Abstract
Butterfly eyespots may have evolved from the recruitment of pre-existent gene circuits or regulatory networks into novel locations on the wing. Gene expression data suggests one such circuit, the Hedgehog (Hh) signaling pathway and its target gene engrailed (en), was recruited from a role in patterning the anterior-posterior insect wing axis to a role patterning butterfly eyespots. However, while Junonia coenia expresses hh and en both in the posterior compartment of the wing and in eyespot centers, Bicyclus anynana lacks hh eyespot-specific expression. This suggests that Hh signaling may not be functioning in eyespot development in either species or that it functions in J. coenia but not in B. anynana. In order to test these hypotheses, we performed functional tests of Hh signaling in these species. We investigated the effects of Hh protein sequestration during the larval stage on en expression levels, and on wing size and eyespot size in adults. Hh sequestration led to significantly reduced en expression and to significantly smaller wings and eyespots in both species. But while eyespot size in B. anynana was reduced proportionately to wing size, in J. coenia, eyespots were reduced disproportionately, indicating an independent role of Hh signaling in eyespot development in J. coenia. We conclude that while Hh signaling retains a conserved role in promoting wing growth across nymphalid butterflies, it plays an additional role in eyespot development in some, but not all, lineages of nymphalid butterflies. We discuss our findings in the context of alternative evolutionary scenarios that led to the differential expression of hh and other Hh pathway signaling members across nymphalid species.
Introduction
The field of evolutionary developmental biology has revealed that complex traits that are homologous at the morphological level do not necessarily have the same developmental basis [1]. Vulva development in worms [2], and head development in insects [3]–[5] are two examples where conservation of morphology is not underlaid by conservation of developmental mechanisms. These phenomena offer exciting opportunities for investigating the relationship between morphology and underlying genetic circuitry, and gaining insight into how genes get co-opted, redeployed, and gain and lose functionality in gene regulatory networks underlying the development of complex traits.
Nymphalid butterfly eyespots are complex traits that originated once within the nymphalid butterfly clade, roughly 90 million years ago and are, thus, homologous at the morphological level [6]. At the level of gene expression, however, eyespots from different nymphalid species express a very different complement of genes during their early development [6], [7]. The differential gene expression across lineages appears to originate predominantly via a shared and basal gene co-option event followed by lineage-specific gene expression losses [6].
hedgehog (hh) is one of the genes differentially expressed in eyespots across nymphalid species. Transcripts of this gene were originally visualized flanking the center of the future eyespots in Junonia coenia larval wings [8] (Fig. 1), but recent stainings in a different nymphalid species, Bicyclus anynana, show that hh is not expressed in eyespots at comparable larval stages [9] (Fig. 1). The recruitment of hh to eyespot development in J. coenia was proposed to be part of a larger genetic circuit co-option to the eyespot field [8]. This circuit is the anterior-posterior axis patterning circuit described for fly wings and presumed to play a role in wing patterning and growth across insects [8]. In particular, transcripts of hh and its receptor patched (ptc), and proteins of the presumptive target gene Engrailed (En) and signal transducer Cubitus interruptus (Ci), are all co-localized to the eyespot centers in J. coenia (Fig. 1). These genes share a conserved pattern of expression on the fly and butterfly wing: hh mRNA transcripts and En proteins are present in the posterior compartment, Ci protein is present in the anterior compartment, and ptc mRNA is present along the anterior-posterior boundary [8], [9] (Fig. 1). It is remarkable then, that while some members of this circuit, such as Ci and En are expressed in B. anynana eyespots [8], the Hh receptor ptc and hh itself, are not [9].
Figure 1. Differential expression of Hh signaling pathway members in Junonia coenia and Bicyclus anynana larval hindwings.

Summary of mRNA (italics) and protein expression data from [8], [9]. hedgehog (hh) and its receptor patched (ptc) are expressed in primitive patterns throughout the posterior wing compartment and in a narrow anterior domain abutting that compartment, respectively. These two genes are also expressed in novel domains in J. coenia but not in B. anynana: flanking the developing eyespot centers, and in the centers, respectively. The other depicted genes share a similar expression pattern between J. coenia and B. anynana.
The differential expression of hh and ptc in J. coenia and B. anynana eyespots is intriguing and suggests that Hh signaling may not be functional in either species, or may be functional in J. coenia but not in B. anynana eyespots. In order to test these hypotheses, we provide the first functional tests for the role of Hh signaling in wing and eyespot development in butterflies by directly manipulating Hh function in developing wings of B. anynana and J. coenia.
Instead of taking an RNAi approach, which is proving challenging in Lepidoptera [10], we investigated an alternative method to disrupting protein function using antibodies [11]–[13]. In order to manipulate Hh signaling we first tested whether the 5E1 antibody, designed to target mammalian Sonic hedgehog, could also be used to target Hh in invertebrates. The 5E1 antibody inhibits Hh signaling by binding directly to the Hh ligand, sequestering it, and thus preventing Hh binding to the receptor Patched (Ptc) [14], [15]. The upstream elements of the Hh signaling pathway, including the binding of Hh to Ptc, are known to be highly conserved between vertebrates and invertebrates [16]. In order to investigate the likelihood that the 5E1 antibody also recognizes insect Hh proteins we first performed epitope sequence comparisons between mammalian, Drosophila melanogaster, and butterfly Hh proteins and then tested whether butterfly protein extracts produce the expected number and size of Hh protein fragments known from the conserved autoproteolysis of genes from this family [17] using Western blots. We compared the expected length of Hh protein products with those known from D. melanogaster.
After confirming the specificity of the 5E1 antibody for Hh nymphalid butterfly proteins we subsequently injected the 5E1 antibody (as well as a vehicle, NS1 control medium) into both J. coenia and B. anynana larvae at the developmental stage when hh transcripts have been detected in eyespots. We monitored levels of a known target of Hh signaling, en, in the developing larvae of both species to see if levels of en were altered via the antibody injections. After the butterflies pupated and emerged, we measured adult wing and eyespot size. Our experiments support a role for Hh signaling in overall wing growth in both B. anynana and J. coenia butterflies, but only in eyespot development in J. coenia.
Materials and Methods
Sequence alignment and western blots
In order to test antibody specificity, sonic hedgehog protein sequences of rat (SHH-N, Q63673), D. melanogaster (AAF56102), B. anynana (ADO60878) and J. coenia (AAD08931) were aligned using muscle3.6 [18], Clustal X [19] and Genedoc [20]. Sequence identity and similarity were calculated in SIAS (http://imed.med.ucm.es/Tools/sias.html) using the PID1 identity method, Blossom 62 matrix, and remainder defaults. Western blots were performed on <40 hr old pupal wing discs of B. anynana and band size was compared against blots from 3rd larval wing discs of D. melanogaster, with a previously characterized Hh protein profile [21]. In particular, in D. melanogaster the full-length form of hedgehog protein (Hh-F) is converted to a species of 39 kD (Hh-U), a signal-cleaved form of Hh-F, which further undergoes autoproteolysis to generate two main products, a 19kD amino-terminal fragment (Hh-N), and a 25 kD carboxyl-terminal fragment (Hh-C). The 25-kD Hh-C species further generates the 16-kD C* species in imaginal disks [21]. Discs were resuspended and homogenized in lysis buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1% Triton X-100, 10% Glycerol, 1.5 mM EDTA, 1x protease inhibitor cocktail). Homogenates were centrifuged at 14,000 rpm at 4°C for 10 minutes, and the resulting supernatant was collected. A mix of 20 µl supernatant with 5 µl SDS-PAGE loading buffer was separated on a 4%–20% SDS-PAGE gel and transferred to a PVDF membrane (Millipore Corporation cat # K9PN0097). After blocking, the membrane was incubated with the anti-Sonic hedgehog 5E1 antibody (0.14 µg/mL in wash buffer), washed 3 times with wash buffer, 5 min each time, then incubated with goat anti-mouse IgG antibody conjugated to biotin (Invitrogen cat # 643341), washed 3 times with wash buffer, followed by incubation with a Qdot® 625 streptavidin conjugate (Invitrogen cat # 643341). Signals were detected with a standard UV detection system for ethidium bromide-stained gels. A Western blot with NS1 medium, in which the 5E1 antibody is suspended, diluted 1∶500 in wash buffer, was used as control. The monoclonal anti-Sonic hedgehog 5E1 antibody was developed at the Jessell lab at Columbia University [15] and was obtained from the Developmental Studies Hybridoma Bank (DSHB) developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
Butterfly rearing
B. anynana larvae were reared on young corn plants (Zea mays) in aluminum mesh cages in a 27°C environmental chamber with a 12∶12 light: dark cycle with a gradual “sunrise” and “sunset,” each one hour in duration. J. coenia were reared on narrowleaf plantain (Plantago lanceolata) leaves at room temperature (∼25°C), under a natural photoperiod, and inside large plastic containers. After injections, B. anynana were reared in mesh sleeve cages with no more than 15 larvae per corn plant. J. coenia were reared in the same plastic containers. Butterflies being raised to adults were transferred to hanging net cages after pupation, with no more than 15 pupae per cage, and adults were frozen upon emergence.
Antibody injections
Hh activity was suppressed via injection of the 5E1 antibody into larvae. The 5E1 antibody prevents the Hh signaling ligand from binding to its receptor Patched (Ptc). When this happens, Ptc is able to inhibit Smoothened (Smo) resulting in interactions between Smo, Ci and other protein complexes, which results in the transformation of Ci into a repressor form of Ci (CiR). This repressor form acts as a transcription factor inhibiting the transcription of target genes such as en [22]. NS1 medium was used in control injections. Antibody injections were performed using a #701 Hamilton 10 µL syringe with 26–33 gauge needles. Fifth instar larvae were injected on the left side directly posterior to the third thoracic segment with 5 µL of either NS1 medium or 5E1 antibody solutions, at a concentration of 41 µg/mL (in B. anynana) and 100 µg/mL (in J. coenia). A higher concentration of 5E1 was used for J. coenia larvae, in proportion to their higher weight.
Measurements of en transcript levels following injections
We used semi-quantitative PCR to test whether injections of 5E1 antibody had an effect on levels of the putative target gene en. We injected nine B. anynana and four J. coenia larvae with either 5E1 or with NS1, and then we dissected their fore- and hindwings 24 hrs later. We pooled 2 (J. coenia) or 3 (B. anynana) individuals together before extracting total RNA with a RNeasy Micro kit (Qiagen), and reverse transcribing it with a High-Capacity cDNA Reverse Transcription Kit (Applied biosystems). We used equal amounts of total cDNA in each PCR reaction and primers (Fw: GGA CTG GCC TGC TTG GGT NTA YTG TAC; Rv: TTG AGC CAT CAG TTG CAT AGC NAR NGG RT) that amplified a 313 bp fragment of en, within the homeobox region, and that are likely to pick up both en and invected copies (D. Ramos, pers. comm.). We used Elongation Factor 1-alpha (EF1a) as a control housekeeping gene. Primers for EF1a were Fw: GCY GAR CGY GAR CGT GGT ATY AC and Rv: CAT GTT GTC GCC GTG CCA AC [23]. PCR reactions for each gene were run for 30 cycles. After running the same amount of reaction products on a gel, we quantified the intensity of each PCR band using a digital grayscale image of the gel in Photoshop. We did this by demarcating each gel band inside a constant-size rectangular frame, averaging the intensity of the pixels inside that frame, and collecting the brightness value for the band (100 minus the K-value) using the color picker tool. We corrected the brightness of each band by the correspondent housekeeping gene band brightness by calculating the ratio of the two values. We then used these ratios in a GLM analysis (see below).
Measurements in adult wings
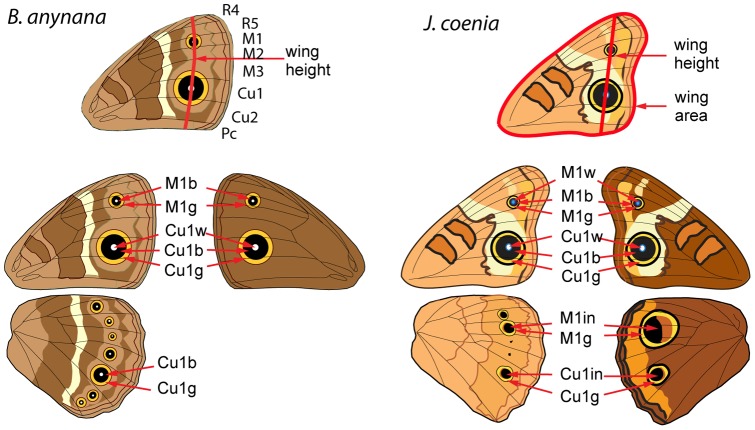
Left and right adult forewings and hindwings from injected larvae were photographed under a Stereo Discovery V8 Carl Zeiss stereomicroscope equipped with an AxioCam MRC with AxioVision AC Rel. 4.5 software. Pictures were taken using an Anchromat S 0.3X FWD 253 mm lens at 1.0X magnification and saved as TIFF files. Object-Image2.21 [24] and ImageJ 1.42q were used for measurements of the adult wings. In B. anynana, we measured the width of each of the wing compartments on the forewing given that Hh-inhibition has been known to result in wing compartment specific effects in D. melanogaster [25]. Wing cell measurements were taken perpendicular to the wing veins, along the same axis that intersects the two eyespot centers. We used the sum of the wing compartment measurements as a measure of forewing height (Fig. 2). In J. coenia, we measured forewing height along the line that crosses the center of both eyespots, and also measured forewing area (Fig. 2). In addition to wing size, we measured the diameter of several eyespot traits on both dorsal and ventral wing surfaces as indicated in Figure 2. All eyespot diameters were taken parallel to the wing veins, along the wing fold.
Figure 2. Measurements taken of adult B. anynana and J. coenia wings.
Wing height and wing area (J. coenia only) and a series of eyespot trait diameters for the M1 and Cu1 eyespots were measured on both ventral (left) and dorsal surfaces (right). R5, R4, M1, M2, M3, Cu1, Cu1+Pc, and 1A+2A refer to the wing compartments that were individually measured in B. anynana wings only. Their combined height defined the B. anynana wing height. w: white center; b: black disc; g: gold ring; in: inner ring (from the distal border of the black patch to the proximal border of the orange patch).
Analysis
SPSS Statistics, version 19, was used for statistical analyses of adult wing measurements and for quantification of en amplification levels after semi-quantitative PCR. Differences in left and right measurements were examined for each of the treatments, but, given no significant differences between left and right sides, we averaged measurements for the two sides and analyzed the average values thereafter. General Linear Model (GLM) analyses were performed on wing size and eyespot size measurements to test for differences in average trait size between the 5E1- and NS1-injected butterflies using both treatment and sex as fixed variables and a full-factorial design. In order to evaluate eyespot trait size independently of wing size, we performed analyses of covariance on eyespot measurements using forewing height as a covariate. We also used GLM analysis to test for differences in relative levels of en amplification in injected animals using treatment (5E1 and NS1) and species (J. coenia and B. anynana) as fixed factors and a full-factorial design.
Results
The 5E1 antibody recognizes butterfly Hedgehog
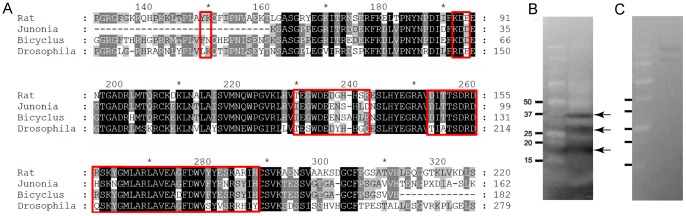
A comparison between the Hh-N terminal sequence of rat (198 a.a., against which the 5E1 antibody was raised), and the comparable sequence in B. anynana (181 a.a.), J. coenia (150 a.a.) and D. melanogaster (241 a.a.) showed 69%/75%, 70%/68%, and 63%/75% amino acid sequence identity/similarity to rat SHH-N, respectively (Fig. 3A). Recent structural and biophysical analysis revealed that 5E1 binds at the pseudo-active site groove of Shh [26], [27]. We compared the 54 residues forming the 5E1 epitope between rat and each of the three other Hh insect sequences (Fig. 3A, red boxes) and found 72%/80% (D. melanogaster), 75%/85% (B. anynana) and 68%/ 83% (J. coenia) amino acid identity/similarity in these regions.
Figure 3. Western blots and similarity of Sonic-Hh and butterfly Hh sequences suggest that 5E1 antibody recognizes Hh in butterflies.
(A) Alignment of sequences corresponding to the Sonic-Hh peptide used to make the 5E1 monoclonal antibody [15]. Areas boxed in red correspond to the 5E1 epitope [26], [27]. (B) Western blot with B. anynana proteins extracted from wing discs showing three potential Hh fragments with the predicted sizes of 19 kD, 25 kD, and 37 kD (arrows) previously characterized from D. melanogaster Hh [21]. (C) No bands were detected with the control NS1 medium. The left lane of each photo is the protein standard.
To confirm that the 5E1 antibody can recognize Hh proteins in insects, proteins from B. anynana and from D. melanogaster wing discs (where we have knowledge of expected Hh protein size) were extracted and subjected to Western blot analysis. The monoclonal anti-Sonic hedgehog 5E1 antibody, designed to recognize the Shh-N protein fragment in rats [15], detected three bands with the expected sizes in the extracts from D. melanogaster (data not shown), and B. anynana (Fig. 3B, arrows). No bands were detected with the NS1 control medium in this region (Fig. 3C). The band around 19 kDa is consistent with the size of the Hh-N protein in D. melanogaster (Fig. 3B). This indicates that the anti-Shh antibody is probably recognizing the Hh-N protein of both B. anynana and D. melanogaster. The larger protein, at∼37Kd, is consistent with the Hh-U fragment. Another protein, at ∼25 kD, was also observed in Western blots with proteins extracted from imaginal discs of D. melanogaster [21] and could represent an alternative Hh-N size fragment containing the 5E1 epitope. Our data suggest that the monoclonal anti-Sonic hedgehog 5E1 is binding to B. anynana Hh, and, by doing so, likely blocking the function of the Hh-N protein as it does for Sonic-Hh.
Quantification of en expression following injections
Injections of 5E1 significantly reduced the amount of en/inv PCR amplicon relative to NS1 injections (F1,6 = 8.635; p = 0.026) (Fig. 4). Both J. coenia and B. anynana responded in a similar way (F1,6 = 3.375; p = 0.116), and there was no significant treatment*species interaction (F1,6 = 0.409; p = 0.546). The reduction of en/inv transcript levels, 24 hrs following the 5E1 antibody injections, suggests that the antibody directly, or indirectly, negatively impacted the transcription of this gene. These experiments, however, do not indicate whether lower en/inv transcript levels result from one or both of the en/inv expression domains (the posterior compartment or eyespot centers expression domains; see Fig. 1).
Figure 4. Injections of 5E1 antibody reduce the levels of en/inv transcripts one day later in both B. anynana and J. coenia.
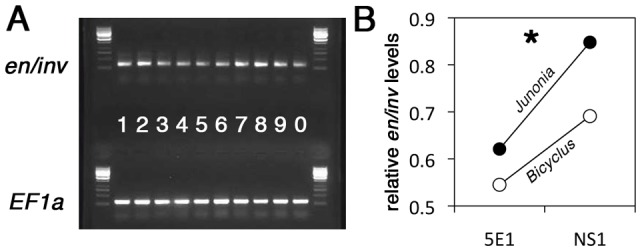
(A) PCR amplification of en/inv (top) and the house-keeping gene EF1a (bottom) from the same samples after injection of either 5E1 antibody or NS1 vehicle. Samples 1–3: B. anynana NS1; 4–6: B. anynana 5E1; 7–8: J. coenia NS1; 9–10: J. coenia 5E1. (B) Quantification of brightness levels of en/inv PCR amplification relative to brightness levels of the EF1a housekeeping gene (averages from data in A). Asterisk (*) indicates a significant difference in en/inv relative levels between 5E1 and NS1 injections.
Adult phenotypes resulting from 5E1 antibody and NS1 medium injections
Injections of 5E1 or NS1 medium performed on one side of the larval body led to symmetrical changes in both left and right wings suggesting that the antibody, once injected, circulates throughout the hemolymph and is able to target both sides of the animal, and probably most tissues.
Wing size
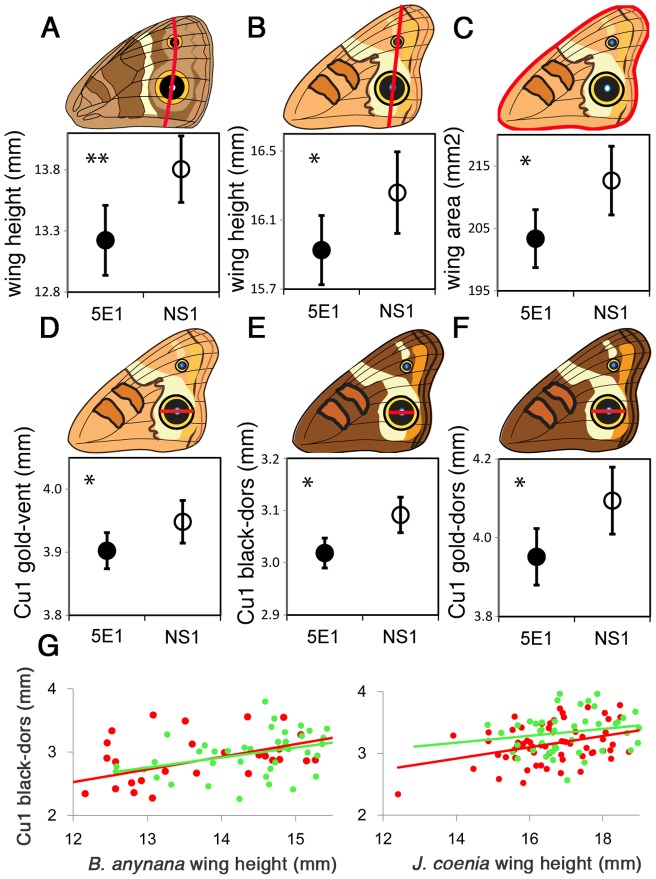
B. anynana and J. coenia adults injected with the 5E1 antibody as larvae had a smaller forewing height relative to NS1-injected controls (B. anynana: F1, 82 = 8.62, p = 0.004; J. coenia F1, 140 = 4.47, p = 0.036) (Fig. 5A, B). J. coenia, where we additionally measured wing area, had a smaller forewing area as well (F1, 140 = 6.48, p = 0.012) (Fig. 5C). Wing height reductions in B. anynana were due to the compound effect of reductions across all wing compartments as there were no specific compartments that were more affected than others (Table 1). These compartment-specific investigations were not undertaken in J. coenia, but the wings appeared also proportionately reduced across the anterior-posterior axis as in B. anynana.
Figure 5. Hh sequestration decreases wing size in both species and relative eyespot size in J. coenia.
Forewing height in B. anynana (A) and J. coenia (B) and forewing area in J. coenia (C) are smaller in 5E1-injected butterflies compared with NS1-injected controls. Relative eyespot size, e.g., the diameter of the black and gold rings of the Cu1 eyespot on both ventral (vent) and dorsal (dors) surfaces of J. coenia is also smaller in 5E1-injected individuals (D-F; mean trait values are displayed for a wing height of 16 mm). GLM analyses use sex as a grouping variable but here sexes are plotted together. (G) Regression of black disc diameter of Cu1 dorsal eyespot on wing height for B. anynana (left) and J. coenia (right). In J. coenia, 5E1-injected individuals (red dots) display significantly smaller Cu1 eyespots relative to NS1-injected individuals (green dots) of comparable size, while B. anynana eyespots are not significantly smaller for an individual of a given size.
Table 1. F statistics and p-values for GLM analysis testing for differences in wing compartment size in B. anynana across treatments.
| wing compartment | N (5E1) | N (NS1) | F | p |
| Forewing R4 | 35 | 48 | 1.40 | 0.216 |
| Forewing R5 | 35 | 48 | 0.015 | 0.982 |
| Forewing M1 | 35 | 48 | 1.219 | 0.231 |
| Forewing M2 | 35 | 48 | 0.158 | 0.651 |
| Forewing M3 | 35 | 48 | 0.402 | 0.627 |
| Forewing Cu1 | 35 | 48 | 2.393 | 0.086 |
| Forewing Cu2+Pc | 35 | 48 | 0.002 | 0.957 |
| Forewing 1A +2A | 35 | 48 | 0.875 | 0.279 |
Individuals were injected with either 5E1 antibody or NS1 control media as larvae. Treatment and sex were used as fixed factors and wing height was used as a covariate. Treatment and sex were not significant across analyses.
Absolute eyespot size
5E1 injected butterflies had overall smaller eyespots than NS1 injected butterflies, but while some differences were significant others were not. In B. anynana the diameter of the black and gold ring of scales of the largest wing eyespot, the Cu1 forewing eyespot on both dorsal and ventral surfaces, was significantly smaller in 5E1- relative to NS1-injected butterflies (Table 2). In J. coenia most eyespots had some trait that was significantly smaller in 5E1-injected individuals relative to controls. This included all eyespot traits measured on the ventral hindwing and dorsal forewing, as well as the white center of the Cu1 eyespot on the ventral forewing, and both measurements for the Cu1 eyespot on the dorsal hindwing (Table 2).
Table 2. F statistics and p-values for GLM analysis testing for differences in eyespot trait sizes across treatments.
| B. anynana | J. coenia | |||||||||
| Surface | Wing | Trait | N (5E1) | N (NS1) | F | p | N (5E1) | N (NS1) | F | p |
| Ventral | Forewing | M1 white | NA | NA | NA | NA | 82 | 59 | 2.67 | 0.10365 |
| M1 black | 40 | 48 | 2.37 | 0.09760 | 82 | 59 | 0.003 | 0.95416 | ||
| M1 gold | 40 | 48 | 0.19 | 0.82595 | 82 | 59 | 0.68 | 0.40793 | ||
| Cu1 white | 40 | 49 | 1.32 | 0.27048 | 82 | 59 | 29.47 | <0.0000 *** | ||
| Cu1 black | 40 | 49 | 3.19 | 0.04455 * | 82 | 59 | 0.38 | 0.53503 | ||
| Cu1 gold | 40 | 49 | 4.59 | 0.01192 * | 82 | 59 | 2.36 | 0.12594 | ||
| Hindwing | M1 inner disc | NA | NA | NA | NA | 82 | 59 | 13.76 | 0.00026 *** | |
| M1 gold | NA | NA | NA | NA | 82 | 59 | 4.75 | 0.03033 * | ||
| Cu1 black/inner | 37 | 49 | 1.44 | 0.23993 | 82 | 59 | 8.97 | 0.00305 ** | ||
| Cu1 gold | 37 | 49 | 0.98 | 0.37855 | 82 | 59 | 6.31 | 0.01271 * | ||
| Dorsal | Forewing | M1 white | NA | NA | NA | NA | 82 | 59 | 23.13 | <0.0000 *** |
| M1 black | 40 | 46 | 0.98 | 0.37734 | 82 | 59 | 13.10 | 0.00036 *** | ||
| M1 gold | 40 | 47 | 2.45 | 0.09086 | 82 | 59 | 12.97 | 0.00039 *** | ||
| Cu1 white | 40 | 48 | 1.62 | 0.20285 | 82 | 59 | 14.98 | 0.00014 *** | ||
| Cu1 black | 37 | 41 | 4.37 | 0.01487 * | 82 | 59 | 5.07 | 0.02529 * | ||
| Cu1 gold | 37 | 41 | 8.37 | 0.00040 *** | 82 | 59 | 9.72 | 0.00206 ** | ||
| Hindwing | M1 inner | NA | NA | NA | NA | 82 | 59 | 0.43 | 0.50912 | |
| M1 gold | NA | NA | NA | NA | 82 | 59 | 1.49 | 0.22313 | ||
| Cu1 inner | NA | NA | NA | NA | 82 | 59 | 3.90 | 0.04938 * | ||
| Cu1 gold | NA | NA | NA | NA | 82 | 59 | 7.98 | 0.00516 ** | ||
Individuals were injected with either 5E1 antibody or NS1 control media as larvae. All significant differences correspond to smaller trait sizes in the 5E1-injected individuals. GLM analyses were performed with sex (always significant across traits with females usually displaying a larger trait size than males, not shown); and interaction between line and sex (not significant in all analyses; not shown). *p<0.05, **p<0.01, ***p<0.001.
Relative eyespot size
The reductions in absolute eyespot size obtained for 5E1-injected butterflies could be due to eyespot differentiation processes having allometrically adjusted to the overall smaller wings. In order to test whether the 5E1 antibody had effects on eyespot size that were independent of its effects on wing size, we performed analyses of co-variance on eyespot traits, corrected for overall wing size (wing height). There was a significant interaction between treatment and wing height for J. coenia's largest eyespot traits, the diameter of the black and gold ring of the Cu1 forewing eyespot on both dorsal and ventral surfaces, but no such interaction in B. anynana (Table 3). The converging (non-parallel) regression lines (Fig. 5G) indicate that smaller wings displayed disproportionately smaller eyespots for the 5E1 treatment relative to the NS1 treatment in J. coenia, whereas treatment had no apparent effect on eyespot size on larger wings in J. coenia (Fig. 5G). This result may simply indicate that stronger effects were seen on smaller animals where the concentration of the antibody was effectively higher (given that the same antibody amount was injected in each animal). In addition, there were significant differences in the relative eyespot size across treatments for J. coenia but not for B. anynana. 5E1-injected J. coenia had significantly smaller eyespots relative to controls for a given wing size (Fig. 5D–F). The wing size data combined with the eyespot size data suggests that Hh signaling promotes wing growth in both butterfly species but promotes larger eyespots only in J. coenia. Hh signaling appears to mediate eyespot size in B. anynana only indirectly through its effects on general wing growth, but does not appear to be playing a direct role in eyespot development in this species.
Table 3. F statistics and p-values for GLM analysis testing for differences in eyespot trait sizes across treatments using wing size as a covariate.
| B. anynana | J. coenia | |||||||||
| Surface | Wing | Trait | N (5E1) | N (NS1) | F | p | N (5E1) | N (NS1) | F | p |
| Ventral | Forewing | M1 white | NA | NA | NA | NA | 82 | 59 | 1.381 | 0.241 |
| M1 black | 40 | 48 | 0.250 | 0.618 | 82 | 59 | 0.657 | 0.419 | ||
| M1 gold | 40 | 48 | 0.490 | 0.486 | 82 | 59 | 1.360 | 0.245 | ||
| Cu1 white | 40 | 49 | 0.066 | 0.797 | 82 | 59 | 0.014 | 0.905 | ||
| Cu1 black | 40 | 49 | 0.115 | 0.735 | 82 | 59 | 5.455 | 0.020 * | ||
| Cu1 gold | 40 | 49 | 0.097 | 0.757 | 82 | 59 | 4.152 | 0.031 * | ||
| Hindwing | M1 inner | NA | NA | NA | NA | 82 | 59 | 1.558 | 0.213 | |
| M1 gold | NA | NA | NA | NA | 82 | 59 | 0.001 | 0.989 | ||
| Cu1 black/inner | 37 | 49 | 0.063 | 0.803 | 82 | 59 | 1.023 | 0.313 | ||
| Cu1 gold | 37 | 49 | 0.125 | 0.725 | 82 | 59 | 1.482 | 0.225 | ||
| Dorsal | Forewing | M1 white | NA | NA | NA | NA | 82 | 59 | 0.418 | 0.519 |
| M1 black | 40 | 46 | 0.049 | 0.826 | 82 | 59 | 0.612 | 0.431 | ||
| M1 gold | 40 | 47 | 0.469 | 0.496 | 82 | 59 | 0.788 | 0.376 | ||
| Cu1 white | 40 | 48 | 2.961 | 0.090 | 82 | 59 | 2.123 | 0.147 | ||
| Cu1 black | 37 | 41 | 0.071 | 0.790 | 82 | 59 | 5.308 | 0.022 * | ||
| Cu1 gold | 37 | 41 | 0.386 | 0.537 | 82 | 59 | 5.337 | 0.0218 * | ||
| Hindwing | M1 inner | NA | NA | NA | NA | 82 | 59 | 0.276 | 0.600 | |
| M1 gold | NA | NA | NA | NA | 82 | 59 | 0.259 | 0.611 | ||
| Cu1 inner | NA | NA | NA | NA | 82 | 59 | 0.094 | 0.797 | ||
| Cu1 gold | NA | NA | NA | NA | 82 | 59 | 0.318 | 0.573 | ||
Individuals were injected with either 5E1 antibody or NS1 control media as larvae. All significant differences correspond to smaller trait sizes in the 5E1-injected individuals. GLM analyses were performed with sex (always significant across traits with females usually displaying a larger trait size than males; not shown); interaction between line and sex (not significant in all analyses; not shown); and wing size as a covariate. *p<0.05.
Discussion
We performed functional experiments in butterflies using an antibody that was developed to target Sonic Hh and inhibit this signaling pathway in rats. We showed that this antibody could potentially target other Hh family members, namely Hh from flies and butterflies, because the homologous epitope sequences of all these proteins were quite conserved across species. By performing a Western blot we showed that the antibody targeted protein fragments of the expected size and number as known Hh fragments from D. melanogaster, as well as similar sized fragments from B. anynana butterflies. Finally, we showed that by injecting the antibody into butterfly larvae, the expression of a known target of Hh signaling in the D. melanogaster wing, en/inv [28], was affected in B anynana and J. coenia larval wings. These results collectively indicate that the 5E1 antibody, once injected into butterflies, is likely inhibiting the Hh signaling pathway.
The Hh signaling pathway is involved in cell proliferation in many tissues [29]–[31], and the uncontrolled activation of the Hh signaling pathway has been linked to the growth of tumors in many forms of cancer (reviewed in [32], [33]). The specific role of Hh signaling in wing growth was demonstrated in D. melanogaster. Mutants lacking hh function had severely reduced wings [25], [34], and reduced en expression [28], whereas hh gain-of-function mutants had enlarged wings with duplicated posterior compartment vein structures [34]–[36]. The hh knock down experiments of Basler and Struhl [34] were performed in D. melanogaster during the first larval instar, which is the time interval when the anterior/posterior wing compartments are being established. The comparatively less drastic reduction in wing size resulting from Hh sequestration in B. anynana is likely in part the consequence of manipulations late in development, during the fifth instar, after the anterior-posterior axis has already been established, but just as eyespots begin to differentiate. Changes in wing size resulting from Hh sequestration in B. anynana and J. coenia during the fifth instar suggest that Hh signaling continues to play a role in wing growth even during later larval development.
While Hh sequestration inhibited wing growth in both butterflies, eyespot trait size reductions independent of wing size were only seen in J. coenia. B. anynana butterflies displayed small wings with proportionately-sized eyespots, whereas J. coenia displayed small wings with disproportionately small eyespots. This indicates that Hh signaling directly affects eyespot development in J. coenia but not in B. anynana, and that Hh signaling promotes larger eyespots in J. coenia.
The functional study done here, directly manipulating Hh availability in B. anynana and J. coenia butterflies, has illuminated some surprising differences in the effect of Hh signaling on wing development and eyespot development in these two nymphalid butterfly species. Our study shows that hh maintains its role in promoting wing growth in butterflies, as it does in D. melanogaster, and that hh acquired a novel functional role in promoting eyespot development in some butterflies, but not in others. We also note that Hh signaling may have had a more generalized effect on tissue growth, beyond wing growth, which was not documented here.
The presence of Hh signaling in J. coenia eyespot development but the absence of such signaling in B. anynana requires interpretation from both a mechanistic as well as an evolutionary perspective, i.e., what these differences represent in terms of the proposed recruited circuit and how they could come about in evolution. Originally, the Hh circuit involved in specifying the anterior-posterior wing axis (including hh, the Hh receptor ptc, the signal transducer ci, and the target gene en) were proposed to have been co-opted, as a unit, to help build the novel eyespot gene regulatory network [8]. All members of this circuit are present in J. coenia butterflies, whereas two of the members are missing in B. anynana (hh and ptc; [9]). In addition, as shown here, disrupting Hh signaling in B. anynana does not affect eyespot development. Given these data, it is particularly intriguing that En is being expressed at high levels in the eyespot centers in B. anynana, when the gene proposed to activate its transcription (hh) is missing. Several explanations for this observation are possible. First, a different member of the Hh family of proteins may activate en transcription in B. anynana. Presence of additional Hh family members can be tested once the completed B. anynana genome becomes available. In arthropods, however, only a single hh copy is currently known [37]. Second, en transcription in B. anynana eyespot centers (and possibly also in J. coenia), is being activated by transcription factors unconnected to the Hh signaling pathway. Note that our semi-quantitative PCR experiment cannot distinguish which domains of en/inv expression on the wing were actually targeted by the 5E1 antibody injections. It is likely that the lower levels of en/inv expression observed following Hh signal inhibition result primarily from the response of cells localized in the posterior compartment of the wing in both species, because this domain is much larger and is also the domain known to be under the control of Hh signaling in D. melanogaster wings [28]. If en/inv transcription in eyespots is being activated by transcription factors unconnected to the Hh signaling pathway, then either the gene circuit co-opted for eyespot development is different from the one proposed by Keys et al. [8], or the co-opted circuit replaced some of its members in the B. anynana lineage but not in the lineage leading to J. coenia.
A broader phylogenetic sampling of multiple species for presence and absence of hh and ptc expression is required to clarify when these genes became associated with eyespots during evolutionary history and to elucidate how and when differential hh expression emerged between B. anynana and J. coenia. Recent comparative gene expression data across 21 nymphalid species and two outgroups showed that the origin of expression of four genes in the eyespot centers (including en) happened in a very basal branch of the nymphalid tree, concurrently with the origin of eyespots [6]. Subsequently, many of these gene expression patterns were lost from eyespots in a lineage-specific fashion without loss of eyespots. We proposed that this pattern of rapid, perhaps simultaneous, gene expression gains in association with eyespots, could indicate a gene network co-option event that was followed by the elimination of genes that did not play a role in the development of the novel trait [6]. The same could apply to members of the Hh signaling pathway. All members being co-opted at the same time, as part of a larger network, and some members, such as hh and ptc, being lost in the lineage leading to B. anynana. This gene loss would imply that Hh signaling was not critical for eyespot development in the early nymphalid ancestors. The retention of the whole pathway in J. coenia could result from the pathway having been secondarily co-opted to function in eyespot development later in this lineage. An alternative scenario to the single origin of multiple eyespot-associated genes via gene network co-option is a more gradual process of eyespot network modification via lineage-specific additions. Under this scenario, hh and ptc are co-opted to the J. coenia lineage allowing Hh signaling to become functional in this lineage but not in B. anynana. Comparative work showed that late additions to the cluster of genes associated with eyespot origins are possible as the gene Antennapedia was co-opted into the eyespot centers late and independently in two nymphalid lineages [6], [7]. Only future comparative work involving several more species, however, will determine how exactly hh and ptc expression in butterfly eyespots evolved.
In conclusion, this work documents an example of a conserved wing pattern, the eyespot, with a single origin within nymphalid butterflies [6] that displays a different developmental basis in different lineages. In one lineage Hh signaling influences adult eyespot size, whereas in another lineage it does not. This example adds to others in the evo-devo literature [2]–[5], [38], where different genes and developmental mechanisms pattern homologous traits.
Acknowledgments
We thank Fred Nijhout and Laura Grunert for J. coenia eggs, Diane Ramos for engrailed primer sequences, Jeffrey Oliver, Diane Ramos, and two anonymous reviewers for comments on the manuscript, and Chris Bollick, Robert Rak, and Eric Larson for growing the corn plants to feed B. anynana larvae.
Funding Statement
A.L. was supported by an REU DO 1357 supplement to National Science Foundation (NSF) award IOB -0653399 to AM and XT was partly supported by NSF award IOS 1146933 to AM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. True JR, Haag ES (2001) Developmental system drift and flexibility in evolutionary trajectories. Evolution & Development 3: 109–119. [DOI] [PubMed] [Google Scholar]
- 2. Wang XY, Sommer RJ (2011) Antagonism of LIN-17/Frizzled and LIN-18/Ryk in Nematode Vulva Induction Reveals Evolutionary Alterations in Core Developmental Pathways. PLoS Biology 9: e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C (2006) Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439: 728–732. [DOI] [PubMed] [Google Scholar]
- 4. Schroder R (2003) The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature 422: 621–625. [DOI] [PubMed] [Google Scholar]
- 5. McGregor AP (2006) Wasps, beetles and the beginning of the ends. BioEssays 28: 683–686. [DOI] [PubMed] [Google Scholar]
- 6. Oliver JC, Tong X, Gall LF, Piel WH, Monteiro A (2012) A single origin for nymphalid butterfly eyespots followed by widespread loss of associated gene expression. PLoS Genet 8(8): e1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shirai LT, Saenko SV, Keller RA, Jeronimo MA, Brakefield PM, et al. (2012) Evolutionary history of the recruitment of conserved developmental genes in association to the formation and diversification of a novel trait. BMC Evol Biol 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, et al. (1999) Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283: 532–534. [DOI] [PubMed] [Google Scholar]
- 9. Saenko SV, Marialva MSP, Beldade P (2011) Involvement of the conserved Hox gene Antennapedia in the development and evolution of a novel trait. EvoDevo 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. (2011) RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57: 231–245. [DOI] [PubMed] [Google Scholar]
- 11. Aste-Amezaga M, Zhang NY, Lineberger JE, Arnold BA, Toner TJ, et al. (2010) Characterization of Notch1 Antibodies That Inhibit Signaling of Both Normal and Mutated Notch1 Receptors. PLoS ONE 5: e9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukasawa H, Yamamoto T, Suzuki H, Togawa A, Ohashi N, et al. (2004) Treatment with anti-TGF-beta antibody ameliorates chronic progressive nephritis by inhibiting Smad/TGF-beta signaling. Kidney International 65: 63–74. [DOI] [PubMed] [Google Scholar]
- 13. Stockwin LH, Holmes S (2003) Antibodies as therapeutic agents: vive la renaissance! Expert Opinion on Biological Therapy. 3: 1133–1152. [DOI] [PubMed] [Google Scholar]
- 14. Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, et al. (2000) Receptor-mediated endocytosis of soluble and membrane-tethered Sonic hedgehog by Patched-1. Proc Nat Acad Sci USA 97: 12044–12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM (1996) Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 87: 661–673. [DOI] [PubMed] [Google Scholar]
- 16. De Rivoyre M, Ruel L, Varjosalo M, Loubat A, Bidet M, et al. (2006) Human receptors patched and smoothened partially transduce hedgehog signal when expressed in Drosophila cells. J Biol Chem 281: 28584–28595. [DOI] [PubMed] [Google Scholar]
- 17. Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, et al. (1995) The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 374: 363–366. [DOI] [PubMed] [Google Scholar]
- 18. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholas KB, Jr NHB, Deerfield DWI (1997) GeneDoc: Analysis and Visualization of Genetic Variation. EMBNETnews. 1–4.
- 21. Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, et al. (1994) Autoproteolysis in hedgehog Protein Biogenesis. Science 266: 1528–1537. [DOI] [PubMed] [Google Scholar]
- 22. Zhao Y, Tong C, Jiang J (2007) Hedgehog regulates smoothened activity by inducing a conformational switch. Nature 450: 252–258. [DOI] [PubMed] [Google Scholar]
- 23. Monteiro A, Pierce NE (2001) Phylogeny of Bicyclus (Lepidoptera: Nymphalidae) inferred from COI, COII and EF-1a gene sequences. Mol Phylogen Evol 18: 264–281. [DOI] [PubMed] [Google Scholar]
- 24.Vischer NOE, Huls PG, Woldringh CL (1994) Object-Image: An interactive image analysis program using structured point collection. Binary (Bioline) 6.
- 25. Strigini M, Cohen SM (1997) A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124: 4697–4705. [DOI] [PubMed] [Google Scholar]
- 26. Bosanac I, Maun HR, Scales SJ, Wen XH, Lingel A, et al. (2009) The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nature Struct Mol Biol 16: 691–697. [DOI] [PubMed] [Google Scholar]
- 27. Maun HR, Wen XH, Lingel A, de Sauvage FJ, Lazarus RA, et al. (2010) Hedgehog Pathway Antagonist 5E1 Binds Hedgehog at the Pseudo-active Site. J Biol Chem 285: 26570–26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bossing T, Brand AH (2006) Determination of cell fate along the anteroposterior axis of the Drosophila ventral midline. Development 133: 1001–1012. [DOI] [PubMed] [Google Scholar]
- 29. Duman-Scheel M, Weng L, Xin S, Du W (2002) Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature. 417: 299–304. [DOI] [PubMed] [Google Scholar]
- 30. Rowitch DH, S-Jacques B, Lee SM, Flax JD, Snyder EY, et al. (1999) Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J Neurosci 19: 8954–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hervold K, Martin A, Kirkpatrick RA, Mc Kenna PF, Ramirez-Weber FA (2007) Hedgehog Signaling Pathway Database: a repository of current annotation efforts and resources for the Hh research community. Nucleic Acids Res 35: D595–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruiz i Altaba A (1999) Gli proteins and Hedgehog signaling: development and cancer. Trends Genet 15: 418–425. [DOI] [PubMed] [Google Scholar]
- 33. Pasca di Magliano M, Hebrok M (2003) Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer 3: 903–911. [DOI] [PubMed] [Google Scholar]
- 34. Basler K, Struhl G (1994) Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 368: 208–214. [DOI] [PubMed] [Google Scholar]
- 35. Bejarano F, Perez L, Apidianakis Y, Delidakis C, Milan M (2007) Hedgehog restricts its expression domain in the Drosophila wing. EMBO Rep 8: 778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson RL, Grenier JK, Scott MP (1995) patched overexpression alters wing disc size and pattern: transcriptional and post-transcriptional effects on hedgehog targets. Development 121: 4161–4170. [DOI] [PubMed] [Google Scholar]
- 37. Burglin TR (2008) The Hedgehog protein family. Genome Biol 9: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matson CK, Zarkower D (2012) Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Genet 13: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]