Abstract
Viruses that replicate in the nucleus need to pass the nuclear envelope barrier during infection. Research in recent years indicates that the nuclear envelope is a major hurdle for many viruses. This review describes strategies to overcome this obstacle developed by seven virus families: herpesviridae, adenoviridae, orthomyxoviridae, lentiviruses (which are part of retroviridae), Hepadnaviridae, parvoviridae and polyomaviridae. Most viruses use the canonical nuclear pore complex (NPC) in order to get their genome into the nucleus. Viral capsids that are larger than the nuclear pore disassemble before or during passing through the NPC, thus allowing genome nuclear entry. Surprisingly, increasing evidence suggest that parvoviruses and polyomaviruses may bypass the nuclear pore by trafficking directly through the nuclear membrane. Additional studies are required for better understanding these processes. Since nuclear entry emerges as the limiting step in infection for many viruses, it may serve as an ideal target for antiviral drug development.
Keywords: disassembly, nuclear entry, nuclear transport, virus entry, viruses
Introduction
Viruses, being the most rapidly evolving biological entities, are an amazing reflection of evolution and genetic diversity. Their genetic material is either DNA or RNA, either single stranded or double stranded. Viral genomes vary in size from thousands to million base pairs or nucleotides. To accomplish their entire life cycle, including cell entry, replication, assembly and egress, viruses usurp cellular factors and machinery. Thus viruses are attractive probes for investigating cellular processes.
During cell entry viruses need to overcome a number of barriers. The first barrier is the cell membrane. Cell entry from the external environment is usually accomplished by endocytosis. Receptors on the cell surface determine the tropism of different viruses. For the many viruses that replicate in the cell nucleus, the nuclear envelope (NE) is another major barrier.
Many viruses never enter the nucleus. These include most RNA viruses (with the exception of retroviruses and orthomyxoviruses) that carry their own RNA polymerase and replicate and assemble their genome in the cytoplasm. Some examples are the Reoviridae (rotavirus), Picornaviridae (poliovirus), Flaviviridae (dengue, west Nile virus, hepatitis C virus), Rhabdoviridae (Rabies virus) Filoviridae (Ebola virus, Marburg virus) and paramyxoviridae (measles, mumps). Three families of large DNA viruses, Poxviridae, Asfarviridae and Mimiviridae, also conduct their complete life cycle in the cytoplasm.1 These giant viruses carry a complete replication and transcription machinery, and do not need the host nuclear enzymes for propagation.1
Other viruses avoid crossing the NE by limiting their infection to dividing cells. Retroviruses, after undergoing reverse transcription and formation of the pre-integration complex in the cytoplasm, await NE disassembly during mitosis in order to reach the host chromosomes and integrate into the cellular genome. It was recently suggested that infecting papillomaviruses also await cell division to achieve nuclear entry of their DNA genomes.2 This simple strategy for circumventing the NE barrier harbors the obvious disadvantage of the need to wait for cell division. Thus, these viruses bypass the necessity to cross the nuclear membrane barrier by compromising their ability to infect non-diving cells.
This review will focus on the different mechanisms developed by viruses to enter the non-dividing nucleus. Most of these are too large to pass through the nuclear pore, the canonical gate to the cell nucleus. Throughout evolution viruses evolved various strategies to overcome the NE barrier. The various mechanisms are summarized in Figure 1.
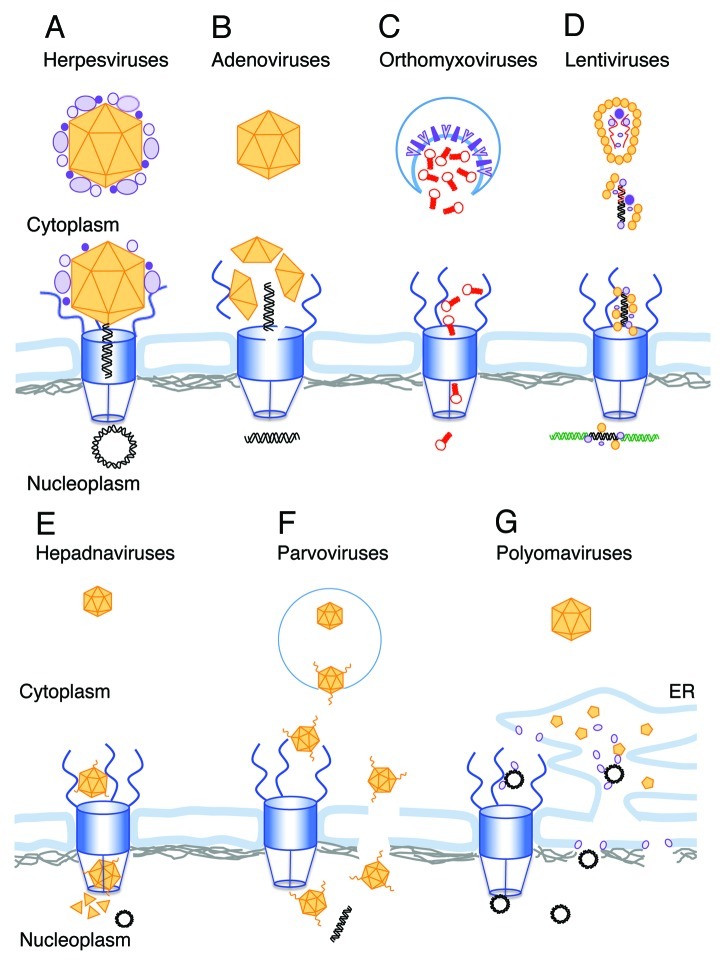
Figure 1. Schematic illustrations of the different strategies used by viral genomes for nuclear entry. The following color scheme is used through the figure: Viral DNA, black; host DNA, green; viral RNA, red; NPC, blue; Lamins, gray; viral capsid and capsid proteins, orange and other viral proteins are in different shades of purple. (A) The herpesvirus capsid arrives at the NPC with the internal tegument proteins attached. Following conformational change and opening the portal ring at the capsid vertex the DNA is ejected into the nucleus. (B) After release from the endosome the adenovirus capsid docks to the NPC, where molecular motors disrupt both the capsid and the NPC structure allowing the viral DNA to enter the nucleus. (C) Orthomyxovirus RNPs are released from the endosome into the cytoplasm after fusion of the viral envelope (enriched with viral glycoproteins) with the endosome membrane (shown as a blue line). The RNPs freely diffuse toward the NPC, where they are actively transported as karyopherin cargo into the nucleus. (D) Following uncoating in the cytoplasm, the lentivirus RNA genome is reverse-transcribed into a double stranded DNA. The PIC, containing the viral DNA and several viral proteins including CA, promotes nuclear entry by interaction with NPC proteins. After passing through the NPC the viral DNA integrates into the host chromosome. (E) Hepadnavirus capsids enter the NPC but are too big to pass intact through the basket and into the nucleus. Mature, DNA-containing capsids, disassemble in the basket, releasing the circular viral genome into the nucleoplasm. (F) Parvovirus particles enter the nucleus intact. The N-terminal domain of the minor capsid parvoviruses is extruded in the endosome, exposing phospholipase A activity that facilitates its release to the cytoplasm. Extrusion of the N-terminus also exposes four NLS domains that appear to function in nuclear entry through the NPC. An alternative model suggests direct nuclear entry from the cytoplasm through local disruptions in the NE. G. Polyomaviruses disassemble in the ER. The exposed genomes exit the ER via viroporins created by the internal capsid proteins by either of two proposed mechanism. One is directly from the ER lumen through the inner nuclear membrane, and the other is via the cytoplasm and the NPC.
Structure of the Nuclear Envelope
The NE is a continuous barrier between the cytoplasm and the nucleoplasm, interrupted only by the nuclear pores. The outer nuclear membrane (ONM) is a continuum with the ER membrane. The inner nuclear membrane (INM) is associated with the lamina at the nucleoplasmic surface. The lamina is composed of four lamin proteins that are subdivided into A- and B-types, both of which belong to the type V intermediate filament family. The lamin genes are highly conserved in metazoans throughout evolution, but are not found in plants or unicellular organisms.3 Lamin A and lamin C are alternative splice variants of the LMNA gene,4 while lamin B1 and lamin B2 are encoded by two different genes, LMNB1 and LMNB2.3 The lamin proteins exhibit extensive amino acid sequence similarity and have analogous general structure, with N-terminal globular “head,” central α-helical rod, and C-terminal “tail” domain. The tail domains of all lamins contain a nuclear localization signal and an Ig-fold motif.5 Maturation of lamins A, B1 and B2 involves a sequential series of post-translational modifications.3 The four lamin isoforms are required for the structure and integrity of the NE, suggesting physical interactions between the individual lamin networks.
Lamins are associated with various NE proteins in a lamina. Emerin is a lamin A/C associated NE protein, implicated in Emery-Dreifuss muscular dystrophy.6 Other proteins are LAP’s (lamin associated protein), which also interact with chromatin and possibly mediates mechano-transductional signals from the NE to the transcriptional machinery.7 Interestingly the cellular factor BAF (barrier-to-autointegration factor), which inhibits retroviral autointegration,8 is another essential lamina component. BAF was recently reported to be a novel epigenetic regulator, functioning through its influence on specific histone H3 modifications.9
Multiple nuclear pore complexes (NPCs), the gateway between nucleus and cytoplasm, are embedded in the NE. The relationship between NPCs and lamins is not fully understood. The NPC is a huge proteinaceous structure with a molecular mass of ~100 MDa composed of about 30 different proteins, collectively known as nucleoporins (Nups), which are conserved across eukaryotes from yeast to human. Each Nup is present at multiple copies, accommodating the 8-fold rotational symmetry of the NPC.
The NPC is composed of three rings—the cytoplasmic ring, the spoke ring and the nuclear ring, which together form a cylindrical structure containing the NPC main channel. Eight filaments are connected to both the cytoplamic and nuclear rings. On the cytoplasmic face of the NPC, the filaments extrude into the cytoplasm. These cytoplasmaic filaments are composed of Nup358 (RanBP2), Nup214 (CAN), Nup98, Nup88, hCG1, Rae1 and ALADIN. On the nuclear face of the NPC these filaments converge to create the nuclear basket of the NPC. This structure is composed of Nup153, Nup50 and TPR. A comprehensive review on the structure of the NPC was recently published by Hoelz et al.10
Many Nups contain multiple phenylalanine-glycine (FG) repeats. At the central region of the NPC, several Nups form a cohesive meshwork of filaments through hydrophobic interactions, which involve phenylalanines in FG repeats.11 The FG repeats within the central pore are hypothesized to confer selectivity to the NPC. The peripheral filamentous Nups also contain FG repeats. The main channel allows free diffusion of small molecules, whereas larger cargos (up to approximately 39 nm in diameter12) may be translocated across by an active process involving transport receptors such as importin β. Proteins that carry nuclear localization signals (NLS) are transported through the NPC by binding to the adaptor molecule importin α, which in turn binds importin β to initiate nuclear import. Other proteins carry a non-classical NLS that can directly bind to importin β. Interactions between importin β and the FG repeats are essential for nuclear import of NLS-containing cargo. Importin β coupled to the cargo protein is able to pass freely through the central meshwork in either direction. The small GTPase Ran in its GTP-bound form (RanGTP) is enriched in the nucleus, where it interacts with and dissociates the complex of cargo-importin receptors.
Virus Strategies to Overcome the NE Barrier
Herpesviruses
Herpesviridae is a large family of enveloped large DNA viruses that infect many species of mammals and birds. There are 8 known human herpesviruses distributed among the three subfamilies of the herpesviridae: α, β and γ. All members of this family share a set of 44 genes, termed the core genes, and a similar virion structure.13 The complex virion is composed of more than 90 different viral and host proteins.14,15 The large double stranded DNA genome (~125- ~250kb) is present inside an icosahedral capsid (T number 16). The capsid is surrounded by a loosely structured layer of proteins known as the tegument layer. The tegument is divided to the denser inner tegument layer that is associated with the capsid, and to the outer tegument layer. A lipid bilayer envelope containing viral glycoproteins encapsulates the tegument. Although there are small differences in the entry and replication processes among different viruses of this family, in this review we will focus on the best studied entry mechanism of the human herpes simplex virus-1 (HSV-1) as a representative of this viral family.
The viral capsid enters the infected cell by direct fusion of the viral envelope with the host cell membrane (or by endocytosis and fusion of the viral envelope with endosomal membranes). The inner tegument proteins remain associated with the capsid and interact with cellular microtubule motor proteins that transport the capsid toward the nucleus.16,17 Upon reaching the nucleus the capsid and some of the inner tegument proteins are docked to the NPC.18 Following docking to NPC, the capsid undergoes a conformational change (known as “uncoating”) creating an opening at a single vertex while the rest of the capsid remains intact.19 The DNA is released from the opening in the capsid into the NPC and is translocated to the nucleus in a process that is not fully understood. Several viral gene products were suggested to facilitate these processes. Importantly, all the HSV-1 genes described in this section are part of the 44 core genes that are present in all the Herpesviridae family.13
The interaction between the capsid and the NPC was suggested to require energy and to depend on the major tegument protein VP1/2 (encoded by the gene UL36) and the host proteins importin-β and Nup358.18,20 Binding assays showed direct interaction between the capsid-associated protein UL25 and the nucleoporins Nup214 and hCG1.21 Nup214, Nup358 and hCG1 are found on the cytoplasmic face of the nuclear pore, and are therefore accessible to the incoming capsids.
The current view of nuclear entry of the herpes genome appears to include NPC docking, uncoating (or capsid conformational change) and nuclear entry. The trigger for initiating the conformational change is yet to be identified. However, roles of several viral proteins in this process have been clarified. The function of the tegument protein VP1/2 in the process has been known for a long time, since a temperature sensitive mutation (tsB7) mapped to its gene allows binding to the nuclear membrane but prevents genome release into the nucleus at the nonpermissive temperature.22 Recently, this tsB7 mutation was characterized as a single amino acid change, 1453Y-H, in the VP1/2 protein.23 Further evidence has shown that proteolytic cleavage of VP1/2 is necessary for DNA release into the nucleus.24 The cleavage occurs only after capsid docking to the NPC, which presumably initiates the conformational change needed for VP1/2 cleavage and DNA release.24 The capsid-associated DNA-packaging protein UL25 has also been implicated in the uncoating process, as a temperature sensitive mutation (ts1249) in the UL25 gene prevents nuclear entry of viral genomes at the nonpermissive temperature.25
In a recent paper, Rode et al. were able to uncouple docking and uncoating from genome entry to the nucleus.26 The investigators found that overexpression of UL25 constructs impaired DNA nuclear entry, although EM studies of the infected cells indicated that it did not interfere with docking and uncoating. To explain their data the authors proposed that the ectopically expressed UL25 competes with the native, virus-bound UL25 for interacting with an unidentified cellular factor, an interaction that facilitates import of the herpes genome into the nucleus. Thus it appears that following Herpes simplex docking to nuclear pore, at least two viral proteins, VP1/2 and UL25, as well as host factors, participate in its uncoating and genome release to the nucleus.
Capsid assembly and DNA packaging of Herpesviridae are both structurally and functionally analogous to the same processes in double-stranded DNA bacteriophages, such as lambda and T4. Like bacteriophages, herpes packages its DNA into preformed shells and releases the DNA through a small opening in the capsid into the nucleus, while otherwise remaining essentially intact. This analogy led to the current model of herpes capsid formation and DNA release. The complex process of capsid assembly occurs in the infected nucleus and involves at least 11 viral proteins (recently reviewed in ref. 27). One of the 12 vertices of the icosahedral capsid, termed the portal vertex, is unique. This vertex contains a ring structure composed of 12 copies of UL6, the portal protein. The UL6 ring structure binds the scaffold proteins and initiates assembly of the major capsid protein to form the viral shell. In analogy to bacteriophages, the portal channel is thought to be the entry gate for DNA into the capsid.28 Since DNA release into the nucleus occurs through a single vertex, and since the last end of the genome to be packed is the first to be ejected, it was suggested that the UL6 portal is also the exit way for the viral DNA.19,29
Translocation of the viral genome through the NPC is considered to be a fast process. In vitro uncoating studies visualized by either fluorescent or electron microscopy suggested that the DNA is ejected from the capsid in a linear fashion.29 Atomic force microscopy indicated, however, that the viral genome is densely packaged inside the capsid30 and that the genome passes through the NPC as a condensed rod-like structure.31 The authors hypothesized that the structural changes that the capsid undergoes when interacting with the NPC lead to an opening at the portal vertex. They further suggested that the packaged viral DNA exerts high pressure on the capsid walls and that the internal capsid pressure pushes the viral DNA into the nucleus through the NPC.32 This mechanistic model is analogous to DNA ejection by bacteriophages into bacterial cells, reviewed by Roos et al.33
Adenoviruses
Adenoviridae is a family of nonenveloped double stranded DNA viruses that infect many vertebrate species. Currently there are 56 known types of human adenoviruses (HAdV), classified into 7 species (A–G).34 There are at least 16 common genes to this family that function in DNA replication, DNA encapsidation and virion structure formation.35 The adenovirus (Ad) genome is ~36 kb double stranded DNA. The icosahedral capsid (T number 25) is characterized by 12 fibers (one projecting from each vertex) decorating the capsid. The virion contains 12 viral proteins, 5 proteins inside the capsid (including the 3 core proteins VII, X and V that bind the viral DNA and form nucleosome-like structures) and 7 capsid structural proteins. A detailed structure of the capsid was obtained by cryo-EM and by crystal structure studies.36,37 Most of the results described in this review were obtained using the most studied adenoviruses Ad2 and Ad5, both of subtype C.
Ad infection starts by binding of the capsid to the coxsackievirus Ad receptor (CAR)38 and to the co-receptors integrins αVβ3 and αVβ5.39 The virion is internalized mainly by clathrin-mediated endocytosis.40 Next, the virion escapes the early endosome and traffics on microtubules toward the nucleus.41,42 At the NPC, the final steps of uncoating occur and the viral DNA is transported into the nucleus.
Uncoating of the Ad DNA is a multi-step process that initiates at the cell membrane and ends in the NPC. Burckhardt et al.43 have suggested that the first step of uncoating, the release of the fibers, occurs when the fibers are pulled away from the capsid by the drifting CAR receptors, while the capsid is immobilized by its binding to integrins. The release of the fibers from the capsid is not necessary for the endocytic process, but enhances escaping from the early endosomes.44,45
The escape from the endosomes is mediated by the Ad lytic factor protein VI, present inside the capsid.46 The released capsid binds directly to the dynein motor and moves along microtubules toward the microtubule-organizing center (MTOC).47,48 From the MTOC the capsid is transported to the NPC in a process that requires the nuclear export factor CRM1 (chromosome region maintenance 1). It is not clear if the CRM1 factor interacts with the Ad capsid directly, or whether it functions by exporting from the nucleus other proteins required for capsid transport to the NPC.49
The NPC has three distinct roles in viral nuclear entry: it docks the capsid, dissociates the DNA from the capsid and imports the viral DNA.50 The nucleoporin Nup214 was suggested as the docking protein.51 Recently, participation of Kinesin-1 in nuclear import of Ad DNA was characterized and a detailed model for viral DNA entry was proposed.52 The model proposes that the viral capsid docked to Nup214 binds to the kinesin-1 light-chain, while the nucleporin Nup358 associates with kinesin-1 heavy-chain. The movement of the bound Kinesin-1 motor on the microtubules disassembles the viral capsid, and at the same time increases NE permeability by dislocating Nup214, Nup358 and Nup62.52 According to this model, disruption of the NPC promotes entry of the viral DNA into the nucleus.
The highly charged viral DNA needs to be masked by proteins in order to pass through the NPC. This function seems to be fulfilled by the viral core protein VII (but not protein V), which is attached to the viral DNA during nuclear entry53,54 In addition, cellular proteins including histone H1, importin 7, importins α and β, transportin and hsp70 were suggested to participate in the nuclear import of Ad DNA.51,55,56 It appears that Ad DNA passage into the nucleus is facilitated by a large number of proteins as well as disruption of the NPC.
Orthomyxoviruses
Orthomyxoviridea is a family of enveloped segmented negative strand RNA viruses. This family includes five genera, three of which are human pathogens: influenza A, B and C. Unlike other RNA viruses, orthomyxoviruses replicate their 6–8 RNA segments inside the nucleus. Most segments encode one or more proteins, a total of 7–12 proteins per virus. Influenza A virus (IAV) is the most intensively studied member of this viral family.
Each of the eight RNA segments of IAV is coated with four viral proteins, NP (nuclear protein) and the three subunits of the viral encoded RNA-dependent RNA polymerase (PA, PB1 and PB2), forming a rod-like ribonucleoprotein (RNP). All eight RNPs are selectively packed together in a tight arrangement in each virion.57 The matrix protein (M1) surrounds the RNPs and interacts with the viral membrane that contains the viral glycoproteins.
Influenza A virion initiates its entry process by binding through its hemagglutinin (HA) to the host cell sialic acid. Influenza virus may enter the cell by clathrin-mediated endocytosis as well as by macropinocytosis.58,59 Acidification at the endocytic vesicle opens the viral M2 ion channel, leading to acidification of the virus interior and release of the RNPs from the M1 protein.60,61 The acidified endosome initiates the HA-dependent fusion step that leads to cytoplasmic release of the RNPs (reviewed in ref. 62). The RNPs freely diffuse toward the NPC, followed by their active transport into the nucleus as importin cargo.63,64 While all four RNP proteins have nuclear localization signals (NLS), the NP NLS signals are sufficient for nuclear entry of the RNPs.65 NP carries two NLSs: NLS1 at the N terminus and NLS2 in the middle of the protein.66,67 The intake of the RNP’s into the nucleus is predominantly by NLS1,68,69 and mediated by the host importin α.66
Recent RNAi screen studies performed in recent years identified several host genes that might participate in the nuclear import of the RNPs including importin β1, Nup153 and Nup98 as reviewed in ref.70 It is not known whether these factors participate in the nuclear import of the viral RNPs, or in the nuclear import of newly synthesized influenza viral proteins or in both processes.
It was recently observed that NP proteins from avian and mammalian influenza interact specifically with different isoforms of importin α, suggesting a role for NP specificity in determining the host range of the virus.71 These findings may be of utmost importance for understanding of possible new flu pandemics originating from avian influenza.
Lentiviruses
Lentiviruses are a subgroup of the retroviridae family. Retroviruses are enveloped positive strand RNA viruses that pass through a DNA precursor stage. The DNA, synthesized in the cytoplasm, enters the nucleus where it integrates into the host cell chromosome, producing progeny viruses. Most retroviruses, such as MLV (murine leukemia virus), enter the nucleus during mitosis, depending on cell proliferation for nuclear entry. Lentiviruses are unique among retroviruses in their ability to enter the nucleus of non-dividing cells. The most intensively investigated lentivirus is the human immunodeficiency virus-1, HIV-1. A set of HIV-1/MLV chimeric viruses were instrumental in revealing the mechanism of the nuclear entry process as described below.72,73
Each mature HIV particle contains two copies of the genomic RNA, complexed with the nuclear capsid (NC) protein. The RNA-protein complex is present inside a conical capsid, composed of a single capsid protein, CA. In addition the capsid contains the viral reverse transcriptase (RT). The matrix protein MA surrounds the capsid core and is connected to the viral membrane. NC, CA and MA proteins are cleavage products of the Gag polypeptide.74
The HIV-1 core is released into the cytoplasm following receptor-mediated membrane fusion. The RNA genomes are uncoated and the RNA is reverse-transcribed into a double stranded DNA by the viral RT. In the cytoplasm the HIV-1 DNA is present in complex with viral proteins, known as the preintegration complex (PIC). The PIC carries the viral genome to the nucleus via the nuclear pore, and participates in its integration into the host genome. The PIC complex includes a number of viral proteins: integrase (IN), MA, RT, NC, Vpr75-78 and CA.79 The CA protein was not considered as part of the PIC for many years; however recent studies suggest that presence of low levels of CA in PIC is essential for nuclear entry of the HIV genome, as discussed below.79,80
Early studies identified potential NLS domains in the PIC proteins IN81 MA82 and Vpr.83 These proteins were hypothesized to contribute to the PIC nuclear entry via their respective NLSs.81-85 Thus in the late 90s MA and Vpr were considered to be the main players in the PIC nuclear import. An additional element implicated in nuclear entry was a triple-stranded DNA structure in the viral genome known as the central DNA flap,86 which results from strand displacement during reverse transcription.87
Later genetic dissection of these elements raised doubts regarding their role in PIC nuclear entry. HIV mutants that were either deleted or mutated in the NLS elements, or mutants that did not form the DNA flap, retained the ability to infect non-dividing cells.88-92 Firm evidence came from experiments with an HIV-1/MLV chimeric mutant virus lacking all the putative nuclear import elements, including the three NLS domains and the DNA-flap (the IN and MA HIV genes were replaced by IN and MA of MLV; Vpr and the DNA flap were mutated). In spite of missing all those elements, the chimeric virus infected dividing and non-dividing cells at a similar level.73
Additional experiments based on HIV-1/MLV chimeric viruses have revealed that CA is a key factor in HIV nuclear entry.72 In these experiments chimeric viruses that carried the CA protein of MLV instead of the HIV CA lost the ability to infect non-dividing cells. Furthermore, substantial amounts of the CA protein remained associated with PIC after the uncoating process.79,80 Interestingly, the amount of CA that remains associated with PIC was found to be critical for nuclear entry: mutations in the CA protein that either decreased or increased uncoating rate impaired the process.79,93 These studies provided strong support to the key role of CA in nuclear entry.
NPC proteins and transport receptors that participate in HIV-1 infection were identified by high throughput siRNA and two-hybrid screens. These include Nup153, Nup214, Nup358, and transportin 3 (TNPO3).94,95 The CA protein was shown to interact with Nup153 and TNPO3. A single point mutation in CA, N74D, changed the interaction between the HIV-1 PIC and nuclear pore proteins. The mutation modified the viral requirement for specific nucleoporins, as the mutant virus shifted its dependence from NUP153 to NUP155.96 Recently, CA was found to also bind to NUP358, and CA mutations that affected NUP358 binding altered the targeting of the integration site to genomic regions sparse in transcriptional activity, thereby affecting the efficiency of replication of this RNA virus.97
CA binds to TNPO3, which is part of the importin β superfamily of the nuclear transport receptors.98 CA in PIC complex binds tRNA species that promote HIV-1 entry into the nucleus.99 A recent study has suggested that TNPO3 might play a role in displacing any CA and tRNA that remain bound to the pre-integration complex after nuclear entry, thus facilitating efficient integration.100
The current model proposes that following uncoating residual amounts of CA remain associated with PIC, promoting nuclear entry by interaction with NPC proteins. It is anticipated that further studies, which will reveal specific details of these processes, may identify potential targets for HIV drug therapy.
Hepadnaviruses
Hepadnaviruses are small, enveloped DNA viruses that propagate in the liver. Carriers of the human pathogen hepatitis B virus, HBV, are at high risk to develop cirrhosis and hepatocellular carcinoma. Human HBV studies are limited by poor cellular entry in available cell cultures. Therefore, investigators of nuclear entry bypass this first cell entry stage by using either digitonin-permeabilized cells or by direct cytoplasmic injection into Xenopus oocyes, a classical experimental system for studies on NPC fucntion. Some researchers investigate the duck hepatitis B virus as a model for the human virus.
Within the envelope, the HBV nucleocapsid is composed of a single capsid (or core) protein, which surrounds the viral genome. Most capsids are 34 nm in diameter, composed of 120 dimers of the single capsid protein in a T = 4 symmetry. The minority are assembled as 30 nm capsids composed of 90 dimers, arranged in a T = 3 geometry.101 The genome encodes for the viral polymerase, the capsid protein, the regulatory protein HBx and the three surface antigens. These DNA viruses have an amazing replication strategy, through an RNA intermediate, which is reverse-transcribed within the viral capsid.102 Furthermore, the closed, circular 3–3.4 kb DNA genome is partly double-stranded and partly single-stranded.
DNA is released into the nucleus where gaps in the DNA are filled-in by the host machinery. From the resulting covalently closed circular DNA (cccDNA), full size (3.2 Knt) pregenomic RNA, is transcribed in the nucleus by the host enzymes. The pregenomic RNA is transported to the cytoplasm and assembles into nascent viral capsids together with the virally encoded reverse transcriptase and a cellular protein kinase, tentatively identified as serine arginine protein kinase (SRPK) 1 and 2.103 The RNA containing assembled particles are immature virions. Maturation occurs when the packaged RNA is reversed transcribed inside the immature capsid into genomic DNA. Studies on duck hepatitis B showed that viral maturation is associated with phosphorylation and dephosphorylation104 at a number of sites located at the C-terminus of the core proteins.105 The phosphorylation state was found to affect the conformational state of the C-terminus of the core protein.106 Furthermore, S245 phosphorylation was suggested to play a central role in nuclear targeting and DNA release from capsids during viral infection.107 Recent in vitro studies on HBV demonstrated binding of SRPK to the C-terminus, leading the authors to propose that the kinase functions as a non-canonical chaperone, preventing premature self-assembly and packaging of nonspecific RNA.108
The cytoplasm of an HBV-infected cell contains both immature and mature particles. The mature, DNA-containing capsids may be enveloped, released from the cell and spread to initiate infection of other cells,109 or they may re-enter the nucleus for additional rounds of transcription and viral propagation.
The virus enters cells via receptor mediated endocytosis and is taken up by the endosome. Following processing in the endosomal compartment the genome-containing nucleocapsid is released into the cytoplasm.110 The capsid then travels in a microtubule dependent motion toward the nucleus.111
Nuclear entry of Hepadnaviruses is via the nuclear pore. In human HBV, an NLS sequence of the capsid protein is located at the arginine-rich C-terminal domain (CTD, aa 141–185), which, in the mature virus, is facing the interior of the capsid.112 In contrast, duck HBV does not contain an arginine-rich CTD. Nevertheless, an NLS was identified in the core protein of DHBV.113 Experiments with HBV core proteins synthesized in E. coli have suggested that phosphorylation of the internal domain of the capsid protein leads to exposure of the NLS114 by extrusion of the C-terminus through a flexible linker hinge.115 However, it is equally possible that the CTD transiently extrudes from holes in the capsid, allowing exposure of nuclear localization signals, phosphorylation sites, and dephosphorylation sites.108
It has been demonstrated that HBV capsomers bound RNA at high affinity, forming T = 4 capsids, whereas binding to DNA was poor, resulting in a mixture of non-capsid complexes.116 The authors proposed that capsids that assemble on RNA are stabilized by the negatively charged, flexible nucleic acid scaffold. However as they mature by reverse transcription they reach a metastable state, due to the double stranded DNA coiled spring.116 As opposed to the flexibility RNA, double-stranded DNA is a relatively stiff polymer that does not readily bend or circularize. The stiffness of DNA, measured experimentally as its persistence length, is ~50 nm, about twice the internal diameter of the HBV capsid, in contrast to the ~0.7 nm persistence length of ssRNA. The metastable state of mature particles, in which the mechanical force exerted by the DNA is comparable to the capsid protein-protein interactions, may contribute to extrusion of the C-terminus of the core protein,116 in addition to the phosphorylation step proposed earlier.117
Association of the exposed NLS with importins α and β mediates attachment of the capsid to the nuclear pore.117 Electron microscopy studies in Xenopus oocytes demonstrated that although the diameter of the capsid is barely smaller than the upper limit for transport through the pore,12 the karyophilic capsids enter the NPC and into the basket. Up to this stage the process seems to follow the canonical nuclear entry pathway. However, from this step on HBV has evolved a unique nuclear transport mechanism. Additional studies in digitonin-permeabilized cells demonstrated that while both mature and immature capsids dock at the NPC, the mature capsids (DNA-containing) disassemble, releasing the viral genome into the karyoplasms, while the immature capsids (RNA-containing) remain arrested in the basket.118 The different fate of mature and immature capsids after entering the nuclear pore indicates that the outcome of a nuclear import event may be regulated within the nuclear basket. This is consistent with a destabilizing effect of dsDNA.
In search for a mechanism Schmitz et al.119 have recently revealed that the selective arrest of immature capsids is controlled by Nup153, an essential protein of the nuclear basket. HBV capsids were shown to co-immunoprecipitate with Nup153 from rat liver cells as well as with recombinant Nup153 expressed in E. coli. Evidence for participation of Nup153 in HBV arrest at the nuclear basket was obtained from partial silencing of Nup153. While immature capsids are fully retained in the basket, partial silencing of Nup153 allowed a small, yet significant proportion of immature capsids, to enter the nucleoplasm. The selective arrest did not appear to depend on differential interaction of Nup153 with the two capsid types, as the protein bound both immature and mature capsids directly and specifically. Interaction occurred at more than one site, including the Nup153 importin β binding site, implying that importin β and the capsids compete for Nup153 binding. This finding suggests that in the case of HBV Nup153 is responsible for releasing importin β from the capsids, consistent with previous observation that RanGTP, which functions in dissociating importins from their cargo, is dispensable for HBV nuclear entry.114
Evidence for the requirement for disassembly in the basket was obtained from experiments with UV cross-linking of mature capsids, which prevented disassembly. Cross-linking did not interfere with capsids binding to the NPC but prevented entry into the karyoplasm.119 The authors proposed that mature capsids disintegrate in the basket into core protein homodimers, and that the numerous core proteins titrate all available Nup135 sites, allowing release of the DNA and the remaining free core proteins into the nucleus. As mentioned above, the mechanism of selective disassembly of mature capsids is presumably based on their metastable state that is caused by internal mechanical force of the coiled spring dsDNA.116
It appears that the unusual replication mechanism of HBV, which results in a mixture of mature and immature capsids in the cytoplasm, has imposed the development of a unique nuclear entry strategy that allows selective access of the viral DNA genome into the nucleoplasm, excluding the viral RNA.
Parvoviruses
Members of the Parvoviridae are among the smallest DNA viruses known. They are non-enveloped with a linear single stranded DNA genome. Their ~5 kb genome is encapsidated by a 26 nm capsid arranged in a T = 1 symmetry. The capsid is composed of 2–4 capsid proteins (depending on the specific virus) termed VP1 to VP4. These are translated from the same mRNA by different starting codons.
This broad group is divided into two subfamilies—the Parvovirinae that infects vertebrate hosts and the Densovirinae that infects arthropods. The Parvovirinae are further divided into five genera, four of which are autonomous (parvoviruses, amdoviruses, betaparvoviruses and erythroviruses), i.e., propagate in infected cells without a helper. The fifth genus is dependoviruses, which cannot complete the replication cycle in the absence of a helper virus, usually the unrelated adeno or herpes.
Parvoviruses enter cells by receptor mediated endocytosis. Due to the wide interest in adeno-associated viruses (AAV) for gene delivery applications, a number AAV strains with different cellular/tissue tropism have been recently characterized. The general theme observed is that disparate loop structures at the external surface of their capsids confer differential receptor recognition.120 Following clathrin-mediated endocytosis, parvoviruses are internalized and traffic through the endosomal/lysosomal system to the periphery of the nucleus (reviewed in refs. 121 and 122). The low endosomal pH leads to capsid conformational changes and extrusion of N-terminal sequence of the minor protein VP1, exposing viral encoded elements that are critical for further trafficking and nuclear entry.
Since parvoviruses are non-enveloped, they cannot escape from the endosomal system by membrane fusion, as enveloped viruses do. Instead, parvoviruses deploy a phospholipase A2 (PLA2) domain located at the N-terminus of VP1. Studies on the prototypic minute virus of the mouse, MVM, revealed that the PLA2 domain, which is hidden in the interior of the capsid, becomes exposed in the endosome when the N-terminus of VP1 is extruded, but remains capsid-tethered. PLA2 activity was found to be required for virus escape from the endosome into the cytoplasm.123 First, a point mutation in the PLA2 domain of the VP1 protein inactivated the phospholypase activity and at the same time also abrogated infection. Second, the mutant could be rescued by co-infection with wt MVM virus, or by co-infection with adenovirus that harbors its own endosome escape machinery. Finally, the mutant was also rescued by polyethyleneimine treatment, which caused endosomal disruption. Hence it was proposed that MVM PLA2 activity leads to ruptures in the endosomal membrane through its lypolytic activity.123 Once in the cytoplasm parvoviruses traffic to the perinucleus in a microtubuli dependent motion.124
AAV2 and MVM were found to enter the nucleus as intact virus particles, as seen by electron microscopy and by using antibodies that specifically recognize the full capsid.125-127 Proteosome inhibitors blocked nuclear entry of MVM, CPV (canine parvovirus) and BPV (bovine parvovirus), suggesting that proteosome activity participates in the process, although its role is not known.128 On the other hand, nuclear entry of AAV is enhanced by proteosome inhibition.126,129 These findings suggest that parvoviruses enter the nucleus by divergent pathways.128
It has been generally assumed that since parvoviruses are only 26 nm in diameter, they enter through the NPC. VP1 of parvoviruses contains 4 putative NLS sequences, termed basic clusters 1–4 (BC1–4) located at the N-terminus of VP1, encompassing the PLA2 domain described above.130 These sequences are normally located in the interior of the capsid and become exposed in the endosome together with the PLA2 domain. Extrusion of the N-terminus of VP1 was also shown for AAV2.125 Analyses of point mutations in MVM that abolished NLS activity of these elements indicated that they were required by the incoming MVM particle for the onset of infection.130 However since the PLA2 domain, paramount for endosomal escape, lies in the same region, the possibility that the NLS mutations affected the PLA2 activity through improper protein folding cannot be excluded.
Other studies on the autonomous parvovirus MVM suggest that it enters the nucleus through local disruptions of the NE, bypassing the nuclear pore. When the virus was injected into the cytoplasm of Xenopus oocytes, or during infection of mouse fibroblasts, it caused disruptions of about 100–200nm in the ONM that were clearly visible by electron microscopy.131,132 The disruptions were not dependent on PLA2 activity, as shown by the PLA2 inhibitor manoalide as well as by a PLA2 mutant MVM, which was directly injected into the cytoplasm of Xenopus oocytes to circumvent the need for PLA2 activity for endosomal escape.127
The disruptions were found to depend on caspase-3 activity, but not on caspase-6. Importantly, this was seen not only in Xenopus oocytes and semi-permeabilized HeLa cells, but also in infected mouse fibroblasts.127 A caveat in experiments using microinjection or permeabilized cells rather than endocytic infection is that the particles may bypass the normal endosomal pathway, keeping the N-terminus of VP1 and the NLS domains hidden, hence precluding the NPC entry pathway. The experiments suggested that caspase-3 did not participate in upstream events prior to nuclear import. In cells treated with a caspase-3 inhibitor, MVM capsids were frequently observed at the cytoplasmic side of the nuclear envelope, suggesting that cytoplasmic trafficking to the nuclear envelope was not affected by the inhibitor. Importantly, infection of mouse fibroblasts by MVM showed that treatment with a caspase-3 inhibitor reduced not only nuclear entry but also the number of infected cells, each by ~50%, linking the two processes.127 Furthermore, although caspase-3 activity remained at a basal level following the infection, the enzyme appeared to re-locate to the vicinity of the disruptions in the nuclear lamina.127 In addition, a 16 kDa lamin B fragment, consistent with caspase-3 cleavage of lamin B2, appeared in the MVM infected cells. The investigators proposed that caspase-3 cleavage of lamin B2 leads to NE deformation and lamina disruption.127 However direct passage of virus particles through the NE disruption remains to be seen.
It should be noted that the two mechanisms proposed for parvovirus entry are not mutually exclusive. Furthermore, it is possible that members of this diverse family use varied nuclear entry pathways, which also depend on the tissue of target. Future research is anticipated to resolve whether parvoviruses enter the nucleus through the nuclear pore, by disruption of the NE or by both ways.
Polyomaviruses
The polyomaviridea is a family of nonenveloped small double stranded DNA viruses. Members of the family are known human pathogens: JC virus, cause progressive multifocal leukoencephalopathy, BK virus, which leads to rejection of transplanted kidneys and the recently identified Merkel cell polyomavirus, MCPyV, suspected as an emerging pathogen in Merkel cell carcinoma.133 The related primate polyomavirus, SV40, is easiest to propagate and is usually used as a representative of the family. SV40 has been a valuable tool for molecular genetics and cell biology research for many years.
The SV40 viral capsid, surrounding the viral minichromosome, is a T = 7d icosahedral lattice ~50 nm in diameter. It is composed of three viral-encoded proteins, VP1, VP2 and VP3. VP1 forms the outer shell while VP2 and VP3 bridge between the VP1 shell and the chromatin core. VP3 is translated from an internal initiation codon of the same mRNA as VP2. VP1 monomers are tightly bound in pentamers through interdigitating β-strands,134 formed immediately following translation.135 The capsid of SV40 is built of 360 VP1 molecules, arranged as 72 pentamers. A single molecule of VP2 or VP3 is tightly anchored to each pentamer at its inward facing cavity, through a region close to the C-terminus of VP2/3.136,137 The viral genome is a circular double-stranded DNA, 5.2 kb, complexed in nucleosomes in a chromatin-like structure, the minichromosome.
Polyomaviruses are capable of infecting dividing as well as non-dividing cells. They share an unusual cell entry mechanism via binding to glycolipids and glycoproteins in caveolar/lipid raft domains at the plasma membrane.138 During the first 3–6 h of infection, the virus traffics via the endosomal pathway to the ER, where disassembly occurs at 5–6 h post-infection. The viruses utilize the cellular machinery for intracellular trafficking,139,140 and disassembly.138,140-142 Disassembly is seen as separation of the outer capsid protein, VP1, from the inner proteins VP2 and VP3 (Butin-Israeli, unpublished). The viral DNA is thought to remain attached to VP2 and VP3, through their identical DNA-binding domains (DBDs) (note however that in contrast to SV40, murine polyoma VP2 and VP3 are shorter and do not contain DBDs). The 5.2 kb viral minichromosome enters the nucleus at ~8 h post adsorption.
The internal capsid proteins of SV40 (and other members of the family including murine polyoma), VP2 and VP3, contain NLS signals. As SV40 particles were found to be karyophilic, it was initially proposed that an intact virus enters the nucleus via the canonical gate, the NPC.143 However when the size of the NPC was determined12 it became clear that the virus capsid is too large and must undergo some sort of rearrangement or disassembly before entering the nucleus. The model was modified accordingly, suggesting that the viral genome enters the nucleus accompanied by one or more of the NLS-containing capsid proteins.144
After capsid disassembly within the ER lumen, the SV40 genome associated with some viral capsid proteins exists from the ER to the cytoplasm via the ER-associated degradation (ERAD) of misfolded prtoeins pathway.142,145 Viral proteins, DNA and full capsids are present in the cytoplasm at 10–12 h post infection.146 Exit from the ER to the cytoplasm would be required for genome entry into the nucleus through the NPC. However recent studies (Butin-Israeli and Oppenheim, unpublished) demonstrated presence of VP1 aggregates in the cytoplasm starting at 6 h post infection, some of which were co-localized and co-immunoprecipated (following cross-linking) with vimentin, suggesting that at least some of those are aggresomes. These findings raise the possibility that the ERAD pathway is part of the host defense protein quality control rather than a step in the infection pathway.
SV40147 and murine polyomavirus148 appear to exit the ER via porins created by the internal capsid proteins VP2 and/or VP3. It was shown that VP2/3 integrate into the ER membrane, creating local disruption in the integrity of the membrane that enable disassembled viral particle, or the minichromosome, to exit the ER. Both groups pointed out that exit from the ER may be either to the cytosol, or directly into the nucleus via the INM, since the ER and the ONM are contiguous.
SV40 infection is very inefficient with only one in several hundred of DNA-full virions able to lead to a productive infection.149 The efficiency of infection varies between different cell types, presumably due to variations in cellular factors employed by the infecting virus.150 A recent study151 demonstrated that infection is severely obstructed by the nuclear envelope: most of the DNA of the infecting virions never enters the nucleus. Furthermore, downregulation of lamin A/C by siRNA was found to enhance infection (measured as T-antigen expression) by at least 7-fold, indicating that the infection is hindered by the INM.151 These findings are not compatible with nuclear pore entry and suggest that the viral genome is transported directly from the ER to the nucleus through the INM bypassing the cytoplasm.151 Supportive evidence for this model was provided by experiments demonstrating dramatic alterations of the INM that peaked at 6 h post infection, just prior to nuclear entry.151 Furthermore those alterations were completely inhibited by caspase-10 and -6 inhibitors. Notably, the same inhibitors also blocked the infection. The alterations included sequential dephosphorylation and re-phosphorylation of a specific lamin A/C epitope, leakage of lamin A/C to the cytoplasm and dramatic deformations of the nuclear envelope. The NE components lamin A/C, lamin B1 and NPC remain associated with the deformed nuclear envelope. The temporal correlation with nuclear entry, and the requirement of caspase-6 activity for lamina conformational changes as well as for T-antigen expression, strongly suggest that the two processes are linked and that the DNA enters the nucleus via the inner nuclear envelope.151
Is it possible that nuclear entry occurs by both pathways, via the NPC and directly through the INM? Further studies are needed to answer this question. However, since over 99% of the virions that enter a cell are non-infectious, this issue may be difficult to resolve.
Summary and Concluding Remarks
Many viruses use the nucleus for replicating their genomes and assembly. However, most are larger than the ~39 nm central pore of NPC.12 Therefore viruses have evolved a range of strategies for nuclear entry of their genomes. It is not surprising that the prevailing strategy is transport of the genome after its release from the too-large capsid. The emerging exceptions are members of the parvoviridae and polyomaviridae.
The mechanism of NPC entry of different viruses varies (Fig. 1). Lentiviruses and orthomyxoviruses uncoat in the cytoplasm and their genomes, attached to viral proteins (PIC or RNPs respectively), enter through the NPC. Polyomaviruses were suggested to use a similar mechanism following disassembly in the ER. Herpesviruses and adenoviruses also transport their genomes into the nucleus through the NPC. However these viruses uncoat only after docking at the nuclear pore and release their DNA following the required conformational changes. Hepadnaviruses and parvoviruses are sufficiently small to enter the NPC intact. However this necessitates exposure of NLS signals present at internal domains of their respective capsid protein. Both virus families evolved mechanisms for extruding those domains to facilitate NPC transport. Interestingly polyomaviruses and parvoviruses appear to be able to transverse the nuclear membrane and enter the nucleoplasm directly, bypassing the NPC.
Research on viruses that enter the nucleus through NPC has identified a number of host proteins that participate in this process. The intimate interaction of viral elements with these host factors is a reflection of their evolutionary adaptation to the host cellular machinery. An exciting possibility is that MVM and polyomaviruses, which appear to directly traverse the NE, will be used as probes for future discovery of unknown cellular processes.
Nuclear entry of viruses has been so far investigated to only a limited extent. For many virus families a single or few representatives have been studied. In view of the wide range of nuclear entry strategies, it will not be surprising if other members of the same families have evolved different nuclear entry mechanisms. For example, lentiviruses, which are part of retroviridea, have developed their own strategy for entering the nucleus of non-dividing cells.
Some viruses have developed a few nuclear entry mechanisms, probably adapting to different organisms or cell types. The different preferences of avian influenza and mammalian influenza for the importin α isoform of their specific host species is an example.71 Therefore, some of the controversies in the field may be attributed to the particular system being studied. For example, there may be a difference in the entry mechanism in cell lines as compared with primary cells, or in cells derived from the virus natural host vs. cells derived from a different species. Additional studies are anticipated to present a fuller understanding of these mechanisms and their differences from one host system to another.
Most capsid proteins carry NLS signals that allow import of the newly synthesized proteins for assembly of progeny viral particles inside the nucleus. This was thought in the past to be an indication of their role in nuclear entry. However recent studies showed that presence of an NLS signal does not necessarily implies a role of the respective protein in nuclear entry of the infecting particle. A striking example is the study of the NLS of IN, MA and Vpr proteins of lentiviruses, which were shown to be dispensable for nuclear entry. Therefore the role of NLS in nuclear entry of an infecting virus should be evaluated with caution.
Infectivity of many mammalian viruses, measured as the ratio of plaque forming units (pfu) to particle number, is very low. This inefficiency may be due to the presence of a large proportion of defective particles, or to the difficulty in passing through cellular barriers such as the nuclear envelope. These factors are not mutually exclusive, and they likely contribute to a different extent in diverse virus families. For example, Varicella Zoster virus (VZV, a herpesvirus, the causative agent of chicken-pox) was shown to produce up to 85% light particles (particles that do not contain DNA),152 suggesting that its inefficiency is due to defective, “empty” particles. On the other hand, following disassembly of SV40 in the ER, only few SV40 genomes are able to cross the nuclear membrane barrier, reaching the nucleus and initiating replication.151 Similarly, most MVM particles are unable to escape from the endosomes, therefore only few genomes enter the nucleus and initiate replication.153 In contrast, 40% of adenovirus particles bound to the cell surface were suggested to release their genomes into the nucleus.154
In addition to nuclear entry, additional factors may limit viral efficiency. Recent studies with herpesviruses and HIV have indicated that only a limited number of genomes that enter the cell serve as template for progeny viruses.155,156 Possibly, limitation of nuclear factors or constrains in nuclear architecture lead to this limitation, at least for Herpes.157 It appears that viruses compensate for their inefficient infectivity by producing a large number of progeny, many thousands, from every productively infected cell. During their cell binding and entry viruses induce a wide array of signaling networks, to facilitate their intracellular trafficking and to combat host defense response. It is possible that the excess amount of virus particles that seem to be non-infective are needed for induction of the myriad of signals required for entry and propagation of the few.
The inefficient nuclear entry imposes a serious problem in uncovering entry mechanisms. As many of the infecting particles do not lead to productive infection, it is difficult to ascertain which ones represent the genuine entry pathway. We anticipate, however, that present and future technological development, such as single molecule detection, will be instrumental in resolving these intriguing issues.
Finally, as viral entry to the nucleus seems to be a major bottleneck in the infection cycle, it is attractive to propose that antiviral drugs targeted against this step will be highly effective. Hopefully, future research in this field will open the way for development of such potential therapies.
Acknowledgments
We thank Adam Zlotnick for his valuable input, Amos Panet for helpful comments and David Kobiler for critical reading of the manuscript. Apologies are extended to colleagues whose studies were not included because of space limitation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/21979
References
- 1.Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–92. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010;2:a000547. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268:16321–6. [PubMed] [Google Scholar]
- 5.Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimi T, Butin-Israeli V, Goldman RD. The functions of the nuclear envelope in mediating the molecular crosstalk between the nucleus and the cytoplasm. Curr Opin Cell Biol. 2012;24:71–8. doi: 10.1016/j.ceb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 2002;16:533–47. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 8.Cai M, Huang Y, Zheng R, Wei SQ, Ghirlando R, Lee MS, et al. Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat Struct Biol. 1998;5:903–9. doi: 10.1038/2345. [DOI] [PubMed] [Google Scholar]
- 9.Montes de Oca R, Andreassen PR, Wilson KL. Barrier-to-Autointegration Factor influences specific histone modifications. Nucleus. 2011;2:580–90. doi: 10.4161/nucl.2.6.17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 11.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 12.Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–34. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Loret S, Guay G, Lippé R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82:8605–18. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer T, Greco TM, Enquist LW, Cristea IM. Proteomic characterization of pseudorabies virus extracellular virions. J Virol. 2011;85:6427–41. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–21. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfstein A, Nagel CH, Radtke K, Döhner K, Allan VJ, Sodeik B. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7:227–37. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 18.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20:4922–31. doi: 10.1128/MCB.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb WW, Booy FP, Brown JC. Uncoating the herpes simplex virus genome. J Mol Biol. 2007;370:633–42. doi: 10.1016/j.jmb.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copeland AM, Newcomb WW, Brown JC. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol. 2009;83:1660–8. doi: 10.1128/JVI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83:6610–23. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abaitua F, Daikoku T, Crump CM, Bolstad M, O’Hare P. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1-2 protein of herpes simplex virus. J Virol. 2011;85:2024–36. doi: 10.1128/JVI.01895-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jovasevic V, Liang L, Roizman B. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol. 2008;82:3311–9. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J Virol. 2008;82:6654–66. doi: 10.1128/JVI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rode K, Döhner K, Binz A, Glass M, Strive T, Bauerfeind R, et al. Uncoupling uncoating of herpes simplex virus genomes from their nuclear import and gene expression. J Virol. 2011;85:4271–83. doi: 10.1128/JVI.02067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baines JD. Herpes simplex virus capsid assembly and DNA packaging: a present and future antiviral drug target. Trends Microbiol. 2011;19:606–13. doi: 10.1016/j.tim.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–71. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb WW, Cockrell SK, Homa FL, Brown JC. Polarized DNA ejection from the herpesvirus capsid. J Mol Biol. 2009;392:885–94. doi: 10.1016/j.jmb.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liashkovich I, Hafezi W, Kühn JE, Oberleithner H, Kramer A, Shahin V. Exceptional mechanical and structural stability of HSV-1 unveiled with fluid atomic force microscopy. J Cell Sci. 2008;121:2287–92. doi: 10.1242/jcs.032284. [DOI] [PubMed] [Google Scholar]
- 31.Shahin V, Hafezi W, Oberleithner H, Ludwig Y, Windoffer B, Schillers H, et al. The genome of HSV-1 translocates through the nuclear pore as a condensed rod-like structure. J Cell Sci. 2006;119:23–30. doi: 10.1242/jcs.02705. [DOI] [PubMed] [Google Scholar]
- 32.Liashkovich I, Hafezi W, Kühn JM, Oberleithner H, Shahin V. Nuclear delivery mechanism of herpes simplex virus type 1 genome. J Mol Recognit. 2011;24:414–21. doi: 10.1002/jmr.1120. [DOI] [PubMed] [Google Scholar]
- 33.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJ. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci. 2007;64:1484–97. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, et al. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology. 2011;409:141–7. doi: 10.1016/j.virol.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenoviruses. J Gen Virol. 2003;84:2895–908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–43. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 A resolution. Science. 2010;329:1071–5. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–3. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 39.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–19. doi: 10.1016/0092-8674(93)90231-E. [DOI] [PubMed] [Google Scholar]
- 40.Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2003;5:451–62. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- 41.Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP, Greber UF. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–72. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–65. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 43.Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, Hemmi S, Greber UF. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe. 2011;10:105–17. doi: 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J Virol. 2000;74:7085–95. doi: 10.1128/JVI.74.15.7085-7095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog. 2010;6:e1000959. doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey CJ, Crystal RG, Leopold PL. Association of adenovirus with the microtubule organizing center. J Virol. 2003;77:13275–87. doi: 10.1128/JVI.77.24.13275-13287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bremner KH, Scherer J, Yi J, Vershinin M, Gross SP, Vallee RB. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe. 2009;6:523–35. doi: 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strunze S, Trotman LC, Boucke K, Greber UF. Nuclear targeting of adenovirus type 2 requires CRM1-mediated nuclear export. Mol Biol Cell. 2005;16:2999–3009. doi: 10.1091/mbc.E05-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greber UF, Suomalainen M, Stidwill RP, Boucke K, Ebersold MW, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat Cell Biol. 2001;3:1092–100. doi: 10.1038/ncb1201-1092. [DOI] [PubMed] [Google Scholar]
- 52.Strunze S, Engelke MF, Wang IH, Puntener D, Boucke K, Schleich S, et al. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe. 2011;10:210–23. doi: 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Xue Y, Johnson JS, Ornelles DA, Lieberman J, Engel DA. Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J Virol. 2005;79:2474–83. doi: 10.1128/JVI.79.4.2474-2483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puntener D, Engelke MF, Ruzsics Z, Strunze S, Wilhelm C, Greber UF. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J Virol. 2011;85:481–96. doi: 10.1128/JVI.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saphire AC, Guan T, Schirmer EC, Nemerow GR, Gerace L. Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J Biol Chem. 2000;275:4298–304. doi: 10.1074/jbc.275.6.4298. [DOI] [PubMed] [Google Scholar]
- 56.Hindley CE, Lawrence FJ, Matthews DA. A role for transportin in the nuclear import of adenovirus core proteins and DNA. Traffic. 2007;8:1313–22. doi: 10.1111/j.1600-0854.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, et al. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat Commun. 2012;3:639. doi: 10.1038/ncomms1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76:10455–64. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Vries E, Tscherne DM, Wienholts MJ, Cobos-Jiménez V, Scholte F, García-Sastre A, et al. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7:e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–30. doi: 10.1016/0092-8674(91)90576-K. [DOI] [PubMed] [Google Scholar]
- 61.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 63.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–44. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babcock HP, Chen C, Zhuang X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J. 2004;87:2749–58. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem. 1995;270:22701–4. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 66.Wang P, Palese P, O’Neill RE. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–6. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weber F, Kochs G, Gruber S, Haller O. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology. 1998;250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 68.Cros JF, García-Sastre A, Palese P. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic. 2005;6:205–13. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- 69.Wu WW, Sun YH, Panté N. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol J. 2007;4:49. doi: 10.1186/1743-422X-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–39. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, et al. Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat Commun. 2011;2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78:5670–8. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamashita M, Emerman M. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Adv Exp Med Biol. 2012;726:441–65. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci U S A. 1993;90:6125–9. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–5. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol. 2001;75:3626–35. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–90. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80:3712–20. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, et al. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–37. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci U S A. 1997;94:9825–30. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubel A, Spitz L, et al. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–9. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heinzinger NK, Bukinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, et al. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci U S A. 1994;91:7311–5. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, et al. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J Mol Biol. 2000;299:359–68. doi: 10.1006/jmbi.2000.3768. [DOI] [PubMed] [Google Scholar]
- 85.de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, et al. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science. 2001;294:1105–8. doi: 10.1126/science.1063957. [DOI] [PubMed] [Google Scholar]
- 86.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–85. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 87.Charneau P, Clavel F. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J Virol. 1991;65:2415–21. doi: 10.1128/jvi.65.5.2415-2421.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–9. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kootstra NA, Schuitemaker H. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology. 1999;253:170–80. doi: 10.1006/viro.1998.9482. [DOI] [PubMed] [Google Scholar]
- 90.Limón A, Nakajima N, Lu R, Ghory HZ, Engelman A. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J Virol. 2002;76:12078–86. doi: 10.1128/JVI.76.23.12078-12086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petit C, Schwartz O, Mammano F. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J Virol. 2000;74:7119–26. doi: 10.1128/JVI.74.15.7119-7126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reil H, Bukovsky AA, Gelderblom HR, Göttlinger HG. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–10. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 95.König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7:221–33. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, et al. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol. 2010;84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zaitseva L, Myers R, Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 2006;4:e332. doi: 10.1371/journal.pbio.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, Fassati A. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 2011;7:e1002194. doi: 10.1371/journal.ppat.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Crowther RA, Kiselev NA, Böttcher B, Berriman JA, Borisova GP, Ose V, et al. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–50. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 102.Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–15. doi: 10.1016/0092-8674(82)90157-X. [DOI] [PubMed] [Google Scholar]
- 103.Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J, et al. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol. 2002;76:8124–37. doi: 10.1128/JVI.76.16.8124-8137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pugh J, Zweidler A, Summers J. Characterization of the major duck hepatitis B virus core particle protein. J Virol. 1989;63:1371–6. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schlicht HJ, Bartenschlager R, Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989;63:2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu M, Summers J. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J Virol. 1994;68:2965–9. doi: 10.1128/jvi.68.5.2965-2969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kock J, Kann M, Putz G, Blum HE, Von Weizsacker F. Central role of a serine phosphorylation site within duck hepatitis B virus core protein for capsid trafficking and genome release. J Biol Chem. 2003;278:28123–9. doi: 10.1074/jbc.M300064200. [DOI] [PubMed] [Google Scholar]
- 108.Chen C, Wang JC, Zlotnick A. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 2011;7:e1002388. doi: 10.1371/journal.ppat.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70:4269–74. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stoeckl L, Funk A, Kopitzki A, Brandenburg B, Oess S, Will H, et al. Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proc Natl Acad Sci U S A. 2006;103:6730–4. doi: 10.1073/pnas.0509765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Funk A, Mhamdi M, Lin L, Will H, Sirma H. Itinerary of hepatitis B viruses: delineation of restriction points critical for infectious entry. J Virol. 2004;78:8289–300. doi: 10.1128/JVI.78.15.8289-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci U S A. 1997;94:9556–61. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mabit H, Breiner KM, Knaust A, Zachmann-Brand B, Schaller H. Signals for bidirectional nucleocytoplasmic transport in the duck hepatitis B virus capsid protein. J Virol. 2001;75:1968–77. doi: 10.1128/JVI.75.4.1968-1977.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rabe B, Vlachou A, Panté N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci U S A. 2003;100:9849–54. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Watts NR, Conway JF, Cheng N, Stahl SJ, Belnap DM, Steven AC, et al. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 2002;21:876–84. doi: 10.1093/emboj/21.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dhason MS, Wang JC, Hagan MF, Zlotnick A. Differential assembly of Hepatitis B Virus core protein on single- and double-stranded nucleic acid suggest the dsDNA-filled core is spring-loaded. Virology. 2012;430:20–9. doi: 10.1016/j.virol.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rabe B, Delaleau M, Bischof A, Foss M, Sominskaya I, Pumpens P, et al. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids. PLoS Pathog. 2009;5:e1000563. doi: 10.1371/journal.ppat.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, Hoellenriegel J, et al. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 2010;6:e1000741. doi: 10.1371/journal.ppat.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Halder S, Ng R, Agbandje-McKenna M. Parvoviruses: structure and infection. Future Virol. 2012;7:253–78. doi: 10.2217/fvl.12.12. [DOI] [Google Scholar]
- 121.Cotmore SF, Tattersall P. Parvoviral host range and cell entry mechanisms. Adv Virus Res. 2007;70:183–232. doi: 10.1016/S0065-3527(07)70005-2. [DOI] [PubMed] [Google Scholar]
- 122.Harbison CE, Chiorini JA, Parrish CR. The parvovirus capsid odyssey: from the cell surface to the nucleus. Trends Microbiol. 2008;16:208–14. doi: 10.1016/j.tim.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 123.Farr GA, Zhang LG, Tattersall P. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci U S A. 2005;102:17148–53. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vihinen-Ranta M, Yuan W, Parrish CR. Cytoplasmic trafficking of the canine parvovirus capsid and its role in infection and nuclear transport. J Virol. 2000;74:4853–9. doi: 10.1128/JVI.74.10.4853-4859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J Virol. 2006;80:11040–54. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnson JS, Samulski RJ. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol. 2009;83:2632–44. doi: 10.1128/JVI.02309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cohen S, Marr AK, Garcin P, Panté N. Nuclear envelope disruption involving host caspases plays a role in the parvovirus replication cycle. J Virol. 2011;85:4863–74. doi: 10.1128/JVI.01999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ros C, Kempf C. The ubiquitin-proteasome machinery is essential for nuclear translocation of incoming minute virus of mice. Virology. 2004;324:350–60. doi: 10.1016/j.virol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 129.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Invest. 2000;105:1573–87. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lombardo E, Ramírez JC, Garcia J, Almendral JM. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J Virol. 2002;76:7049–59. doi: 10.1128/JVI.76.14.7049-7059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cohen S, Behzad AR, Carroll JB, Panté N. Parvoviral nuclear import: bypassing the host nuclear-transport machinery. J Gen Virol. 2006;87:3209–13. doi: 10.1099/vir.0.82232-0. [DOI] [PubMed] [Google Scholar]
- 132.Cohen S, Panté N. Pushing the envelope: microinjection of Minute virus of mice into Xenopus oocytes causes damage to the nuclear envelope. J Gen Virol. 2005;86:3243–52. doi: 10.1099/vir.0.80967-0. [DOI] [PubMed] [Google Scholar]
- 133.zur Hausen H. Novel human polyomaviruses--re-emergence of a well known virus family as possible human carcinogens. Int J Cancer. 2008;123:247–50. doi: 10.1002/ijc.23620. [DOI] [PubMed] [Google Scholar]
- 134.Stehle T, Gamblin SJ, Yan Y, Harrison SC. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165–82. doi: 10.1016/S0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 135.Li PP, Nakanishi A, Clark SW, Kasamatsu H. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc Natl Acad Sci U S A. 2002;99:1353–8. doi: 10.1073/pnas.032668699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen XS, Stehle T, Harrison SC. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998;17:3233–40. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gordon-Shaag A, Ben-Nun-Shaul O, Roitman V, Yosef Y, Oppenheim A. Cellular transcription factor Sp1 recruits simian virus 40 capsid proteins to the viral packaging signal, ses. J Virol. 2002;76:5915–24. doi: 10.1128/JVI.76.12.5915-5924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tsai B, Qian M. Cellular entry of polyomaviruses. Curr Top Microbiol Immunol. 2010;343:177–94. doi: 10.1007/82_2010_38. [DOI] [PubMed] [Google Scholar]
- 139.Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 140.Butin-Israeli V, Drayman N, Oppenheim A. Simian virus 40 infection triggers a balanced network that includes apoptotic, survival, and stress pathways. J Virol. 2010;84:3431–42. doi: 10.1128/JVI.01735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J Virol. 2002;76:5156–66. doi: 10.1128/JVI.76.10.5156-5166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schelhaas M, Malmström J, Pelkmans L, Haugstetter J, Ellgaard L, Grünewald K, et al. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–29. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 143.Clever J, Yamada M, Kasamatsu H. Import of simian virus 40 virions through nuclear pore complexes. Proc Natl Acad Sci U S A. 1991;88:7333–7. doi: 10.1073/pnas.88.16.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nakanishi A, Shum D, Morioka H, Otsuka E, Kasamatsu H. Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. J Virol. 2002;76:9368–77. doi: 10.1128/JVI.76.18.9368-9377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, et al. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol. 2011;13:1305–14. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- 146.Inoue T, Tsai B. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog. 2011;7:e1002037. doi: 10.1371/journal.ppat.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Daniels R, Rusan NM, Wadsworth P, Hebert DN. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: implications for DNA translocation out of the ER. Mol Cell. 2006;24:955–66. doi: 10.1016/j.molcel.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 148.Rainey-Barger EK, Magnuson B, Tsai B. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J Virol. 2007;81:12996–3004. doi: 10.1128/JVI.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Black PH, Crawford EM, Crawford LV. The Purification of Simian Virus 40. Virology. 1964;24:381–7. doi: 10.1016/0042-6822(64)90175-8. [DOI] [PubMed] [Google Scholar]
- 150.Drayman N, Kler S. Ben-nun-Shaul O, Oppenheim A. Rapid method for SV40 titration. J Virol Methods. 2010;I164:145–7. doi: 10.1016/j.jviromet.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 151.Butin-Israeli V, Ben-Nun-Shaul O, Kopatz I, Adam SA, Shimi T, Goldman RD, et al. Simian virus 40 induces lamin A/C fluctuations and nuclear envelope deformation during cell entry. Nucleus. 2011;2:320–30. doi: 10.4161/nucl.2.4.16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Carpenter JE, Hutchinson JA, Jackson W, Grose C. Egress of light particles among filopodia on the surface of Varicella-Zoster virus-infected cells. J Virol. 2008;82:2821–35. doi: 10.1128/JVI.01821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mani B, Baltzer C, Valle N, Almendral JM, Kempf C, Ros C. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J Virol. 2006;80:1015–24. doi: 10.1128/JVI.80.2.1015-1024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Greber UF, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–86. doi: 10.1016/0092-8674(93)90382-Z. [DOI] [PubMed] [Google Scholar]
- 155.Kobiler O, Lipman Y, Therkelsen K, Daubechies I, Enquist LW. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat Commun. 2010;1:146. doi: 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Del Portillo A, Tripodi J, Najfeld V, Wodarz D, Levy DN, Chen BK. Multiploid inheritance of HIV-1 during cell-to-cell infection. J Virol. 2011;85:7169–76. doi: 10.1128/JVI.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kobiler O, Brodersen P, Taylor MP, Ludmir EB, Enquist LW. Herpesvirus replication compartments originate with single incoming viral genomes. MBio. 2011;2 doi: 10.1128/mBio.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]