Abstract
Temporal and spatial organization of the nucleus is critical for the control of transcription, mRNA processing and the assembly of ribosomes. This includes the occupancy of specific territories by mammalian chromosomes, the presence of subnuclear compartments such as the nucleolus and Cajal bodies and the division of chromatin between active and inactive states. These latter are commonly associated with the location of DNA within euchromatin and heterochromatin respectively; critically these distinctions arise through modifications to chromatin-associated proteins, including histones, as well as the preferential localization of heterochromatin at the nuclear periphery. Most research on nuclear organization has focused on metazoa and fungi; however, recent technical advances have made more divergent eukaryotes accessible to study, with some surprising results. For example, the organization of heterochromatin is mediated in metazoan nuclei in large part by lamins, the prototypical intermediate filament proteins. Despite the presence of heterochromatin, detected both biochemically and by EM in most eukaryotic organisms, until this year lamins were thought to be restricted to metazoan taxa, and the proteins comprising the lamina in other lineages were unknown. Recent work indicates the presence of lamin orthologs in amoeba, while trypanosomatids possess a large coiled-coil protein, NUP-1, that performs functions analogous to lamins. These data indicate that the presence of a nuclear lamina is substantially more widespread than previously thought, with major implications for the evolution of eukaryotic gene expression mechanisms. We discuss these and other recent findings on the organization of nuclei in diverse organisms, and the implications of these findings for the evolutionary origin of eukaryotes.
Keywords: evolution, trypanosoma, antigenic variation, lamina, nuclear envelope
Overview
Even in the earliest explorations of eukaryotic ultrastructure it was clear that the nucleus, the defining organelle of eukaryotes, can and does differ widely in size and morphology between different species. These nuclei do, however, seem to share a number of certain features in the electron microscope: every nucleus is bounded by a double-membraned nuclear envelope (NE), perforated by nuclear pore complexes (NPCs) and often underlayered on the nuclear periphery by a dense lamina.1-4 Electron dense bodies such as the nucleolus are commonly found in the nuclear interior; and even the earliest light microscopic studies recognized that chromatin, the nucleus’ packaged DNA, seemed to come in less and more densely packaged forms, termed euchromatin and heterochromatin respectively.5 Though all these features are clearly present in the model metazoan organisms, such as vertebrate, fly, and nematode cells, stark differences emerge when we look at another favorite group of organisms, the fungi. The model yeast Saccharomyces lacks both a lamina and obvious heterochromatin, and the NPCs seem far smaller than those in metazoa.6,7 Other major differences appear during cell division. Thus, metazoan organisms generally disassemble their NE and NPCs completely at the onset of mitosis to allow the spindle to assemble and segregate the chromosomes. A new NE and the NPCs reassemble around the daughter chromosomes at the end of mitosis. By contrast, both the NE and NPCs remain intact throughout the entire cell cycle of yeast, with the spindle forming inside the unbroken nucleus (see refs. 8 and 9 for reviews).
Yeast and metazoa are actually rather close cousins from the perspective of the entire eukaryotic evolutionary tree, both sharing the opisthokont branch or supergroup10 (Fig. 1). However, many unicellular parasitic protozoa are highly divergent eukaryotes, and many sources have suggested that their nuclear organization is markedly different, albeit lacking much in the way of a molecular characterization.11 Thus, our normal opisthokont-centric view, focusing on those cells and organisms most closely related to our selves, i.e., animals and fungi, may be inadequate in attempting to understand these economically important and human-impacting disease organisms. Further, as many protists are major components of the biosphere, their overall impact on the environment is immense. Here we use one such divergent organism, Trypanosoma brucei, as a perspective on this issue. Remarkably, recent advances have revealed that many of the underlying structures and systems in nuclei seem far more conserved than appearances suggested from the rather divergent transcriptional mechanisms in trypanosomes, but that there are crucial differences. These advances allow us not only to view the whole tree of eukaryotic evolution anew, but shed new light on how our own cells are organized and function, with therapeutic implications not only for these divergent parasites but also for ourselves.
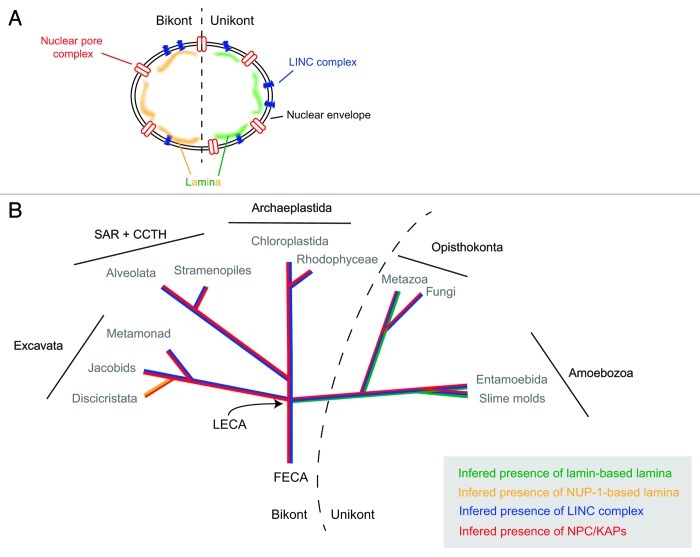
Figure 1. Evolutionary histories of known nuclear envelope components. (A) Cartoon of structures at, and associated with, the nuclear envelope. Shown are the nuclear pore complex/karyopherins (red), Sun/Kash (LINC complex) proteins (blue) and known components of a nuclear lamina (lamins, green; NUP-1, yellow). (B) The evolutionry distributions of the nuclear pore complex/karyopherins, Sun/Kash proteins and known nuclear lamina proteins are shown colorized and overlaid upon a schematic phylogeny of the eukaryotes, emphasizing the five contemporary-recognized supergroups. FECA, First eukaryotic common ancestor; LECA, Last eukaryotic common ancestor; KAP, karyopherin; SAR + CCTH, stramenopiles alveolates, and Rhizaria + cryptomonads, centrohelids, telonemids and haptophytes.
Transcription: Not All the Same
Trypanosomes are members of the Euglenozoa, a group of organisms that belongs to the Excavata supergroup (Fig. 1). They most likely branched very early during eukaryotic evolution, which in part may explain some of their more unusual biology.10,12 Euglenozoa have a truly global impact on the environment, economy and public health. Species include the photosynthetic Euglenids, which populate most fresh water environments, free living phagotrophs, such as Bodo saltans, and plant and animal parasites, the Phytomonads and Trypanosomatids, respectively.13 Trypanosoma brucei has emerged as the model organism for this group, on account of both its importance as the causative agent of human African trypanosomiasis, and from technical advances that include robust RNAi systems, excellent cytology and, most recently, forward genetics based on whole genome RNAi screening.14 Accordingly, our understanding of transcription and chromatin organization is comparatively advanced for T. brucei, although major gaps remain in our knowledge.15-17
As far as we know, all trypanosomatids exploit primarily polycistronic transcription, where large (tens) numbers of open reading frames are transcribed in tandem. There is little evidence for functional association between ORFs in individual polycistronic transcription units (PTUs), although evidence suggests that the position within the PTUs is associated with mRNA copy number.18 The nascent RNAs that are produced from the PTUs are processed by a combination of trans-splicing and polyadenylation to yield mature mRNAs that, bar an unusual 5′ cap structure,19,20 are organizationally indistinguishable from higher eukaryotes. However, this mode of transcription does mean that much of the genome is constitutively transcribed, and mRNA abundance is primarily controlled at the post-transcriptional level,21-24 in sharp contrast to metazoan and yeast model systems, where the transcription of each gene is usually highly regulated by a dedicated promoter.
Notable exceptions to this mechanism are the loci encoding the superabundant surface antigens, the variant surface glycoprotein (VSG) and procyclin; these molecules are expressed at very high levels, and rather remarkably, their transcription is driven by RNA polymerase I, rather than the more usual RNA pol II which is responsible for transcribing the majority of PTUs. Both VSG and procyclin are developmentally regulated, with the former expressed exclusively in mammalian infective forms and the latter in insect vector stages. While procyclin ORFs are embedded within the body of the chromosomes and consist of small loci of a handful of ORFs, VSG transcription is substantially more complex.15,25
VSG is the molecular basis for antigenic variation, the primary immune evasion mechanism in these organisms. The sequential expression of distinct VSGs, in a semistochastic manner, serves to allow escape from immune destruction at the population level, and which appears to rely on monoallelic expression, i.e., a single VSG expressed in any one cell at any one time. Key to this is the restriction of transcriptionally active VSG ORFs to subtelomeric expression sites (ES), where the VSG is the most telomere-proximal ORF in a pol I-driven PTU. Selection of a single VSG is achieved by repression of most VSG ESs within heterochromatin, and the translocation of the active expression site into a specialized region of the nucleus called the expression site body (ESB).15,26,27 It is likely that the positioning of VSG ESs at the nuclear periphery, by virtue of their subtelomeric location, places the inactive ESs within the heterochromatin. Both telomere suppression and VSG-specific repression mechanisms appear to operate (see below). Further, the procyclin locus, which is also transcribed by Pol I, is also subjected to control via positioning within the nucleus; when active, i.e., in the insect, the locus resides peripherally to the nucleolus.28 While definitive data are lacking, all evidence points to a peripheral nuclear position for the locus in the mammalian infective stages, where it is (relatively) inactive.
While polycistronic transcription is certainly not unique to trypanosomes, the high proportion of the genome transcribed by this mechanism is highly unusual. However, taken with the presence of pol I-mediated transcription of protein-coding genes, exploitation of 4D positioning of loci encoding super-abundant surface antigens whose expression is modulated in a life cycle dependent manner and the divergent evolutionary position of trypanosomes suggests the presence of unusual mechanisms at the nuclear periphery.
Conservation and Divergence at the Nuclear Periphery
Although it had been suggested from in silico analysis that the nuclear transport system of trypanosomes had little in common with that of opisthokonts,29 a combination of proteomics, structural studies, and comparative genomics has revealed that the NPC and associated karyopherin transport factors are an ancient feature, and that the overall configuration of this organelle and the transport mechanism and diversity of pathways were established in the last eukaryotic common ancestor (termed LECA). By contrast the lamina and associated macromolecules appear to be more divergent.
The basic NPC architecture, in terms of division into a structural scaffold, consisting predominantly of protocoatomer-related proteins, together with a gating system comprising about ten disordered FG repeat-containing nucleoporins, is conserved across the eukaryotes.30,31 Further, the karyopherin family is likewise ancient and, perhaps more remarkably, based on an ability to reconstruct the majority of karyopherin clades as being widely represented across the eukaryotes, the level of diversity and specificity in nucleocytoplasmic transport is both well conserved across the eukaryotic lineage and a feature of the LECA.32
What these studies imply is that the plural mechanisms of nucleocytoplasmic transport, which are intimately wrapped up with mRNA processing and translocation into the cytoplasm and hence gene expression, are extremely ancient. Minor differences in the compositions between mammalian, yeast, plant and protozoan NPCs may signal some diversification/adaptation of mechanism that reflects the differing transcriptional systems in place in these various lineages, but at a level that is quite detailed and one that is not immediately obvious or accessible from a study of the proteins comprising the NPC and in silico approaches alone. In part, this is due to our rather limited grasp of the precise functions of many individual nucleoporins themselves, and it may well be that some functionality is redundant, such that the presence of a specific subunit from one organism has little or no clear impact on functionality; this is certainly an area where more investigation is warranted. Remarkably, the conservation within NPC functionality even extends to moonlighting nucleoporins in trypanosomes. We have evidence for a nucleoporin, TbNUP92, that appears functionally similar to the mitotic spindle-associated TPR nuclear basket proteins, despite an absence of obvious sequence-based homology and also an FG-repeat nucleoporin, TbNUP53b, potentially involved in transcriptional control and hence analogous to the NUP98 proto-oncogene from Drosophila, albeit again with no obvious primary structural similarity (Holden, J, MPR and MCF, unpublished data, and refs. 9, 33 and 34). It is unclear if these functional similarities in the absence of evidence for sequence relatedness are a result of convergent evolution or a failure in search algorithms to detect such similarity.
In sharp contrast to the NPC/karyopherin system, there is good evidence for significant divergence in the nuclear lamina. While a nuclear lamina has been clearly detectable by ultrastructural analysis in various eukaryotes, and might be expected, in the sense of a basic requirement for structural support of the nucleus (though yeast does manage without apparent support), a knowledge of the molecular basis was restricted to metazoa, where the lamin proteins have been well characterized.35,36 Despite extensive searches of the genomes of yeasts and many other organisms, no evidence for lamin orthologs had been forthcoming, leading to the suggestion that lamins represent a metazoan innovation.
Two studies published in 2012 have, however, overturned this paradigm. First, through a serendipitous route, a clear lamin ortholog was identified for Dictyostelium discoideum, which even includes a C-terminal prenylation signal, heptad repeat signatures and a potential CDK-1 phosphorylation site consensus;37,38 orthologs are also present in additional Amoebozoa genomes. The significance of this discovery is far reaching. With lamins restricted to Metazoa, lineage-specific innovation is the obvious explanation, but the new data suggest that in fact lamins arose several hundred million years earlier, at least as early as the origin of the unikont lineage, which includes the Opisthokonta and Amoebozoa. Clearly then, the absence from fungi can now be interpreted as a secondary loss, and has profound implications for our views on gene expression mechanisms and lifestyles of rather early eukaryotes close to the radiation of the modern supergroups. This study also provides a salient lesson as well—identification of divergent proteins and detecting an ortholog is often challenging; the tools available have especial difficulty with coiled coil proteins. This topic is discussed further in a recent Extra View article in these pages to which the interested reader is referred.37
In the remaining supergroups, the bikonts, there is so far only one example of a lamina-associated protein for which convincing functional evidence is available. This is the trypanosome NUP-1 protein, which actually has structural similarities with and fulfills many of the reported functions of lamins. NUP-1 forms a fenestrated, apparently fibrillar, network on the inner face of the nucleus, and is implicated in functions including maintaining the structural integrity of the nucleus, positioning of NPCs and telomeres, and mediating telomeric silencing. Interactome analysis also indicates a physical association between the NPC and NUP-1 (Obado S, MCF, MPR and Chait BT, unpublished). In trypanosomes, the role in telomere positioning is particularly critical as monoallelic VSG expression is dependent on heterochomatinization of subtelomeric VSG expression sites, and indeed there is good evidence that this process is disrupted by knockdown of NUP-1.39
While the parallels between NUP-1 and unikont lamins are striking, there are some critical differences. First, there is no obvious sequence relatedness and the amino acid repeats in T. brucei are 144 residues, significantly larger than the heptad repeats of the lamins. Second, NUP-1 is a huge protein of 407 kDa, compared with ~60 kDa for lamins, which means that NUP-1 can reach across a substantial proportion of the trypanosome nuclear volume, and hence may indicate a rather distinct structural organization to the lamins (Fig. 2). However, this may in part be specific to trypanosomes as the orthologs in Leishmania are considerably smaller, albeit still predicted to contain over 2,000 residues. Hence, while there is no evidence for any relationship or common ancestry between NUP-1 and the lamins, we remain agnostic on this issue.
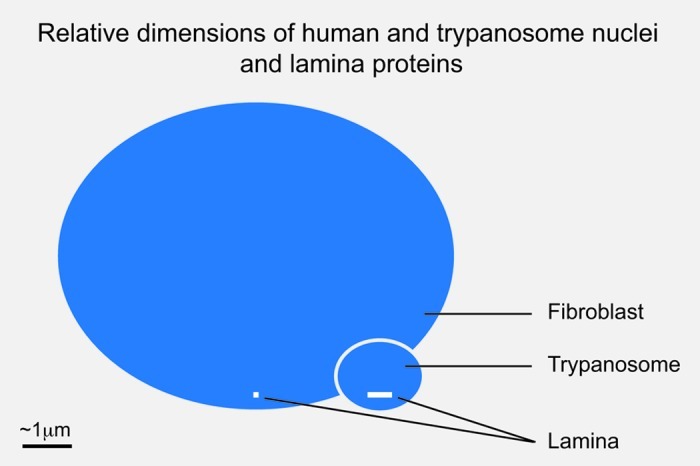
Figure 2. Relative sizes of metazoan and trypanosomatid nuclei and lamina monomers. Blue ovals indicate the relative sizes of a fibroblast and trypanosome nucleus, at 10 μm and 1.5 μm in diameter. The molecular weight of the unikont lamins is approximately 60 kDa while NUP-1 is 450 kDa; these are indicated as white bars close to the indicator lines for each nucleus. The inference is that the structural arrangement of the lamina in these systems is potentially very different.
Also, as mitosis is closed in trypanosomes, the NUP-1 network does not break down as does the mammalian lamin network, but rather it is maintained throughout mitosis, where it may participate in chromosomal segregation (Holden J and MCF, unpublished data). The absence of a massive re-assortment of the nuclear envelope during mitosis is quite significant in terms of maintaining epigenetic marks and the mechanisms by which specific DNA sequences are incorporated into heterochromatin, as in metazoa several lamin-dependent mechanisms that deliver loci to heterochromatin occur as a component of mitosis and reassembly of the nuclear envelope at late anaphase, which clearly cannot be the case for trypanosomes.40 Hence it is unclear just how many of the proteins that control entry, exit and maintenance of heterochromatin are shared across the eukaryotes. However, it seems to us a key point that peripheral organization of developmentally-regulated chromatin (kept inactivated as heterochromatin until needed) at a nuclear lamina composed of coiled-coil proteins is a shared feature of representative unikont and bikont organisms.
A final aspect here are the LINC complex proteins.41 LINC complexes are comprised of trimers of SUN and KASH domain proteins; these trimers then interact within the lumenal space of the nuclear envelope.42,43 These trans-membrane domain proteins are responsible for providing a physical tether for the lamina to the nuclear envelope, as well as a connection between the nuclear lamina and the cytoskeleton. Significantly, SUN and/or KASH domain proteins with the same architecture as those present at the nuclear envelope of metazoa are found in all eukaryotic lineages (including Excavata lineages such as the diplomonads and heterolobosids), with the exception of the trypanosomatids (Fig. 3). By contrast, SUN-like proteins, have different architecture to classical SUN proteins (Fig. 3) and their function is likely distinct from the classical SUN proteins of the LINC complex,44 and are found in trypanosomatids as well as elsewhere. This is perhaps consistent with the restriction of the NUP-1/NUP-2 lamina to Trypanosoma and perhaps argues against convergent evolution from a common lamin/SUN/KASH system. If this is the result of loss from a LECA possessing a lamin-based system, or reflects differential evolution of the lamina in the trypanosomatid and unikont lineages remains to be determined. It is also possible that, as trypanosomes likely branched early from the eukaryotic lineage,11 that evolution of the lamin/SUN/KASH system postdates their speciation event. However, the discovery of the D. discoideum lamin is a further salient lesson in relying too heavily on in silico data alone, if any more are really needed at this juncture, and resolution of this issue will require deeper experimental probing. In summary, the nuclear periphery of the trypanosomes appears to be a mixture of highly conserved elements, together with other elements that may be lineage specific, or are at the very least, extremely divergent (Fig. 1).
Figure 3. Maximum likelihood phylogenetic tree of the SUN domains. The values supporting the separation into two main subfamilies are: approximate likelihood ratio test calculated with PhyML 3.0/non-parametric bootstrap calculated in RAxML 7.2.6/posterior probabilities from PhyloBayes 3.3b. The right panel depicts the domain structures of SUN-domain containing proteins of the representative species used in the phylogenetic analyses. Position of the SUN domain is symbolized by the blue box. Red rectangles represent hydrophobic patches predicted as transmembrane domains by TMHMM Server v. 2.0. Dashed red rectangles represent hydrophobic patches, which were not predicted as transmembrane domains. Red taxa are species where considerable characterization of the SUN proteins has been achieved.
Telomeres and Gene Silencing
Given the evidence that NUP-1 is involved in the positioning of telomeres, NPCs and also the repression of both the VSG and procyclin loci, it is important to consider how the T. brucei lamina impacts singular, mono-telomeric VSG expression and organizes chromatin in a broader context. It seems unlikely that a lamina itself would directly control allelic exclusion, but tethering could certainly be important for maintaining the silencing of all but one of the telomeric VSG genes. For example, a single telomeric VSG may "untether" as part of the process that allows transcriptional activation. A swap in tethering could then allow transcription to switch from one telomeric VSG to another, a process known as in situ switching.45 It is worth considering what is known about the sub-nuclear context of the active and silent telomeric VSGs in this regard. Silent VSGs are thought to be associated with the nuclear periphery while the active VSG is found in the ESB.26 However, no ESB-specific factor has been described, and cells depleted for the telomeric-repeat DNA binding protein, RAP1, that is required for full telomeric VSG silencing, form multiple ESBs.46 This then provides support for the idea that RNA Pol I recruitment and ESB formation are consequences of activation. Thus, current observations are consistent with a tethering-untethering model as outlined above, but it remains to be seen whether activation and untethering, if it happens, occur in a specific order or if they occur concomitantly. Significantly here, no increase in the number of ESBs was observed following NUP-1 knockdown, suggesting a distinct mechanism to RAP1.
There is increasing evidence that basal telomeric silencing in T. brucei and VSG gene silencing are mechanistically distinct. The first evidence in support of this distinction came from studies on telomeres themselves, which could be removed, disrupting basal telomeric silencing, but without disrupting VSG silencing.47 A second piece of evidence derives from studies on the histone deacetylase, Silent Information Regulator 2 related protein 1 (SIR2rp1), which is required to maintain basal telomeric silencing in many organisms but had no major impact on VSG silencing.48 Subsequent reports, including the NUP-1 study, have reinforced this distinction.45 Interestingly, studies on NUP-1 suggest that it is only VSG silencing that is dependent on the intact nuclear lamina.39 It remains possible that basal telomeric silencing is dependent upon another component of the nuclear lamina but characterization of these other components will be required to determine whether this is indeed the case.
It is clear that the identification of regulatory factors provides a “handle” to start to dissect the process in question. The lamina component, NUP-1, is now known to play a role in VSG silencing, and other observations are consistent with the idea that the T. brucei nuclear lamina is important for organizing and reducing the mobility of many subtelomeric VSG genes within the confines of the nuclear space. It will be important to identify lamina-associating sequences and lamina-associated domains49 in T. brucei and structural or regulatory RNAs should probably also be considered in possible models of VSG expression control.50 Ultimately, any model that seeks to explain mono-telomeric and switchable VSG transcription, a system that requires communication among telomeric genes, will likely require the identification of new key regulators and will also need to deal with issues relating to cause and effect. Whatever comes next, we suspect that further characterization of the T. brucei lamina will make an important contribution to improving our understanding of this complex process.
Chromosome Positioning and Genome Segregation
The structure and segregation of the T. brucei nuclear genome, suggests the possibility of differential interactions with the nuclear lamina, with implications for recombination and gene expression. The 32 Mbp haploid nuclear genome of T. brucei is distributed across more than 120 linear chromosomes,51 divided into three classes: 11 pairs of megabase chromosomes (MBC) of at least a megabase in size, one to five intermediate sized chromosomes (IC), and more than 100 minichromosomes (MC) of 50 to 150 kbp.52,53 The MBCs contain all of the housekeeping genes, as well as about 15 telomere-proximal VSG ESs.54 The MCs are highly repetitive in nature and constructed around a palindromic arrangement of a characteristic 177 bp direct repeat;53 many also contain VSG genes, providing a library of distinct variants. The intermediate chromosomes also carry VSG genes, though in contrast to the MCs, these are present in ESs.55 In addition to the linear chromosomes, the T. brucei nucleus contains an unknown number of circular extra-chromosomal DNAs termed NlaIII repeat (NR)-elements of up to 400 kbp in size.56 The NR-elements contribute approximately 6% of the total DNA content of the nucleus,57 but have no known function. In common with the MCs, they are highly repetitive, being made up of short repeats of variable size.
These different classes of nuclear DNA exhibit significant variation in segregation during mitosis and their subsequent organization in the interphase nucleus. The MBCs are segregated by a spindle-kinetochore interaction via microtubules peripheral to the main central spindle,58 so enabling the telomeric regions of the MBCs to maintain their association with the nuclear lamina during mitosis and remain within the established heterochromatin throughout the cell cycle, although to date components of the trypanosome kinetochore remain uncharacterized. This arrangement may serve to prevent interaction with the active VSG ES, which localizes to the extranucleolar expression site body, the site of RNA polymerase I driven expression of the active VSG,36,59 and which replicates later during the cell cycle.27
By contrast the MCs and NR-elements segregate along the central spindle and rapidly migrate to the poles of the daughter nuclei, after which the MCs maintain an asymmetric distribution during interphase, and the NR-elements disperse throughout the interphase nucleus.54,58 Compared with the MBCs, the interaction of the MCs with the nuclear lamina therefore appears episodic, with the association breaking down during mitosis as they migrate along the central spindle, and then becoming re-established upon completion of mitosis, as indicated by a persistent asymmetric distribution during interphase. The VSG genes on the MCs are not in ESs meaning that they cannot be expressed from these loci, so any potential loss of their heterochromatin state during mitosis will presumably not lead to expression, negating a need for this level of repression. Instead, periodic dissociation from the nuclear lamina may allow for recombination between MCs, potentially serving to generate further VSG variants. Establishment of putative MC-lamina interactions may also allow recombination with silent VSG ESs also present in the peripheral heterochromatin, but likely also prevents disruption of the active VSG during interphase on account of the differential positioning and kinetics of replication of the ESB. In contrast to the MBCs and MCs, there is no evidence that the NR-elements interact with the nuclear lamina. Indeed, their circular structure and consequent lack of telomeres may preclude any such interaction, resulting in their dispersal during interphase.56
Perspectives
Our understanding of the evolution of the NE has advanced considerably in the past five years, with 2012 being a particularly good year for moving forward in our understanding of the evolutionary biology of the lamina. The old paradigm of lamins as metazoa specific is now clearly obsolete, and there is robust evidence for a lamina in at least one branch of the Excavata. Many gaps remain, and we currently lack definitive information on the compositions of the lamina of plants, Chromalveolates, Stramenopiles and several Excavata lineages, despite the clear evidence for heterochromatin in these taxa. In the case of Plasmodium, a member of the chromalveolates, a role for telomeric positioning in antigenic variation and with gross similarities to African trypanosomes is well known, but the absence of a molecular definition of a lamina is a significant gap in comprehending the virulence mechanisms of a major pathogen.60-62 Much novel biology and many surprises likely await the investigations of the lamina in these organisms. The rather unusual minichromosomes, which contribute to antigenic variation, are an important aspect of the biology of trypanosomes, and may have had a strong influence of the evolution of the nuclear lamina in these taxa, potentially to avoid promiscuous recombination, reactivation and other defects that could disrupt antigenic variation. However, genome structure alone cannot explain the high divergence between lamin-based and NUP-1-based lamina as, for example, minichromosomes are absent from T. cruzi, a related trypanosomatid that has a clear NUP-1 ortholog.
In terms of defining the functions of the trypanosomatid NUP-1 lamina, a pre-requisite for deeper comprehension of how related the mechanisms of organization of chromatin truly are in trypanosomes and unikonts, it will be essential to probe the interactions of these proteins in greater detail. This will necessitate interrogation of how trypanosome lamina components operate in the context of specific DNA sequences, determination of the nature and distribution of lamina-targeting sequences (if such exist), how the NUP-1 lamina organizes chromatin-modifying activities, and how this system integrates with the nuclear envelope and spindle. Answering these questions is most likely a long journey, but the identification of NUP-1 as a lamina component has finally provided a doorway through which we can begin that journey.
Lastly, we can ask: Why do eukaryotes seem to share, as a most fundamental mechanism of genetic control, developmental regulation by packaging of huge blocks of chromatin at the nuclear periphery, a periphery organized in many cases by an extensive coiled-coil protein network? Such developmental heterochromatin in “higher” eukaryotes is used to store the genetic information needed to build tissues. Cohorts of tissue-specific genes are packaged away in lineages where expression is not needed, or perhaps more critically, would be detrimental if allowed to occur; the dynamic range of heterochromatin suppression is very much greater than simple promoter-based inactivation. Hemoglobin genes are thus normally inactive in the nuclear peripheral heterochromatin of all but hemopoetic cells while cell adhesion molecules that specify cell type and position are similarly inactivated during metazoan embryogenesis.63,64 But LECA was undoubtedly unicellular, and therefore these sophisticated multicellular organism-related processes cannot explain the need for such tight regulation. Likewise, the parasitic lifestyle of trypanosomes necessitates massive remodeling to accommodate drastically different insect and vertebrate host environments, but again LECA predates multicellularity, and hence such parasitic lifestyles. However, the inference that LECA possessed such a developmental regulation mechanism indicates that it could undergo significant changes in its lifestyle. The primordial environment LECA faced was potentially much more unstable and chaotic than most organisms face nowadays, as it is our extensive biosphere that contributes to stabilizing Earth’s modern climates.65 It is also possible that the specific location that LECA found itself in had specific challenges that necessitated periodic modulations of gene expression patterns, for example salinity or seasonal changes. We speculate that perhaps LECA evolved to change its form and function to adapt to these shifting environments. An ability to store great amounts of genetic information to be used upon need could have been the path to the eukaryotic colonization of diverse land and sea environments, and the basis for the multicellularity required for the complex lifeforms that enabled such colonization.
Acknowledgments
We thank Sam Obado and Jenny Holden for discussions and access to unpublished data. Work described in this article that has arisen from the authors’ laboratories has been funded by the Wellcome Trust (082813 to M.C.F., 093010 to D.H.), the National Institutes of Health (R21 AI096069, U54 GM103511 to M.P.R.) and by a Marie Curie fellowship (to L.K.). This support is gratefully acknowledged.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/22167
References
- 1.Watson ML. The nuclear envelope; its structure and relation to cytoplasmic membranes. J Biophys Biochem Cytol. 1955;1:257–70. doi: 10.1083/jcb.1.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pappas GD. Helical structures in the nucleus of Amoeba proteus. J Biophys Biochem Cytol. 1956;2:221–2. doi: 10.1083/jcb.2.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappas GD. The fine structure of the nuclear envelope of Amoeba proteus. J Biophys Biochem Cytol. 1956;2(Suppl):431–4. doi: 10.1083/jcb.2.4.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leeson TS, Bhatnagar R. Microfibrillar structures in the nucleus and cytoplasm of amoeba proteus. J Exp Zool. 1975;192:265–70. doi: 10.1002/jez.1401920216. [DOI] [PubMed] [Google Scholar]
- 5.Moses MJ. Studies on nuclei using correlated cytochemical, light, and electron microscope techniques. J Biophys Biochem Cytol. 1956;2(Suppl):397–406. doi: 10.1083/jcb.2.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strambio-de-Castillia C, Blobel G, Rout MP. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–55. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutay U, Hetzer MW. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol. 2008;20:669–77. doi: 10.1016/j.ceb.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–30. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 11.Wilson KL, Dawson SC. Evolution: functional evolution of nuclear structure. J Cell Biol. 2011;195:171–81. doi: 10.1083/jcb.201103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–5. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadelha C, Holden JM, Allison HC, Field MC. Specializations in a successful parasite: what makes the bloodstream-form African trypanosome so deadly? Mol Biochem Parasitol. 2011;179:51–8. doi: 10.1016/j.molbiopara.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–24. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro M, Peñate X, Landeira D. Nuclear architecture underlying gene expression in Trypanosoma brucei. Trends Microbiol. 2007;15:263–70. doi: 10.1016/j.tim.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Daniels JP, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiol Mol Biol Rev. 2010;74:552–69. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ersfeld KF. Nuclear architecture, genome and chromatin organisation in Trypanosoma brucei. Res Microbiol. 2011;162:626–36. doi: 10.1016/j.resmic.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Kelly S, Kramer S, Schwede A, Maini PK, Gull K, Carrington M. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012;2:120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem. 1992;267:9805–15. [PubMed] [Google Scholar]
- 20.Zamudio JR, Mittra B, Campbell DA, Sturm NR. Hypermethylated cap 4 maximizes Trypanosoma brucei translation. Mol Microbiol. 2009;72:1100–10. doi: 10.1111/j.1365-2958.2009.06696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koumandou VL, Natesan SK, Sergeenko T, Field MC. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics. 2008;9:298. doi: 10.1186/1471-2164-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics. 2009;10:482. doi: 10.1186/1471-2164-10-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martínez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Biotechnol. 2010;2010:525241. doi: 10.1155/2010/525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alsford S, duBois K, Horn D, Field MC. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert Rev Mol Med. 2012;14:e13. doi: 10.1017/erm.2012.7. [DOI] [PubMed] [Google Scholar]
- 26.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–63. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 27.Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol. 2009;186:243–54. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landeira D, Navarro M. Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J Cell Biol. 2007;176:133–9. doi: 10.1083/jcb.200607174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–37. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 30.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, et al. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–30. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Reilly AJ, Dacks JB, Field MC. Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One. 2011;6:e19308. doi: 10.1371/journal.pone.0019308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–71. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–83. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–53. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–71. doi: 10.1016/j.tig.2012.06.001. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batsios P, Peter T, Baumann O, Stick R, Meyer I, Gräf R. A lamin in lower eukaryotes? Nucleus. 2012;3:237–43. doi: 10.4161/nucl.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krüger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, et al. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell. 2012;23:360–70. doi: 10.1091/mbc.E11-07-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, et al. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10:e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tseng LC, Chen RH. Temporal control of nuclear envelope assembly by phosphorylation of lamin B receptor. Mol Biol Cell. 2011;22:3306–17. doi: 10.1091/mbc.E11-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellad JA, Warren DT, Shanahan CM. Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol. 2011;23:47–54. doi: 10.1016/j.ceb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Méjat A, Misteli T. LINC complexes in health and disease. Nucleus. 2010;1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–47. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimada N, Inouye K, Sawai S, Kawata T. SunB, a novel Sad1 and UNC-84 domain-containing protein required for development of Dictyostelium discoideum. Dev Growth Differ. 2010;52:577–90. doi: 10.1111/j.1440-169X.2010.01189.x. [DOI] [PubMed] [Google Scholar]
- 45.Horn D, McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr Opin Microbiol. 2010;13:700–5. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glover L, Alsford S, Beattie C, Horn D. Deletion of a trypanosome telomere leads to loss of silencing and progressive loss of terminal DNA in the absence of cell cycle arrest. Nucleic Acids Res. 2007;35:872–80. doi: 10.1093/nar/gkl1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsford S, Kawahara T, Isamah C, Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol. 2007;63:724–36. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 49.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–87. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 50.Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–63. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–22. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 52.Melville SE, Leech V, Navarro M, Cross GA. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol Biochem Parasitol. 2000;111:261–73. doi: 10.1016/S0166-6851(00)00316-9. [DOI] [PubMed] [Google Scholar]
- 53.Wickstead B, Ersfeld K, Gull K. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 2004;14:1014–24. doi: 10.1101/gr.2227704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ersfeld K, Melville SE, Gull K. Nuclear and genome organization of Trypanosoma brucei. Parasitol Today. 1999;15:58–63. doi: 10.1016/S0169-4758(98)01378-7. [DOI] [PubMed] [Google Scholar]
- 55.Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alsford NS, Navarro M, Jamnadass HR, Dunbar H, Ackroyd M, Murphy NB, et al. The identification of circular extrachromosomal DNA in the nuclear genome of Trypanosoma brucei. Mol Microbiol. 2003;47:277–89. doi: 10.1046/j.1365-2958.2003.03266.x. [DOI] [PubMed] [Google Scholar]
- 57.Jamnadass RH, Pellé R, Pandit P, Ricard B, Murphy NB. Trypanosoma brucei: composition, organisation, plasticity, and differential transcription of NlaIII repeat elements in drug-resistant and sensitive isolates. Exp Parasitol. 2006;113:244–55. doi: 10.1016/j.exppara.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Ersfeld K, Gull K. Partitioning of large and minichromosomes in Trypanosoma brucei. Science. 1997;276:611–4. doi: 10.1126/science.276.5312.611. [DOI] [PubMed] [Google Scholar]
- 59.Günzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, et al. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003;2:542–51. doi: 10.1128/EC.2.3.542-551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westenberger SJ, Cui L, Dharia N, Winzeler E, Cui L. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics. 2009;10:610. doi: 10.1186/1471-2164-10-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernandez-Rivas R, Pérez-Toledo K, Herrera Solorio AM, Delgadillo DM, Vargas M. Telomeric heterochromatin in Plasmodium falciparum. J Biomed Biotechnol. 2010;2010:290501. doi: 10.1155/2010/290501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, et al. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011;10:451–63. doi: 10.1016/j.chom.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giles KE, Gowher H, Ghirlando R, Jin C, Felsenfeld G. Chromatin boundaries, insulators, and long-range interactions in the nucleus. Cold Spring Harb Symp Quant Biol. 2010;75:79–85. doi: 10.1101/sqb.2010.75.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briones V, Muegge K. The ghosts in the machine: DNA methylation and the mystery of differentiation. Biochim Biophys Acta. 2012;1819:757–62. doi: 10.1016/j.bbagrm.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beyond Weather & the Water Cycle. The Biosphere/ Climate Connection. 2011 May; http://beyondweather.ehe.osu.edu/issue/we-depend-on-earths-climate/the-biosphereclimate-connection