Abstract
Genomes are spatially organized on many levels and the positioning of genes within the nucleus contributes to their proper expression. This positioning can also result in the clustering of genes with similar expression patterns, a phenomenon sometimes called “gene kissing.” We have found that yeast genes are targeted to the nuclear periphery through interaction of the nuclear pore complex with small, cis-acting “DNA zip codes” in their promoters. Our recent study demonstrated that genes with the same zip codes cluster together at the nuclear periphery. The zip codes were necessary and sufficient to induce interchromosomal clustering. Finally, we identified a transcription factor (Put3) that binds to the GRS I zip code. Put3 binds to GRS I and is required for both GRS I-dependent positioning at the nuclear periphery and interchromosomal clustering of GRS I-targeted genes. We speculate that our findings might provide insight into other types of gene kissing, some of which also require cis-acting DNA sequences and trans-acting proteins.
Keywords: nuclear pore complex, transcription, nuclear organization, gene kissing, nuclear body
Introduction
DNA within the nucleus is spatially organized. Folded chromosomes occupy discrete “territories” within the nucleus in interphase metazoan cells.1,2 Chromosome folding, inter- and intra-chromosomal interactions and the association of chromosomes with subnuclear structures have been suggested to create subnuclear environments that both reflect and facilitate different expression states.3 Although the spatial organization of chromosomes and the positioning of individual genes are stereotyped within differentiated cells, they are dynamic and can change during differentiation or in response to an environmental cue.1,4 For example, many genes that are induced during differentiation move from the nuclear periphery (where they associate with the nuclear lamina) to a more nucleoplasmic position, sometimes in association with “transcription factories”5. Disruption of the spatial organization of genes is also associated with disease. For example, the spatial arrangement of genes is disrupted in breast cancer cells.6,7 Thus, the spatial positioning of individual genes is dynamic and either reflects their expression state or contributes to their regulation.
Gene clustering
The clustering of co-regulated genes is a common theme in nuclear architecture.3 Techniques such as fluorescent in situ hybridization (FISH), chromosome painting and variations of the chromosomal conformation capture (3C) technique have illuminated a large number of intra-chromosomal and inter-chromosomal interactions in many organisms. For example, in yeast, FISH revealed that many tRNA genes from throughout the genome cluster together in the nucleolus,8 centromeres cluster near the spindle pole body9,10 and telomeres cluster at the nuclear envelope.11 Data from 3C experiments in yeast reveals the clustering of tRNA genes, genes near early firing origins and genes with Gal4 binding sites.10,12 Polycomb-repressed Hox genes cluster during embryogenesis in Drosophila.13,14 In mouse erythrocytes, during hematopoiesis, the globin genes cluster with each other and with many erythrocyte-specific genes.15 These co-regulated genes cluster in close proximity to foci of active RNA polymerase II called “transcription factories”15-18. Therefore, interchromosomal clustering may represent an additional layer of transcriptional regulation that may either reinforce signals or allow better coordinated expression of co-regulated genes.
Gene targeting to the nuclear pore complex
As a model for these phenomena, we have studied the movement of inducible yeast genes from the nucleoplasm to the nuclear periphery upon activation.19,20 Within the yeast nucleus, there are three major zones: the nucleoplasm, the nuclear periphery and the nucleolus. ChIP experiments using nuclear pore proteins suggests that hundreds of active yeast genes interact with the nuclear pore complex (NPC).19,21 Likewise, thousands of active genes interact with nuclear pore proteins in Drosophila.22,23 However, in Drosophila, most of these genes interact with nuclear pore proteins away from the NPC, in the nucleoplasm.22,23 This suggests that in metazoans, nuclear pore proteins can interact with genes both in the nucleoplasm and at the pore. Inducible yeast genes such as INO1, TSA2, GAL1, HSP104 and SUC2 localize to the nuclear periphery when active. Movement from the nucleoplasm to the nuclear periphery is rapid24 and requires several nuclear pore proteins, mRNA export factors, NPC-associated Mlp proteins and the SAGA histone acetyltransferase.25-28 Targeting of INO1 to the nuclear periphery and interaction of GAL1 with the NPC does not require active transcription, suggesting that it is not mediated by nascent mRNA.24,29 Small cis-acting DNA elements called gene recruitment sequences (GRSs) in the INO1 and TSA2 promoter are necessary for targeting to the nuclear periphery and interaction with the NPC.25 These DNA elements function as DNA zip codes: when inserted at a locus that normally localizes in the nucleoplasm, they are sufficient to induce repositioning to the nuclear periphery.25,30,31
The interaction of active genes with the NPC has been proposed to promote transcription,25 to function as a chromatin boundary,32-34 to facilitate epigenetic transcriptional memory24,30,35 and to provide negative feedback.36 Mutations in the GRSs of INO1 and TSA2 that block interaction with the NPC lead to a defect in transcription.25 Likewise, knockdown of nuclear pore proteins in Drosophila results in decreased expression of many of the genes that bind to nuclear pore proteins.22,23 However, currently it is not clear how interaction with nuclear pore proteins or positioning at the nuclear periphery impacts transcription.
Results
To ask if DNA zip codes play a role in interchromosomal clustering of genes at the nuclear periphery, we adapted a chromatin localization assay to allow simultaneous localization of two genes. An array of 112 Tet repressor binding sites was integrated beside INO1 on chromosome X and an array of 128 Lac repressor binding sites was integrated at several other loci. These strains expressed GFP-TetR and RFP-LacI. The position of the two loci, one red dot and one green dot, with respect to the nuclear membrane and with respect to each other, could be measured. Alternatively, we also utilized strains with either 128 or 256 Lac repressor binding sites at two loci, expressing GFP-LacI. These strains exhibited discernably different-sized green dots. These modifications allowed us to study the behavior of two loci with overlapping or distinct mechanisms of targeting in the same cell.
Gene-specific clustering at the nuclear periphery
To analyze clustering of genes, we measured the distances between the loci in the population. When we compared a gene at the nuclear periphery (INO1 or HSP104) and a gene in the nucleoplasm (URA3), we did not observe significant clustering: the distributions of distances were normal with means of 0.8–1.0 μm (approximately the radius of the yeast nucleus) and the two loci were ≤ 0.5 μm apart in less than 20% of the cells. Similarly, repressed INO1, which localizes in the nucleoplasm, did not cluster with itself in diploid cells (1.0 ± 0.47 μm; 20% ≤ 0.5 μm). However, when active, the two alleles of INO1 clustered together (0.60 ± 0.33 μm, 52% ≤ 0.5 μm; p < 0.0001).
To confirm that this clustering was due to targeting and not related to homology between chromosomes, we inserted an ectopic copy of INO1 at the URA3 locus in a haploid strain. This hybrid locus, URA3:INO1, is targeted to the nuclear periphery normally when INO1 is induced.25,30 When active, but not when repressed, INO1 clustered with URA3:INO1. Thus, the information necessary for both targeting to the nuclear periphery and for interchromosomal clustering is contained within the sequence inserted at URA3.
To confirm that the clustering was gene-specific, we compared the positioning of several genes known to localize to the nuclear periphery. We performed pair-wise comparisons between INO1, GAL1, HSP104 and GAL2 and we found that, although these genes all localize to the nuclear periphery, they did not cluster with each other. Therefore, recruitment to the nuclear periphery is not sufficient to cause genes to cluster, even for genes on the same chromosome: HSP104 and GAL2 are ~290 kb apart on opposite sides of the centromere of chromosome XII. The GAL2 locus is close to the rDNA locus and is positioned within or adjacent to the nucleolus.37,38 This example further highlights how chromosome structure can create micro-domains or sub-compartments of the nucleus.
INO1 clustering requires interaction with the nuclear pore complex
We also examined the role of the NPC on the clustering of genes at the nuclear periphery. INO1 recruitment to the nuclear periphery is blocked in strains lacking Nup2, one of the proteins that make up the nuclear basket-like structure on the nucleoplasmic face of the NPC.24 In strains lacking Nup2, INO1 did not cluster with URA3:INO1. This suggests that targeting to the nuclear pore is required for clustering.
When genes are targeted to the nuclear periphery, they are still mobile and the targeting is not uniform. In a typical experiment, a gene will colocalize with the nuclear periphery in ~65% of the cells in the population (compared with ~30% for a nucleoplasmic locus). This reflects both the continuous motion of genes in living cells9,26,37 and the negative regulation of peripheral targeting during S-phase.39 This allowed us to more rigorously test the idea that clustering only occurs at the nuclear periphery. To do this, we examined the clustering of INO1 and URA3:INO1 separately in three different classes of cells: (1) cells in which either both genes were at the nuclear periphery, (2) cells in which both genes were in the nucleoplasm or (3) cells in which one gene was at the nuclear periphery and the other was in the nucleoplasm. As we expected, when both genes were at the nuclear periphery, they were clustered (72% ≤ 0.5 μm) and when one gene was at the periphery and the other was in the nucleoplasm, they were not (12.5% ≤ 0.5 μm). However, we were surprised to find that, when both genes were nucleoplasmic, they were also clustered (59% ≤ 0.5 μm). Therefore, although clustering required Nup2, it could also be maintained in the nucleoplasm. This raised the possibility that targeting to the nuclear pore complex might be a pre-requisite for clustering. To test this idea, we treated the cells with hydroxyurea to arrest them in S-phase, a moment in the cell cycle in which peripheral targeting is blocked.39 We asked (1) if clustering could be maintained in cells arrested in S-phase and (2) if clustering could be established in cells arrested in S-phase. In cells in which INO1 is expressed and targeted to the nuclear periphery prior to arresting in S-phase, INO1 and URA3:INO1 remained clustered in the nucleoplasm after arrest. Therefore, clustering was maintained in the nucleoplasm in cells arrested in S-phase. However, in cells arrested in S-phase and then shifted to medium to induce INO1, INO1 and URA3:INO1 did not cluster. Therefore, clustering could not be established in the nucleoplasm in cells arrested in S-phase. Together, these results suggest that targeting to the nuclear periphery is a pre-requisite for clustering, but that clustering can persist in the nucleoplasm.
DNA zip codes control interchromosomal clustering
We previously identified two DNA zip codes in the promoter of INO1 (GRS I and GRS II) that are necessary and sufficient for gene targeting to the nuclear periphery.25 We explored the role of these elements in controlling INO1 clustering. Insertion of either GRS I or GRS II beside URA3 is sufficient to reposition URA3 to the nuclear periphery.25 Because URA3:INO1 possesses GRS I but not GRS II,25 we hypothesized that GRS I controls clustering. Indeed, insertion of GRS I, but not GRS II, was sufficient to induce clustering with the endogenous INO1 gene. Furthermore, disruption of GRS I, but not GRS II, within the endogenous INO1 promoter blocked clustering with URA3:INO1. Therefore, the GRS I zip code controls both gene targeting to the nuclear periphery and gene clustering.
The GRS I sequence appears in the promoter of 94 genes, including the stress-inducible TSA2.25 Therefore, we asked if TSA2 clustered with INO1. Uninduced TSA2 did not cluster with INO1 (0.83 ± 0.41 μm; 25% ≤ 0.55 μm). But when both genes were activated, INO1 and TSA2 genes clustered (0.58 ± 0.38 μm; 55% ≤ 0.55 μm; p < 0.0001). When the GRS I in the INO1 promoter was disrupted, clustering was lost (0.91 ± 0.42 μm; 25% ≤ 0.55μm). Therefore, genes from different chromosomes cluster based on a small targeting determinant found in their promoters.
To expand on these results, we asked if other genes cluster at the nuclear periphery and if clustering is similarly controlled by DNA zip codes. HSP104 is recruited to the nuclear periphery upon heat shock or in ethanol.28 We identified a DNA zip code in the HSP104 promoter which we have designated GRS III. Like other zip codes, GRS III is sufficient to target URA3 to the nuclear periphery. Furthermore, active HSP104 clusters with URA3:GRS III at the nuclear periphery. Therefore, DNA zip codes play a general role in targeting to the nuclear periphery and in promoting inter-chromosomal clustering.
The Put3 transcription factor mediates GRS I-dependent gene targeting and clustering
Because the GRS I zip code is critical for gene localization and interchomosomal clustering, we identified the protein that recognizes the GRS I. Using a combination of electrophoretic mobility shift assays (EMSA) and affinity chromatography followed by mass-spec analysis, we identified Put3. Put3 from yeast extracts interacted with the GRS I sequence in EMSA experiments. In strains lacking Put3, GRS I-dependent targeting to the nuclear periphery was blocked. Put3 is a Zn6-Cys6 zinc-finger transcription factor that regulates the expression of genes involved in proline metabolism.40 Put3 binds to PUT1 and PUT2 via the UASPUT element (CGG-N10-GCC) that has no obvious similarity to GRS I (GGGTTGGA). However, using chromatin immunoprecipitation we showed that Put3 binds to the GRS I in vivo and that Put3 is required for the interaction between the GRS I and the nuclear pore. Loss of Put3, like loss of GRS I, leads to a defect in INO1 and TSA2 transcription. Finally, clustering of INO1 and URA3:INO1 requires Put3. Therefore, recognition of a DNA zip code by a transcription factor mediates targeting to the nuclear periphery, interaction with the nuclear pore complex and interchromosomal clustering.
Conclusions
The work summarized here showed that, in yeast, genes that are targeted to the nuclear pore complex upon activation can cluster together. Clustering is mediated by DNA zip codes. We showed that two different zip codes, when inserted at the URA3 locus, induce both targeting to the nuclear periphery and clustering with endogenous genes having the same zip code. In other words, URA3 can be directed to the nuclear periphery and can cluster with two distinct sets of genes when the right zip code is placed at this locus. The focus of our work has been genes that are targeted to the nuclear periphery and cluster in a regulated manner. It remains to be seen whether these lessons will apply to housekeeping genes or repressed genes that cluster together.
Outlook
A full understanding of the molecular mechanism by which gene clustering can be achieved will require the identification of additional proteins involved in the process and a better understanding of the phenomenon in living cells. However, we would like to close with a brief discussion of three other important questions. First, to what extent are these lessons generalizable, either within the yeast genome or in other genomes. Second, how does gene clustering affect the spatial organization of the genome as a whole? And third, how does gene localization at the nuclear periphery, interaction with nuclear pore proteins or clustering with co-regulated genes impact gene expression? Although we cannot answer any of these questions here, we will briefly summarize our thoughts about each of them.
Generalizability
Hundreds of yeast genes interact with the NPC.19 We have tested a handful of genes (INO1, HSP104, GAL1, ACT1 and GAL2) and we were able to identify cis-acting promoter elements that were capable of targeting URA3 to the nuclear periphery for all but one of these genes (data not shown). Genes with a GRS I zip code in their promoters are over-represented among genes that interact with the NPC.25 Although this set of genes may not be representative and there may be other ways that genes are targeted to the NPC, it is reasonable to propose cis-acting DNA zip codes play an important general role in controlling interaction with the NPC.
Not all zip codes are capable of inducing interchromosomal clustering. Both the GRS III element from the HSP104 promoter and the GRS I element from the INO1 promoter, when inserted at URA3, induced clustering of URA3 with HSP104 and INO1, respectively. However, the GRS II element from the INO1 promoter did not induce clustering and was not required for clustering of INO1 with URA3:INO1. Therefore, we conclude that some, but not all, DNA zip codes induce interchromosomal clustering.
Do these lessons provide insight into the spatial organization of the genomes of other organisms, such as metazoans? Do cis-acting DNA elements and transcription factors control gene positioning and interchromosomal clustering? The positioning of metazoan genes often reflects their expression and can change when genes are induced or repressed. There are several well-documented patterns of gene positioning in metazoan nuclei. Many repressed or silenced genes physically interact with nuclear lamina but, if they are induced, they move to a more internal site.41-43 Such genes can also cluster together with co-regulated genes in close association with transcription factories.15 Likewise, Polycomb-repressed loci in flies colocalize with each other at Polycomb bodies.14,44 Both the localization of genes at the nuclear lamina and the clustering of co-regulated genes require cis-acting DNA sequences or trans-acting transcription factors.15,45,46 The interaction of genes with nuclear pore proteins in metazoans can occur both at the NPC and in the nucleoplasm.23,47,48 It is not known if metazoan genes that interact with nuclear pore proteins exhibit interchromosomal clustering or if the interaction of nuclear pore proteins with these genes is mediated by cis-acting DNA elements or trans-acting factors. However, these genes are enriched for certain transcription factor binding sites.48 Therefore, it is plausible that our understanding of NPC-gene interactions in the yeast system will illuminate similar phenomena in multicellular organisms.
The effect of gene clustering on the global organization of the yeast genome
Our observations suggest that yeast genes that share the same mechanism of targeting to the nuclear pore complex cluster together. The most extreme interpretation of this conclusion is that the DNA zip codes encode targeting to a particular portion of the nuclear envelope. Although our data are consistent with this idea, it is equally likely that targeting to the nuclear periphery, interaction between genes and a gene’s position along the chromosome provide sufficient constraints to limit the fraction of the nuclear periphery that it will visit (Fig. 1). Because yeast centromeres are stably associated with the spindle pole body during interphase, the two-dimensional distance between genes and the centromere will likely impact their mobility and their interchromosomal interactions (Fig. 1).
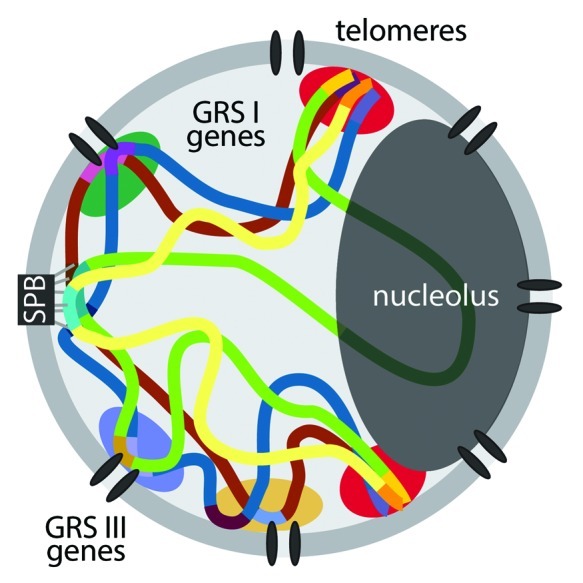
Figure 1. Model for the clustering of co-regulated genes at the nuclear periphery in yeast. Throughout the cell cycle, yeast centromeres remain stably associated with the spindle pole body (SPB), which is embedded in the nuclear envelope. The rDNA locus is positioned within the nucleolus at the opposite pole. Telomeres cluster together at the nuclear periphery and concentrate proteins involved in transcriptional silencing (red clouds). Different sets of genes that are targeted to the nuclear pore complex by different DNA zip codes (GRS I and GRS III) cluster together, potentially resulting in a heterogeneous distribution of factors that promote their expression (green, blue and orange clouds).
If the default position of genes with respect to each other were influenced by their position along the chromosome arm, we would expect the 3D distance between two genes to be influenced by the similarity of their 2D distance from the centromere. The genes that we have examined in detail (INO1, TSA2, URA3 and HSP104) are ~300 kb, 385 kb, 34 kb and 60 kb from their centromeres, respectively. Under conditions where these genes are not targeted to the nuclear periphery (i.e., default positioning), the 3D distances between them are somewhat reflective of the similarity of their 2D distance to the centromere: repressed INO1 and URA3 were further apart (300 kb vs. 34 kb; 1.08 ± 0.43 μm) than repressed INO1 and TSA2 (300 kb vs 385 kb; 0.83 ± 0.41 μm). However, when genes are targeted to the nuclear periphery, we observe clustering between INO1 and TSA2 (300 kb vs. 385 kb), INO1 and URA3:GRS I (300 vs. 34 kb) and HSP104 and URA3:GRS III (60 kb vs. 34 kb). This suggests that, although the distance between a gene and the centromere constrains spatial positioning, DNA zip codes can provide an additional, dominant input. Therefore, we speculate that the clustering of genes at the nuclear periphery may affect the folding and disposition of chromosomes in the yeast nucleus.
Gene clustering and expression
The clustering of INO1 and TSA2 at the nuclear pore complex is required for maximal expression of these genes.25,31 How gene positioning impacts expression is poorly understood. The assumption is that genes move to exploit the heterogeneous nuclear distribution of factors important for transcription or post-transcriptional regulation. For example, the clustering of activated genes during hematopoiesis leads to their co-localization with active RNA polymerase II transcription factories. However, several of the nuclear “bodies” that are observed in mammalian cells can form de novo in association with genes.49-51 Therefore, it is not always necessary to change gene position to establish contact between a gene and a nuclear body.
In yeast, factors that promote transcription or chromatin remodeling do not, in general, exhibit heterogeneous nuclear staining.52 We speculate that genes that share zip codes also share requirements for factors involved in their transcription. If so, then the clustering of co-regulated genes, like the clustering of telomeres at the nuclear envelope,53 might itself serve to compartmentalize the open environment of the nucleoplasm by concentrating these factors and enhancing the efficiency of transcription (Fig. 1). This compartmentalization would be dynamic and the compartments would not necessarily outlive the clusters. Such a model would be consistent with both the functional importance of clustering and the ability of nuclear bodies to form de novo.
The inter- and intra-chromosomal clustering of co-regulated genes is a widespread and fundamental aspect of genome organization. Understanding the molecular basis for this phenomenon will provide new and important insight into how long-range interactions impact gene expression and global organization of the genome.
Acknowledgments
The authors would like to thank the members of the Brickner Lab for stimulating scientific discussions about this manuscript. The work in Brickner et al. (2012) was supported by NIH grant GM080484 and a W.M. Keck Young Scholars in Biomedical Research Award (J.H.B).
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/22663
References
- 1.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–16. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Cavalli G. Chromosome kissing. Curr Opin Genet Dev. 2007;17:443–50. doi: 10.1016/j.gde.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–33. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187:801–12. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–6. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 10.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–63. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehlen LR, Gruenert G, Jones MB, Rodley CD, Langowski J, O’Sullivan JM. Chromosome positioning and the clustering of functionally related loci in yeast is driven by chromosomal interactions. Nucleus. 2012;3:370–83. doi: 10.4161/nucl.20971. [DOI] [PubMed] [Google Scholar]
- 13.Mateos-Langerak J, Cavalli G. Polycomb group proteins and long-range gene regulation. Adv Genet. 2008;61:45–66. doi: 10.1016/S0065-2660(07)00002-8. [DOI] [PubMed] [Google Scholar]
- 14.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–20. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eskiw CH, Cope NF, Clay I, Schoenfelder S, Nagano T, Fraser P. Transcription factories and nuclear organization of the genome. Cold Spring Harb Symp Quant Biol. 2010;75:501–6. doi: 10.1101/sqb.2010.75.046. [DOI] [PubMed] [Google Scholar]
- 17.Carter DR, Eskiw C, Cook PR. Transcription factories. Biochem Soc Trans. 2008;36:585–9. doi: 10.1042/BST0360585. [DOI] [PubMed] [Google Scholar]
- 18.Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta. 2008;1783:2155–60. doi: 10.1016/j.bbamcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–39. doi: 10.1016/S0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 20.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–98. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–83. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–71. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, et al. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–8. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–3. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 27.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, et al. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–9. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 28.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–70. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–91. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–25. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, et al. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell. 2012;22:1234–46. doi: 10.1016/j.devcel.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, et al. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–65. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–62. doi: 10.1016/S0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 34.Kalverda B, Fornerod M. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 2010;9:4812–7. doi: 10.4161/cc.9.24.14328. [DOI] [PubMed] [Google Scholar]
- 35.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–24. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell. 2012;23:1367–75. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger AB, Cabal GG, Fabre E, Duong T, Buc H, Nehrbass U, et al. High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods. 2008;5:1031–7. doi: 10.1038/nmeth.1266. [DOI] [PubMed] [Google Scholar]
- 38.Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, et al. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol. 2009;187:455–62. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brickner DG, Brickner JH. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol Biol Cell. 2010;21:3421–32. doi: 10.1091/mbc.E10-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddiqui AH, Brandriss MC. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989;9:4706–12. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–62. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 42.Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–98. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–25. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 45.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–87. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 46.Vazquez J, Müller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–65. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–39. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–73. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 50.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–7. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 51.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 53.Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–25. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]


