wyatt, 1993 ▶).
2.4. Crystallization
Crystallization screening was performed at 291 K using a Cartesian MicroSys robot (Genomic Solutions) and the sitting-drop vapour-diffusion method in 96-well MRC plates. Nanodrops consisting of 0.2 µl protein solution plus 0.2 µl reservoir solution were equilibrated against 70 µl reservoir solution. Initial screening involved different protein concentrations and commercial crystallization screens (the JCSG+, PACT and Protein Complex Suites from Qiagen). Small crystals were obtained and reproduced in 24-well plates using drops consisting of 1 µl protein solution and 1 µl reservoir solution and were further optimized by varying the protein and precipitant concentrations, the pH and the buffer. The optimal crystallization conditions consisted of 2–3 mg ml−1 protein with 2.5–3 M sodium formate and 0.1 M HEPES pH 6.0 or Tris–HCl pH 7.0 as the crystallization solution. The crystals were transferred to a cryoprotectant solution consisting of the mother liquor plus 10% glycerol and were flash-cooled in liquid nitrogen in a mounted cryoloop.
2.5. Data collection and reduction
The crystals were tested in-house using a Bruker FR-591 X-ray generator and a MAR345 image-plate detector. The crystals displayed diffraction to 3.0 Å resolution. A high-resolution data set was collected on beamline PXI at the SLS synchrotron, Villigen, Switzerland using a PILATUS 6M detector and a wavelength of 1.0 Å. A total wedge of 360° of data was collected with 0.25° oscillation, 0.25 s exposure per frame and a crystal-to-detector distance of 0.3 m. Data processing and scaling were performed using XDS (Kabsch, 2010 ▶). The statistics of the crystallographic data are summarized in Table 1 ▶.
Table 1. Data-collection statistics for the DHO domain of human CAD.
Values in parentheses are for the highest resolution shell.
| Processing software | XDS/XSCALE |
| Source | PXI beamline, SLS |
| Wavelength (Å) | 1.0 |
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 82.1, b = 159.3, c = 61.5 |
| Resolution range (Å) | 48–1.75 (1.79–1.75) |
| Reflections (observed/unique) | 266335/40865 |
| Multiplicity | 6.5 (6.6) |
| Completeness (%) | 99.5 (100) |
| R meas | 0.098 (0.55) |
| 〈I/σ(I)〉 | 13.3 (3.29) |
| Wilson B factor (Å2) | 27.7 |
3. Results and discussion
The DHO domain of human CAD (huDHO) was cloned, expressed, purified and crystallized for structural characterization. The N-terminal (1456MTSQ–) and C-terminal (–RVVL1788) boundaries of a minimal huDHO were defined based on sequence comparison with bacterial monofunctional enzymes, as previously performed for the characterization of hamster CAD (Simmer et al., 1990 ▶). Initial attempts to produce this protein construct or a larger construct with an additional 12 amino acids extending from the C-terminus into the linker region to the ATC domain (amino acids 1456–1800) were unsuccessful. Previous studies showed that the protein could only be produced in an active form by extending the C-terminal boundary of hamster DHO 24–58 amino acids into the linker (Simmer et al., 1990 ▶; Musmanno et al., 1992 ▶; Williams et al., 1993 ▶; reviewed by Davidson et al., 1993 ▶). Indeed, the crystallization of hamster DHO was achieved using a protein that incorporated a C-terminal extension of 33 amino acids (Maher et al., 2003 ▶). In the present work, by extending the C-terminus of huDHO 58 amino acids into the linker sequence (amino acids 1456–1846) we obtained a huDHO construct that was well expressed and soluble (Fig. 1 ▶ b).
The recombinant protein was produced with an N-terminal His6-MBP tag that was removed by digestion with PreScission protease. After two rounds of Ni2+-chelating chromatography separated by a tag-cleavage step and followed by a size-exclusion chromatography step, the huDHO protein could be obtained without tags and with high purity (Fig. 1 ▶ b). SDS–PAGE analysis of the purified huDHO shows that the protein migrates at the expected molecular weight of 43 kDa. The molecular mass of the protein in solution was measured by multi-angle static light scattering (MALS) of the purified sample eluting from an in-line gel-filtration column (Fig. 1 ▶ c). The purified sample contained a single species of average molecular mass 87.50 ± 0.01 kDa. The measured value matches the formation of a homodimer, which is in agreement with previous studies reporting that the DHO domain of hamster CAD is also a dimer in solution (Kelly et al., 1986 ▶; Williams et al., 1993 ▶).
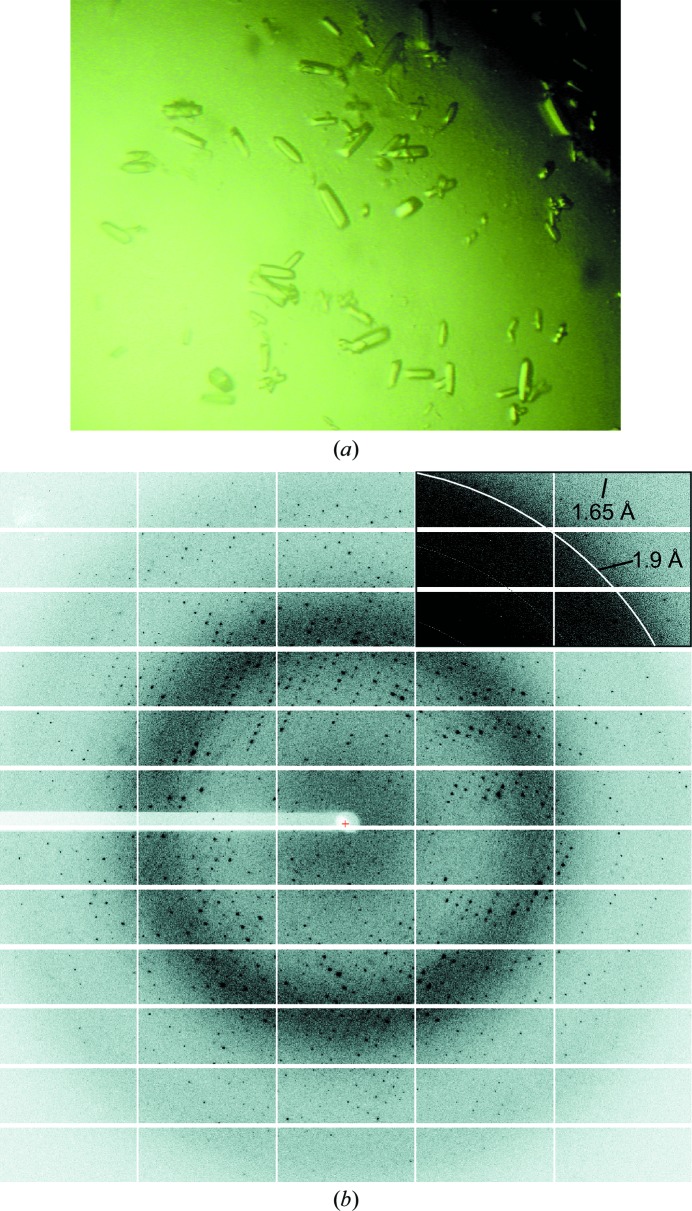
Initial crystallization trials gave crystals of 50 µm in the largest dimension in condition No. 79 of The Protein Complex Suite (0.1 M Tris–HCl pH 7.0, 3 M sodium formate). Following optimization, larger crystals of 0.1 mm in the maximum dimension were obtained at 291 K after 1–2 d (Fig. 2 ▶ a). Initial diffraction tests were performed on an in-house X-ray generator using liquid-nitrogen-cooled crystals. Despite their small size, the crystals diffracted X-rays to 3 Å resolution (data not shown). To obtain higher resolution data, the crystals were studied using synchrotron radiation. The crystals diffracted to a maximum resolution of 1.65 Å on the PXI beamline at SLS at a wavelength of 1.0 Å (Fig. 2 ▶ b). The data set was processed and scaled to 1.75 Å resolution and the statistics are presented in Table 1 ▶. The crystals belonged to space group C2221, with unit-cell parameters a = 82.1, b = 159.3, c = 61.5 Å. Calculation of the Matthews coefficient suggested the presence of one protein molecule per asymmetric unit, with a V M of 2.35 Å3 Da−1 and a solvent content of 48%. Since the protein is a homodimer in solution, according to the calculated unit-cell content the molecular twofold symmetry axis should be coincident with a crystallographic dyad axis.
Figure 2.
Crystallization of the DHO domain of human CAD and X-ray diffraction pattern. (a) Native crystals of recombinant human DHO grown by the hanging-drop method. The typical crystal size is ∼0.1 mm in the largest dimension. (b) X-ray diffraction pattern of a crystal obtained using synchrotron radiation. The upper right corner of the image is shown with different contrast to indicate the diffraction limit.
We have seemingly overcome the cross-linking and crystallization problems that were encountered with the DHO domain of hamster CAD (Maher et al., 2003 ▶). When this protein was produced recombinantly in E. coli, the authors observed a slow transformation of the dimers into tetramers owing to the formation of disulfide bonds between a surface-exposed cysteine, identified as Cys241 (or Cys1696 according to the full-length CAD numbering used in this article), from subunits of different dimers. Interestingly, the crystals of hamster DHO, which appeared after 2–3 months, corresponded to the tetrameric species and diffracted X-rays to a limited resolution of 4 Å. Although the six cysteines that were present in the sequence of hamster DHO are conserved in the human protein, we did not detect the formation of huDHO tetramers under the conditions used for purification (Fig. 1 ▶ c). Since huDHO was kept in the presence of a strong reducing agent and the protein crystals grew quickly (1–2 d), it is most likely that the huDHO crystals are formed by the dimers observed in solution and not by a cross-linked species.
Next, we will attempt to obtain crystallographic phases either by molecular replacement using the available bacterial DHO models or by multi-wavelength anomalous dispersion methods using the signal from the intrinsic Zn2+ ions. The crystal structure of human DHO will provide the first view of a eukaryotic DHO. This should help us to understand the relation between prokaryotic and eukaryotic DHOs and will provide us with the first high-resolution information for use in attempting to decipher the architecture of CAD.
Acknowledgments
This work was initiated in the laboratory of Professor Vicente Rubio (IBV-CSIC), to whom SRM is indebted for his support and advice. We thank Angela de Manzanos for help in cloning human DHO at IBV-CSIC, Dr Ramón Campos-Olivas (CNIO Spectroscopy and NMR Unit) for performing gel-filtration MALS experiments and Dr Guillermo Montoya for synchrotron data collection and critical reading of the manuscript. Financial support was obtained through the Spanish Ministry of Science and Innovation (BFU2010-16812). SRM and RM are researchers of the Ramón y Cajal and Juan de la Cierva programs of the Spanish Ministry of Science and Innovation, respectively.
References
- Ahuja, A., Purcarea, C., Ebert, R., Sadecki, S., Guy, H. I. & Evans, D. R. (2004). J. Biol. Chem. 279, 53136–53144. [DOI] [PubMed]
- Aoki, T. & Weber, G. (1981). Science, 212, 463–465. [DOI] [PubMed]
- Christopherson, R. I., Lyons, S. D. & Wilson, P. K. (2002). Acc. Chem. Res. 35, 961–971. [DOI] [PubMed]
- Coleman, P. F., Suttle, D. P. & Stark, G. R. (1977). J. Biol. Chem. 252, 6379–6385. [PubMed]
- Davidson, J. N., Chen, K. C., Jamison, R. S., Musmanno, L. A. & Kern, C. B. (1993). Bioessays, 15, 157–164. [DOI] [PubMed]
- Evans, D. R. & Guy, H. I. (2004). J. Biol. Chem. 279, 33035–33038. [DOI] [PubMed]
- Fairbanks, L. D., Bofill, M., Ruckemann, K. & Simmonds, H. A. (1995). J. Biol. Chem. 270, 29682–29689. [PubMed]
- Fields, C., Brichta, D., Shephardson, M., Farinha, M. & O’Donovan, G. (1999). Paths Pyrimidines, 7, 49–63.
- Gero, A. M., Brown, G. V. & O’Sullivan, W. J. (1984). J. Parasitol. 70, 536–541. [PubMed]
- Holm, L. & Sander, C. (1997). Proteins, 28, 72–82. [PubMed]
- Jones, M. E. (1980). Annu. Rev. Biochem. 49, 253–279. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kelly, R. E., Mally, M. I. & Evans, D. R. (1986). J. Biol. Chem. 261, 6073–6083. [PubMed]
- Kim, H., Kelly, R. E. & Evans, D. R. (1992). J. Biol. Chem. 267, 7177–7184. [PubMed]
- Krungkrai, J., Cerami, A. & Henderson, G. B. (1990). Biochemistry, 29, 6270–6275. [DOI] [PubMed]
- Lee, L., Kelly, R. E., Pastra-Landis, S. C. & Evans, D. R. (1985). Proc. Natl Acad. Sci. USA, 82, 6802–6806. [DOI] [PMC free article] [PubMed]
- Lee, M., Chan, C. W., Graham, S. C., Christopherson, R. I., Guss, J. M. & Maher, M. J. (2007). J. Mol. Biol. 370, 812–825. [DOI] [PubMed]
- Lee, M., Chan, C. W., Guss, J. M., Christopherson, R. I. & Maher, M. J. (2005). J. Mol. Biol. 348, 523–533. [DOI] [PubMed]
- Lollier, M., Jaquet, L., Nedeva, T., Lacroute, F., Potier, S. & Souciet, J.-L. (1995). Curr. Genet. 28, 138–149. [DOI] [PubMed]
- Maher, M. J., Huang, D. T. C., Guss, J. M., Collyer, C. A. & Christopherson, R. I. (2003). Acta Cryst. D59, 381–384. [DOI] [PubMed]
- Martin, P. D., Purcarea, C., Zhang, P., Vaishnav, A., Sadecki, S., Guy-Evans, H. I., Evans, D. R. & Edwards, B. F. (2005). J. Mol. Biol. 348, 535–547. [DOI] [PubMed]
- Musmanno, L. A., Jamison, R. S., Barnett, R. S., Buford, E. & Davidson, J. N. (1992). Somat. Cell Mol. Genet. 18, 309–318. [DOI] [PubMed]
- Porter, T. N., Li, Y. & Raushel, F. M. (2004). Biochemistry, 43, 16285–16292. [DOI] [PubMed]
- Sigoillot, F. D., Kotsis, D. H., Serre, V., Sigoillot, S. M., Evans, D. R. & Guy, H. I. (2005). J. Biol. Chem. 280, 25611–25620. [DOI] [PubMed]
- Sigoillot, F. D., Sigoillot, S. M. & Guy, H. I. (2004). Int. J. Cancer, 109, 491–498. [DOI] [PubMed]
- Simmer, J. P., Kelly, R. E., Rinker, A. G., Scully, J. L. & Evans, D. R. (1990). J. Biol. Chem. 265, 10395–10402. [PubMed]
- Souciet, J.-L., Nagy, M., Le Gouar, M., Lacroute, F. & Potier, S. (1989). Gene, 79, 59–70. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Thoden, J. B., Phillips, G. N., Neal, T. M., Raushel, F. M. & Holden, H. M. (2001). Biochemistry, 40, 6989–6997. [DOI] [PubMed]
- Williams, N. K., Peide, Y., Seymour, K. K., Ralston, G. B. & Christopherson, R. I. (1993). Protein Eng. 6, 333–340. [DOI] [PubMed]
- Wyatt, P. J. (1993). Anal. Chim. Acta, 272, 1–40.
- Zhang, P., Martin, P. D., Purcarea, C., Vaishnav, A., Brunzelle, J. S., Fernando, R., Guy-Evans, H. I., Evans, D. R. & Edwards, B. F. (2009). Biochemistry, 48, 766–778. [DOI] [PMC free article] [PubMed]
- Zrenner, R., Stitt, M., Sonnewald, U. & Boldt, R. (2006). Annu. Rev. Plant Biol. 57, 805–836. [DOI] [PubMed]