Abstract
Eukaryotic cells adjust their intracellular protein complement as a mechanism to adapt to changing environmental signals. In Saccharomyces cerevisiae the hexose transporters Hxt3 and Hxt7 are expressed and function on the plasma membrane in high and low glucose abundance, respectively. By contrast, Hxt3 is endocytosed and degraded in the vacuole when cells are starved of glucose and Hxt7 in response to rapamycin treatment or when nitrogen is limiting. Yeast uses several signaling pathways, including the TORC1 and Ras/cAMP/Protein Kinase A (PKA) pathways, to adapt to nutrient changes in the environment. The multi-protein Vid30 complex (Vid30c), an E3 ubiquitin ligase required for the degradation of FBPase, assists in this adaptation process in a mechanism that is poorly understood. Here we show the endocytosis and the subsequent degradation of both Hxt3 and Hxt7, in response to different nutrient signals, is dependent on components of the Vid30c. Additionally, we define the signaling events required for the turnover of Hxt3 and Hxt7 by showing that Hxt3 turnover requires Ras2 and PKA inactivation, whereas Hxt7 turnover requires TORC1 and Ras2 inactivation. Further investigation led us to identify Rim15, a kinase that is inhibited by both the TORC1 and Ras/cAMP/PKA pathways, as a key downstream effector in signaling both turnover events. Finally, we show that the turnover of both Hxt3 and Hxt7 is dependent on the essential E3 ubiquitin ligase, Rsp5, indicating that the role of the Vid30c might be indirect of Hxt ubiquitylation.
Introduction
The Target Of Rapamycin (TOR) and Ras/cAMP/Protein Kinase A (PKA) signaling pathways enable Saccharomyces cerevisiae to respond to nutrient availability and stress [1]–[4]. The two TOR kinases, Tor1 and Tor2, are pivotal proteins in the TORC1 signaling cascade that has wide-ranging effects in the cell. Rich nutrient conditions activate TORC1 to promote cell cycle progression and protein synthesis, while preventing autophagy and regulating the expression of metabolic genes in response to nutrient availability, and inhibiting the expression of stress response genes. By contrast, TORC1 is inactivated by nutrient starvation or rapamycin treatment resulting in cell cycle arrest, a decrease in protein synthesis, the activation of autophagy, and the increased expression of stress response and nitrogen-regulated genes [1], [4]–[9]. Similarly, the Ras/cAMP/PKA pathway also antagonizes stress response and promotes cell proliferation in the absence of stress and in the presence of abundant glucose [3], [10]. Glucose limitation and cell stress inactivate this pathway leading to cell cycle arrest, the synthesis of complex carbohydrates, the activation of stress response genes, and the derepression of glucose repressed genes [3], [11], [12]. Interestingly, these two distinct pathways show a level of cross communication, as TOR signaling has been shown to converge on similar targets as the Ras/cAMP/PKA pathway [13], [14].
The activity of PKA is controlled by intracellular cAMP [15]. In the presence of glucose, the two redundant small G proteins Ras1 and Ras2 are activated via the guanine exchange factors Cdc25 and Sdc25 [16], [17]. Active Ras1/2 in turn activates the adenylyl cyclase, Cyr1, to produce cAMP [18]. The presence of cAMP activates PKA by releasing it from its inhibitory interaction with the regulatory subunit Bcy1 [15]. The activity of Ras1/2 is negatively modulated by the GTPase activating proteins Ira1 and Ira2 [19], [20], while the intracellular level of cAMP is controlled by the phosphodiesterases Pde1 and Pde2 [21], [22]. Active PKA prevents cell cycle arrest, post diauxic shift gene expression and glycogen accumulation by phosphorylating and inactivating Rim15, a kinase essential for the activation of these processes [14], [23]. Conversely, in the absence of glucose or in response to stress, the decrease in cAMP allows for Bcy1 to bind and inactivate PKA, resulting in the activation of Rim15 [13].
Hexose transporters are regulated at the transcriptional and post-translational levels to allow yeast to adapt to varying nutrient concentrations in the environment. If conditions become unfavorable for the expression of a specific transporter gene, the cell must repress its transcription and degrade the remaining transporter. This degradation occurs via endocytosis and proteolysis in the vacuole. For example, HXT7 encodes a high affinity hexose transporter and its transcription is induced by low levels of glucose or a non-fermentable carbon source and Hxt7 localizes to the plasma membrane. However, in response to glucose abundance, nitrogen starvation or rapamycin treatment HXT7 transcription is repressed and Hxt7 is degraded in the vacuole [24], [25]. By contrast, HXT3 encodes a low affinity hexose transporter that is actively expressed in glucose abundance, but repressed [26] and the gene product degraded when only a non-fermentable carbon source like ethanol is supplied [27]. Despite much research into the turnover of hexose transporters, the signaling and regulatory mechanisms that govern this process are not fully understood.
The Vid/Gid proteins play an important role in the yeast’s adaptation to different nutrient conditions. These proteins assemble into a multi-component complex termed the Vid30 complex (Vid30c) [28] that functions as an E3 ubiquitin ligase [29], [30] able to facilitate the ubiquitin-dependent degradation of FBPase and Mdh2 following the transition from gluconeogenic to glycolytic growth conditions [31], [32]. Interestingly, at least three of these proteins, Vid30, Gid2 and Vid28, are needed for the turnover of Hxt7 upon nitrogen starvation or rapamycin treatment, and the growth of several vid/gid mutants are sensitive to the presence of rapamycin in the media [25]. Also, the transcription of the VID/GID genes increases in the presence of non-fermentable carbon sources [26]. The function(s) of the Vid30c therefore seems to correlate with the presence of poor carbon and nitrogen sources.
Here we further investigate the link between the Vid30c and the regulatory mechanisms that govern hexose transporter (Hxt) turnover. We expand the known function of the Vid30c in the nitrogen starvation and rapamycin-induced turnover of Hxt7 [25] to include the glucose starvation-induced degradation of Hxt3. Additionally, we show that signaling the condition-specific turnover of both these Hxts requires inactivation of the Ras/cAMP/PKA pathway thereby activating Rim15 to facilitate the turnover process. Finally, we demonstrate that Rsp5, an essential E3 ubiquitin ligase known to directly ubiquitylate nutrient transporters [33]–[35], is critical for the endocytosis and degradation of both Hxt3 and Hxt7.
Materials and Methods
Strains and Growth Conditions
All the yeast strains used in this study are isogenic to BY4742 and listed in Table S1. The chromosomally manipulated strains used in this study were created using the PCR-based integrative transformation procedure described previously [36]. The primers used contained 75 nt homologous to the native chromosomal locus up and downstream of the target integration site and 20 nt homologous to the specific template plasmid. The template plasmids were: pFA6a-GFP(S65T)-His3MX6 for the 3′ chromosomal fusion of the 3′ end of HXT3 to GFP [36]; pYM-N35 (natMX-MET25pro) for the chromosomal fusion of the methionine-repressible MET25 promoter (MET25pro) to the 5′ end of HXT3 [37]; pYM-N4 (natMX-CUP1pro-GFP) for the chromosomal fusion of the copper-inducible CUP1pro-GFP cassette to the 5′ end of HXT7 [37]; pFA6a-hphMX6 for the replacement of VID30 with hphMX6 [38]; and pCW1 (natMX-PGK1pro) for the replacement of the native VID28 promoter with the constitutively active PGK1 promoter. Following transformation, the correct integration events were verified by PCR. It is important to note that HXT7 is a duplicated gene in the yeast genome with HXT6 being its counterpart. The PCR confirmation of HXT7 tagged strains therefore involved the use of an upstream primer in the upstream region of HXT7 that is unique to HXT7. BYtor1-1 was generated as previously described [39]. The proper point mutation was confirmed by sequencing.
YCp50, YCp50-RAS2 and YCp50-RAS2VAL19 [40] were transformed into the indicated strains to determine the effects of the constitutively active RASVAL19 allele on Hxt turnover. Yeast strains used to monitor Hxt3-GFP localization and degradation were pre-cultured in synthetic complete media [0.17% Yeast Nitrogen Base (YNB) without amino acids and ammonium sulfate, 2% glucose, 0.5% ammonium sulfate and the CSM amino acid pool (minus uracil 0.77 g/L, or minus leucine 0.69 g/L, or minus methionine 0.75 g/L, MP Biomedicals)] to generate biomass. Cells were washed and transferred to synthetic media (0.17% YNB without amino acids and ammonium sulfate) containing 2.5% glucose and 0.5% ammonium sulfate. Amino acids were added to complement auxotrophic requirements. Following a three hour incubation to stimulate HXT3 expression, cells were imaged or harvested for protein extraction (time zero). The remaining culture was harvested, washed and transferred to synthetic media (0.17% YNB without amino acids and ammonium sulfate) containing 2% ethanol and 0.5% ammonium sulfate. The 2% ethanol media was used to provide conditions in which glucose repression was alleviated; we will here after refer to its effect as “glucose starvation.” Amino acids were added to complement auxotrophic requirements and/or suppress expression from the MET25 promoter. Samples were subsequently collected at the indicated times and used for fluorescence microscopy or protein extraction.
Yeast strains used to monitor GFP-Hxt7 localization and degradation were cultured as described in Snowdon et al. (2008) with the following exceptions: (1) the four hour pre-shift incubation was performed in raffinose with ammonium media containing 100 µM CuSO4 to stimulate GFP-HXT7 expression from the CUP1 promoter; (2) Cell samples were collected (time zero), and the remaining cells were washed twice with sterile water and resuspended in fresh raffinose with ammonium media devoid of CuSO4 followed by rapamycin treatment.
Fluorescence Microscopy
The monitoring of the subcellular localizations of the Hxt-GFP fusion proteins were performed by preparing slides directly from the indicated cell cultures followed by immediate analysis using the 100× objective lens of a Nikon Eclipse E600 microscope. Images were recorded using a Coolsnapfx monochrome CCD digital camera (Roper Scientific) and processed using Metamorph (Universal Imaging, Version 5.0).
Protein Extraction and Western Blotting
Harvested cells were resuspended in lysis buffer (1% NP40, 0.25% deoxycholate, 150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 1 mM PMSF; plus protease inhibitor cocktail tablets, Roche), added to 0.3 g glass beads and vortexed for two minutes. Lysates were centrifuged at 5,000 rpm for 3 minutes to remove cell debris. Supernatants were collected and the protein concentrations determined using the DC Protein Assay (Biorad) according to the manufacturer’s recommendations. Equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes. Mouse anti-GFP (Roche) and rabbit anti-Aldehyde Dehydrogenase (ADH) (200–4144, Rockland) were used as primary antibodies. ADH was used as the internal control to confirm equal amounts of protein in each lane as previously described [25], [41]. Horse radish peroxidase conjugates of donkey anti-mouse and donkey anti-rabbit immunoglobulin G (GE) were used as secondary antibodies. The ECL Detection kit (GE) was used to detect the secondary antibody according to the manufacturer’s recommendations. Membranes were exposed to autoradiography film for visualization.
Results
Vid30c is Needed for the Ethanol-induced Regulation of Hxt3
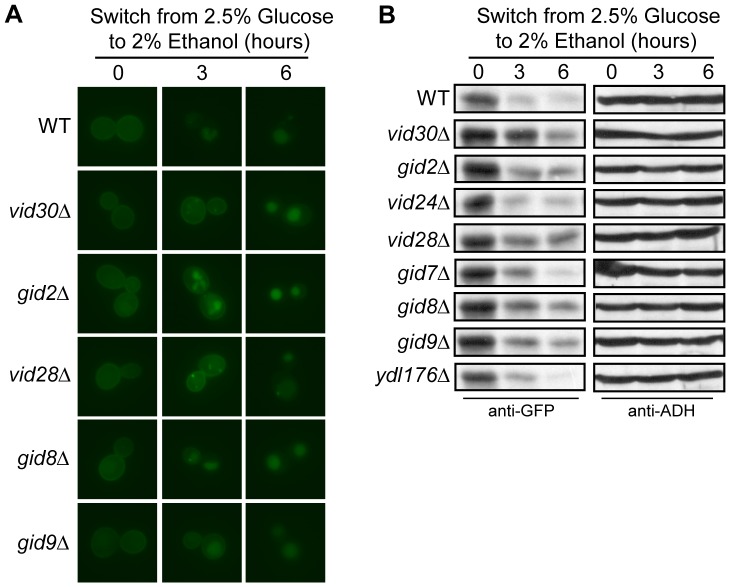
During the adaptation to the absence of glucose in the environment, yeast represses the transcription of HXT3 [26] and targets Hxt3 for endocytosis and ultimately degradation in the vacuole [27]. Since the Vid30c is needed for the nitrogen starvation-induced internalization and degradation of Hxt7 [25] we reasoned that this complex might also participate in the turnover of Hxt3 during glucose starvation, i.e. when glucose is replaced with ethanol as the sole carbon source. Wild type and individual Vid30c component mutant cells carrying HXT3-GFP were grown in glucose media to activate the expression of HXT3, and Hxt3-GFP was monitored upon a switch to ethanol as the sole carbon source. In glucose (time zero) HXT3 transcription was activated and Hxt3-GFP was visible on the plasma membrane in all the strains tested (Figure S1A and Figure 1A). In the wild type strain Hxt3-GFP was internalized with Hxt3-GFP faintly visible on the plasma membrane 3 hours after the switch to ethanol media and almost completely degraded after 6 hours. By contrast, when monitoring the mutant strains we observed delayed internalization and degradation of Hxt3-GFP in the vid30Δ, gid2Δ, vid28Δ, gid8Δ and gid9Δ mutants as Hxt3-GFP displayed a delayed internalization from the plasma membrane (Figure 1A) or was clearly more abundant (Figure 1B) than in the wild type strain following the shift to ethanol media. The turnover of Hxt3-GFP in the vid24Δ, gid7Δ and ydl176Δ mutants was similar to that in the wild type (Figure 1B). Furthermore, our previous findings suggested that Vid28 and Vid30, the proposed core components of the Vid30c [28], had partially overlapping functions since the vid28Δvid30Δ double mutant delayed Hxt7 turnover more efficiently than either of the respective single mutants [25]. Consistently, the glucose starvation-induced internalization and degradation of Hxt3-GFP in the vid28Δvid30Δ strain was severely delayed in comparison to the respective single mutants (Figures 1A and 2A). These results were confirmed by western analyses (Figures 1B and 2B). In combination these data implicate the Vid30c in the turnover of two different Hxts, Hxt3 and Hxt7 [25], in response to different nutrient stimuli.
Figure 1. Components of the Vid30c are required for Hxt3 turnover.
BY4742 (WT) and the indicated Vid30c mutants expressing HXT3-GFP were cultured in glucose media as described in the methods (time 0). Following a switch to ethanol media, samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
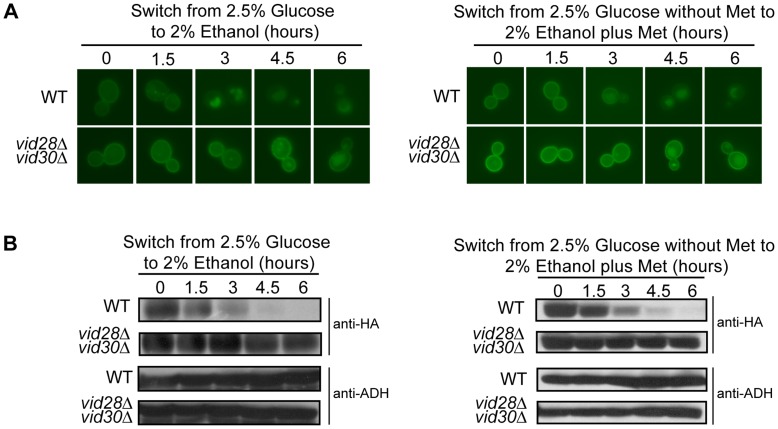
Figure 2. The combined deletion of VID28 and VID30 has an increased effect on Hxt3 turnover, which is maintained when the native promoter is exchanged for the MET25 promoter.
BY4742 (WT) and vid28Δvid30Δ expressing HXT3-GFP from either the native promoter or the MET25 promoter were cultured in glucose media (left) or glucose media minus methionine (right) as described in the methods (time 0). Following a switch to ethanol media (left) or ethanol media plus methionine (right), samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
The delayed protein turnover observed in the vid/gid mutants could be due to a lack of HXT3-GFP transcriptional repression after the shift from glucose to ethanol as the sole carbon source. When analyzing these strains for the transcriptional repression of HXT3-GFP we noticed slight, but reproducibly higher levels of HXT3-GFP mRNA in the vid30Δ, gid2Δ, vid24Δ, vid28Δ, gid8Δ and vid28Δvid30Δ strains compared to the wild type after 3 hours in ethanol media (Figs. S1A and S1B). These findings suggest that the Vid30c may also have a minor impact on the efficient repression of HXT3 transcription, or potentially mRNA stability, in the absence of glucose.
To separate the glucose starvation-mediated transcriptional repression of HXT3 from Hxt3 turnover, we tested Hxt3 turnover with HXT3-GFP expression controlled by the methionine repressible MET25 promoter [37] in the wild type and vid28Δvid30Δ strains. Cells were grown to exponential phase in glucose media devoid of methionine to induce HXT3-GFP expression, washed to remove glucose, and transferred to ethanol media containing methionine to repress HXT3-GFP transcription. We confirmed the severely delayed glucose starvation-induced internalization and degradation of Hxt3 in the vid28Δvid30Δ double mutant were similar regardless of HXT3-GFP being expressed from the native HXT3 or MET25 promoter (Figures 2A and 2B; right panels). The Vid30c is therefore specifically involved in the turnover of Hxt3, independent of transcriptional regulation.
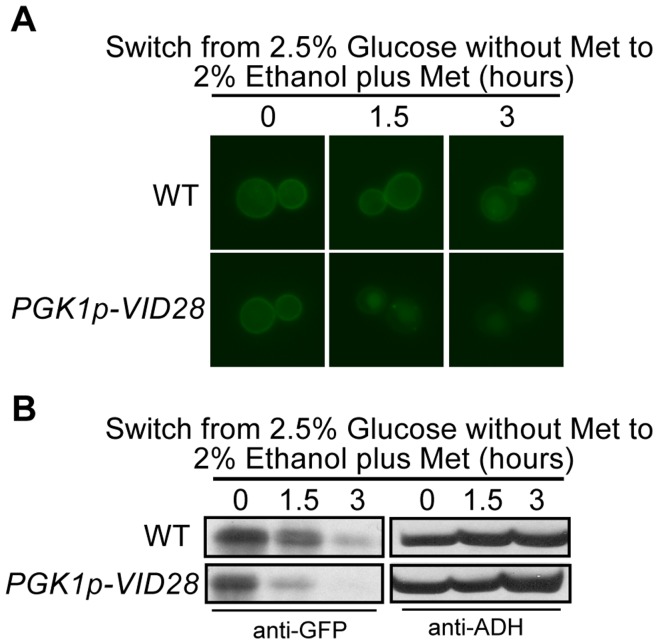
The absence of VID28 had the most severe delay of all the single vid/gid mutants on Hxt3 internalization and degradation (Figure 1). This prompted us to test the impact of the overexpression of VID28 on Hxt3-GFP turnover. The native promoter of VID28 was replaced with the constitutively active PGK1 promoter in the MET25pro-HXT3-GFP strain. The overexpression of VID28 showed a clear accelerated internalization and degradation of Hxt3-GFP following a shift from glucose to ethanol (Figures 3A and 3B). The localization of Hxt3-GFP to compartments of the endocytic pathway appears earlier and the Hxt3-GFP levels decrease quicker in the PGK1pro-VID28 strain than in the wild type strain. Thus, the increased expression of VID28 antagonizes the stability of Hxt3 in the plasma membrane. In combination these results confirm the participation of the Vid30c in the regulation of gene expression and protein turnover of Hxt3 as the cell adapts to ethanol as the sole carbon source.
Figure 3. Overexpression of VID28 accelerates Hxt3 turnover.
BY4742 (WT) and PGK1pro-VID28 expressing MET25pro-HXT3-GFP were cultured in glucose media minus methionine as described in the methods (time 0). Following washing and a switch to ethanol media plus methionine, samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
Active Ras/cAMP/PKA Prevents Ethanol-induced Hxt3 Turnover
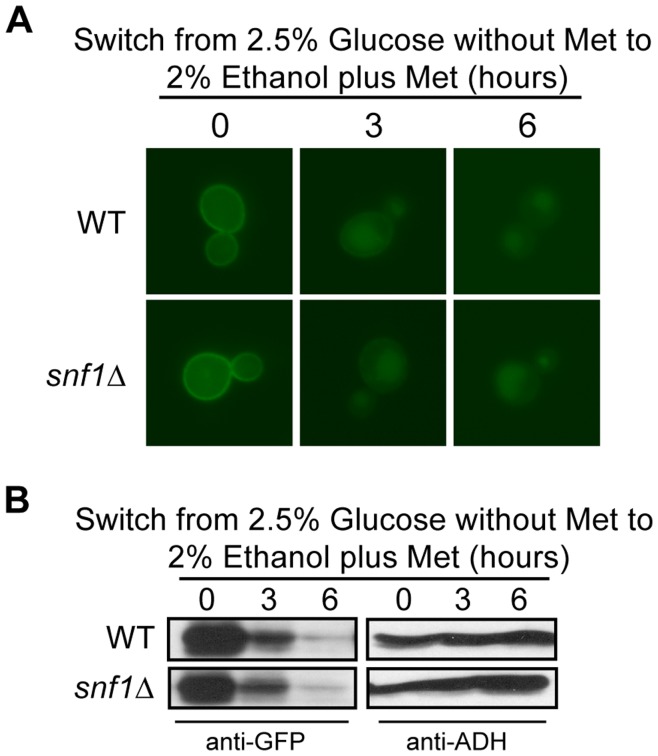
Several signaling pathways enable yeast cells to respond to glucose availability. Snf1 is a central kinase known for its roles in the transcriptional activation of glucose-repressed genes, and the turnover of several target proteins during the adaptation to non-fermentable carbon sources in a glycolytic to gluconeogenic shift [42], [43]. The degradation of Hxt3 upon a switch from abundant glucose to ethanol media [27], led us to investigate the role of Snf1 in this event. Surprisingly, the wild type and snf1Δ mutant strains showed similar patterns of protein internalization and degradation (Figures 4A and 4B). These data indicate that Snf1 is not needed for the internalization and degradation of Hxt3-GFP in response to ethanol as a sole carbon source.
Figure 4. The function of Snf1 is not required for Hxt3 turnover.
BY4742 (WT) and snf1Δ expressing MET25pro-HXT3-GFP were cultured in glucose media minus methionine as described in the methods (time 0). Following a switch to ethanol media plus methionine, samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
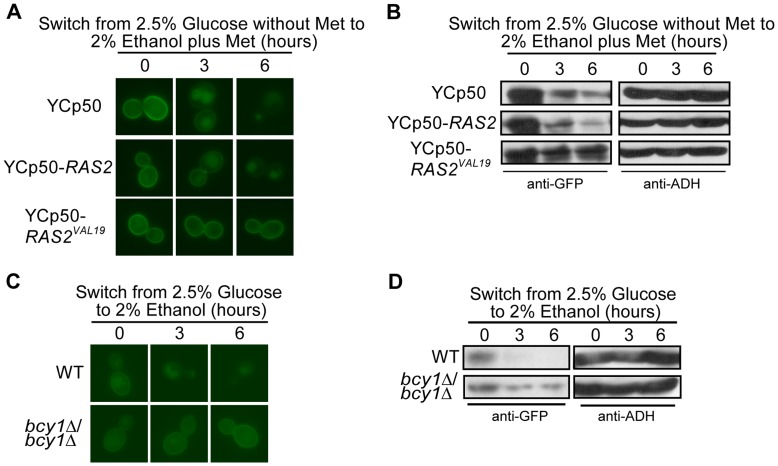
The Ras/cAMP/PKA signaling pathway plays a major role in controlling the response of yeast to glucose in the environment [44]. The activation of Ras1/2 occurs in response to abundant glucose in the environment and stimulates the adenylate cyclase Cyr1 to produce cAMP, which in turn activates PKA [18]. A constitutively active allele of RAS2 (RAS2VAL19) renders PKA constitutively active [45]. We hypothesized that the inactivation of PKA is needed for the turnover of Hxt3-GFP and tested if constitutively active Ras2Val19 would impact the turnover of Hxt3-GFP in a shift to ethanol. Using the MET25pro-HXT3-GFP strain, our fluorescence microscopy and western analysis showed that the native RAS2 allele supported the internalization of Hxt3-GFP at 3 hours and almost complete degradation in the vacuole 6 hours after an ethanol shift, while Ras2Val19 stabilized Hxt3 in the plasma membrane even after 6 hours in ethanol media with little, if any, internalization and degradation visible throughout the entire time course (Figures 5A and 5B). Active Ras2 therefore prevents the turnover of Hxt3.
Figure 5. The turnover of Hxt3 requires inactivation of the Ras2/cAMP/PKA pathway.
BY4742 expressing MET25p-HXT3-GFP was transformed with YCp50, YCp50-RAS2 and YCp50-RAS2VAL19. Cells were cultured in glucose media minus methionine as described in the methods (time 0). Following a switch to ethanol media plus methionine, samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control. BY4743 (WT) and bcy1Δ/bcy1Δ strains expressing HXT3-GFP were cultured in glucose media as described in the methods (time 0). Following a switch to ethanol media, samples were collected at the indicated times and analyzed by (C) fluorescence microscopy, and (D) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
Bcy1 binds and inactivates PKA in the absence of glucose [15]. Consequently, PKA is constitutively active in the absence of BCY1 [46]. We analyzed the turnover of Hxt3 in the bcy1Δ/bcy1Δ mutant and, similar to the observations for the constitutively active Ras2Val19, no internalization or degradation of Hxt3 was observed in the bcy1Δ/bcy1Δ mutant (Figures 5C and 5D). Collectively these results demonstrate that the inactivation of Ras and PKA is integral to the signaling of Hxt3 turnover.
Tor1 and Ras2, but not Npr1, Function in the Rapamycin-induced Turnover of Hxt7
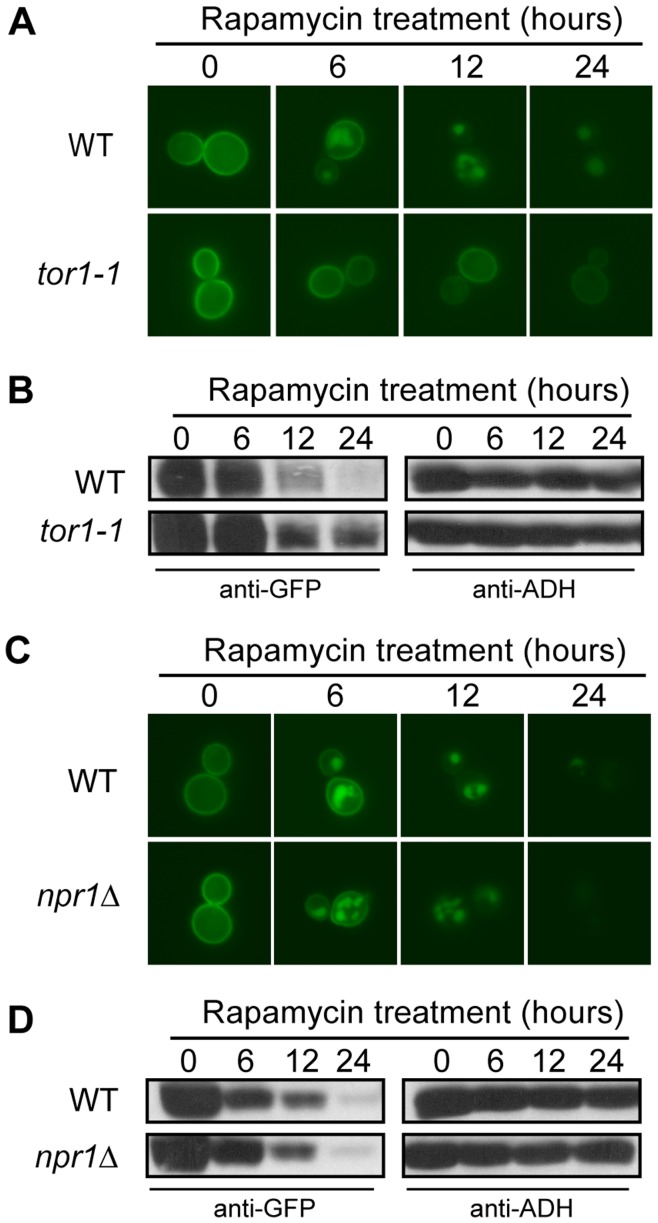
The nitrogen starvation and rapamycin-induced turnover of amino acid permeases is known to be controlled by the Npr1 and Tor1 kinases in the TORC1 pathway [47], [48]. We used the rapamycin-insensitive tor1-1 strain, carrying the CUP1pro-GFP-HXT7 allele, to further analyze the previously reported rapamycin-induced degradation of Hxt7 [25]. The CUP1pro-GFP-HXT7 allele replaces the native HXT7 promoter with the copper inducible CUP1 promoter to eliminate the impact of TOR signaling on HXT7 transcription [25]. Our results indicate that the tor1-1 allele impairs Hxt7 internalization and degradation (Figures 6A and 6B). We next tested if Hxt7 turnover was controlled by Npr1 and found the internalization and degradation profiles in response to rapamycin treatment to be similar in the wild type and npr1Δ mutant strains (Figures 6C and 6D). Collectively, these results suggest that the rapamycin-induced degradation of Hxt7 is TORC1-regulated, but in a manner independent of Npr1.
Figure 6. The rapamycin-induced turnover of Hxt7 signals through Tor1, but is Npr1-independent.
(A) BY4742 and tor1-1 expressing CUP1pro-GFP-HXT7 were cultured in raffinose media plus CuSO4 as described in the methods (time 0). After harvesting and washing, the cells were resuspended in raffinose media devoid of CuSO4 and treated with rapamycin. Samples were collected at the indicated times and analyzed by (A) fluorescence microscopy and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control. BY4742 and npr1Δ strains expressing CUP1pro-GFP-HXT7 were cultured and treated as outlined above. Samples were collected at the indicated times and analyzed by (C) fluorescence microscopy, and (D) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
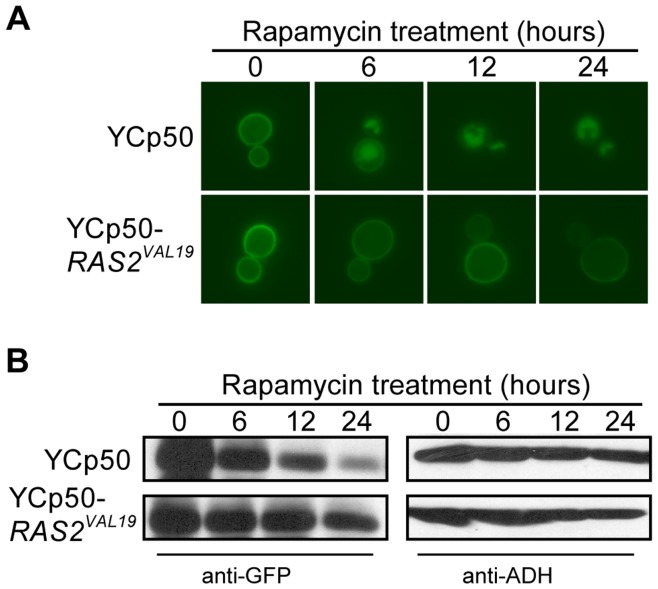
Several studies have shown close interactions between the TOR and Ras/cAMP/PKA pathways ranging from TOR and Ras/cAMP/PKA converging as separate pathways on the same molecular target [14], TOR functioning to control PKA as a downstream effector [49] to TOR and PKA having antagonistic effects in the cell [50]. Since Hxt3 turnover is controlled by Ras/cAMP/PKA (Figure 5) and TOR controls rapamycin-induced Hxt7 turnover in a mechanism independent of Npr1 (Figure 6), we tested the involvement of the Ras/cAMP/PKA pathway in rapamycin-induced Hxt7 turnover. While cells expressing native RAS2 displayed normal rapamycin-induced turnover of GFP-Hxt7, the protein clearly remained in the plasma membrane with limited if any internalization following rapamycin treatment of cells expressing constitutively active RAS2VAL19 (Fig. 7A). Higher levels of GFP-Hxt7 were also detected by western analysis in RAS2VAL19 strains post rapamycin treatment (Figure 7B). These observations suggest that, like glucose starvation-induced Hxt3 turnover, the rapamycin-induced internalization and degradation of Hxt7 is dependent on the inactivation of Ras2.
Figure 7. The rapamycin-induced turnover of Hxt7 requires inactivation of Ras2.
BY4742 expressing CUP1pro-HXT7-GFP was transformed with YCp50 and YCp50-RAS2VAL19. Cells were cultured in raffinose media plus CuSO4 as described in the methods (time 0). After harvesting and washing, the cells were resuspended in raffinose media and treated with rapamycin. Samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
Rim15 Function is Required for the Turnover of Hxt3 and Hxt7
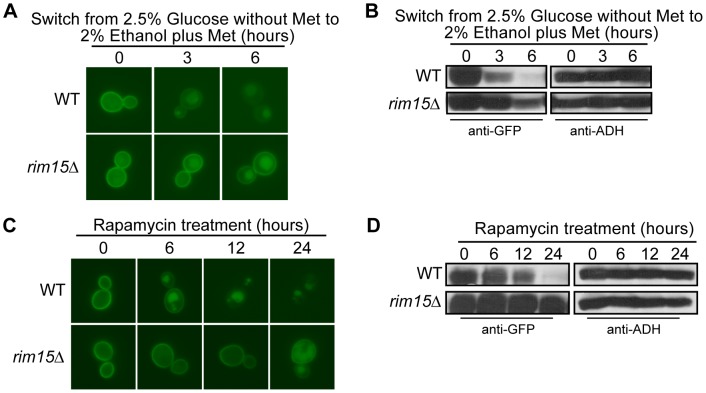
PKA and TOR control the activity of several downstream effectors in the presence of abundant glucose and nitrogen, respectively; a common downstream effector of both pathways is the Rim15 kinase [14], [51]. We investigated the potential role of Rim15 in the turnover of Hxt3 and Hxt7. While the levels of Hxt3-GFP and its localization to the plasma membrane were comparable in the wild type and rim15Δ mutant grown in glucose (time zero), the internalization of Hxt3-GFP in rim15Δ cells was markedly delayed when the cells were shifted to ethanol; the protein was still visible in the plasma membrane and abundantly present after 6 hours in ethanol (Figures 8A and 8B). Similarly, GFP-Hxt7 was expressed at similar levels and localized to the plasma membrane in the wild type and rim15Δ mutant strains grown in raffinose, but the rim15Δ mutant showed a clear delay in internalization and degradation when the cells were treated with rapamycin (Figures 8C and 8D). In combination these results demonstrate that active Rim15 is needed for glucose starvation and rapamycin-induced internalization and degradation of Hxt3 and Hxt7, respectively.
Figure 8. The turnover of both Hxt3 and Hxt7 is dependent on Rim15.
BY4742 (WT) and rim15Δ expressing MET25pro-HXT3-GFP were cultured in glucose media minus methionine as described in the methods (time 0). Following a switch to ethanol media plus methionine, samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control. BY4742 and rim15Δ expressing CUP1pro-GFP-HXT7 were cultured in raffinose media plus CuSO4 as described in the methods (time 0). After harvesting and washing, the cells were resuspended in raffinose media devoid of CuSO4 and treated with rapamycin. Samples were collected at the indicated times and analyzed by (C) fluorescence microscopy, and (B) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
Turnover of Hxt3 is Dependent on Rsp5 and Art8
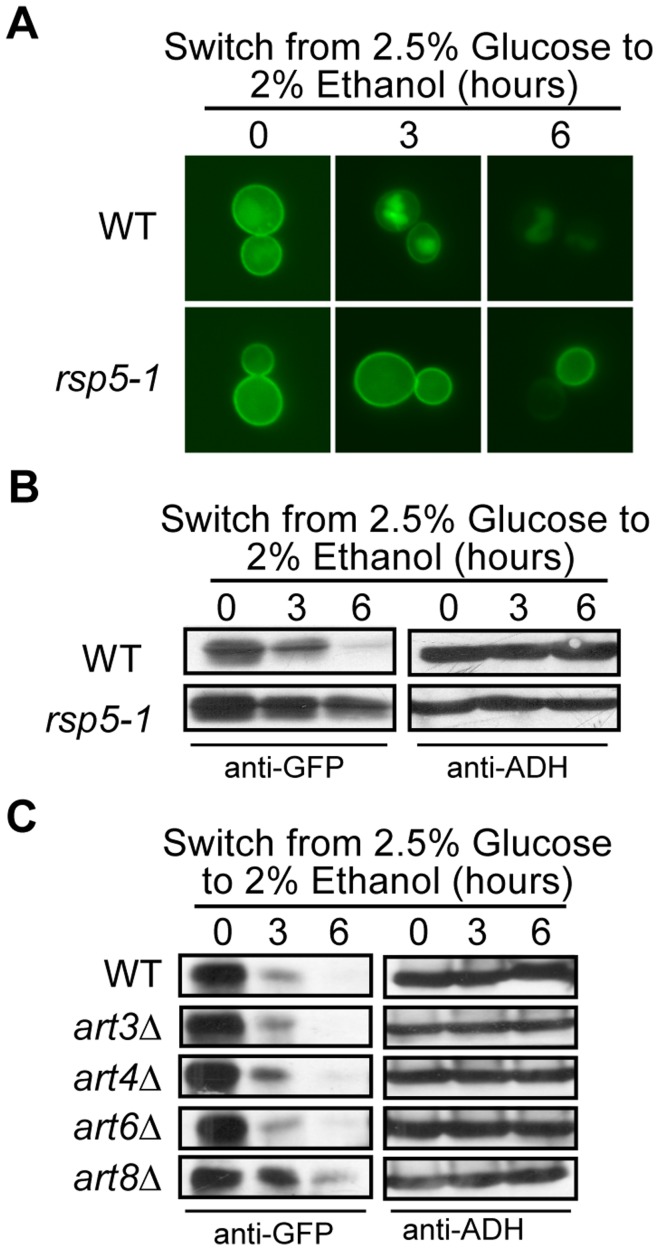
The endocytosis of several amino acid permeases and hexose transporters is dependent on ubiquitylation by the E3 ubiquitin ligase Rsp5 [24], [33], [52], [53]. Members of the Art family of arrestin-like proteins recruit Rsp5 to nutrient transporters targeted for endocytosis [54], [55]. It is not known if Rsp5 or which of the Arts are needed for the internalization and subsequent degradation of Hxt3 and Hxt7. We analyzed the glucose starvation-induced internalization and degradation of Hxt3-GFP in the rsp5-1ts mutant. As this strain dies with prolonged exposure to ethanol at 37°C, we used 30°C as the non-permissive temperature. It was clear that Hxt3-GFP remained on the plasma membrane with no discernible internalization occurring even after 6 hours in ethanol (Figure 9A). The lack of protein degradation was also confirmed by western (Figure 9B). These results confirm Rsp5 as a major E3 ubiquitin ligase needed for the turnover of Hxt3. The glucose-induced turnover of Hxt7 is known to be dependent on Rsp5 [24], [56]. Similarly, we found that the rapamycin-induced endocytosis and subsequent degradation of Hxt7 is also dependent on Rsp5 (Figure S2A).
Figure 9. The turnover of Hxt3 is dependent on Rsp5 and Art8.
(A and B) BY4742 (WT) and rsp5-1, and (C) BY4742 (WT), art3Δ, art4Δ, art6Δ and art8Δ expressing HXT3-GFP were cultured in glucose media as described in the methods (time 0). Following a switch to ethanol media, samples were collected at the indicated times and analyzed by (A) fluorescence microscopy, and (B and C) Western analysis with anti-GFP antibodies. Identical blots were also probed with anti-ADH antibody as a loading control.
We screened four arrestin-like proteins, Art3, Art4, Art6, and Art8 for its potential involvement in Hxt3 and Hxt7 endocytosis and found that only the art8Δ mutant delayed Hxt3 turnover (Figure 9C and Figure S2B). The endocytosis of Hxt3-GFP was still observed in the absence of ART8, but to a much lesser extent than in the wild type strain, suggesting that other Rsp5 adaptor proteins, in addition to Art8, might be needed for the endocytosis of Hxt3. None of these art mutants impacted Hxt7 turnover (Figure S2C).
Discussion
The Vid30c is needed for the adaptation of yeast to glucose replenishment following growth with gluconeogenic carbon sources (such as ethanol or acetate) as it functions as an E3 ubiquitin ligase responsible for the ubiquitylation and subsequent degradation of FBPase and Mdh2 when these enzymes are no longer needed [30], [32], [57]. Here we show components of the Vid30c also play an important role in the adaptation to gluconeogenic growth conditions following growth with glucose as the sole carbon source. HXT3 is actively expressed and Hxt3 localizes to the plasma membrane when glucose is abundant, but HXT3 transcription is repressed [26] and the protein is endocytosed and degraded when ethanol is the sole carbon source [27]. This latter adaptation depends on the Vid30c, more specifically the proposed core components Vid28, Vid30 and Gid8 [28] and the two RING finger proteins Gid2 and Gid9 [29], [30], as the transcriptional repression of HXT3 and more noticeably the turnover of Hxt3 are delayed in the absence of these VID/GID genes (Figure S1; Figures 1 and 2). Components of the Vid30c are also needed for the nitrogen starvation-induced degradation of Hxt7 [25] and nitrogen-regulated gene expression [58]. Several components of the Vid30c therefore do not function solely in the adaptation of yeast to glucose replenishment, but are important for its adaptation to a range of nutrient conditions, indicating a more central role for this complex in the nutrient adaptation of eukaryotic cells.
Several signaling pathways enable the molecular response to the presence or absence of nutrient abundance in the environment. The Ras/cAMP/PKA pathway facilitates the cellular response and proliferation when glucose is abundant [59]–[61]. Similarly, the TORC1 pathway is active and supports cell cycle progression in rich nutrient, including nitrogen, conditions [4], [62], [63]. By contrast, the Snf1 pathway is activated when glucose is depleted and the cell experiences gluconeogenic growth conditions [42], [43], [61]. Through the manipulation of genes required for glucose and nitrogen signaling, we were able to delineate the role of each in the regulatory events required for the degradation of both Hxt3 and Hxt7. Since Hxt3 is endocytosed and degraded in gluconeogenic growth conditions, it was surprising to find that Snf1 did not have a role in this process. Likewise, Npr1 is a TORC1-controlled kinase known to be involved in the endocytosis and subsequent degradation of several amino acid permeases in a nitrogen-dependent manner [33], [52], [53]. We confirm that the previously reported rapamycin-induced degradation of Hxt7 [25] is dependent on Tor1, but surprisingly Npr1 is not involved in this process. Since the rapamycin-insensitive tor1-1 allele prevented the endocytosis and degradation of Hxt7 in response to rapamycin treatment and the activation of the Ras/cAMP/PKA pathway is known to suppress Tor deficiencies [13], we hypothesized that active PKA would similarly prevent the turnover of Hxt3 and Hxt7. The predominant retention of Hxt3-GFP in the plasma membrane in ethanol when PKA is constitutively active in cells either expressing Ras2Val19 or lacking BCY1 supports this hypothesis. Similarly, the rapamycin-induced internalization of Hxt7 is largely prevented in cells expressing Ras2Val19. Furthermore, in rich nutrient conditions in the absence of stress, both TORC1 and PKA prevent cell cycle arrest by inactivating Rim15, the kinase that promotes entry into G0 [14]. Inactivation of either TORC1 with rapamycin or PKA by growth with gluconeogenic carbon sources, results in the activation of Rim15 [14], [51], [64]. Our data show that active Rim15 is needed for the turnover of Hxt3 in ethanol and Hxt7 in response to rapamycin treatment. Collectively, our observations conclude that the Ras/cAMP/PKA pathway needs to be inactivated to enable the turnover of Hxt3 and Hxt7 in response to ethanol and rapamycin treatment. To our knowledge this is the first report of Rim15 participating in nutrient-regulated protein turnover. It is clear that Rim15 has a partial role facilitating Hxt3 and Hxt7 turnover. Other kinases could therefore potentially participate in signaling these turnover events. To this end, the Hal4 and Hal5 kinases are related to Npr1 and have recently been shown to participate in nutrient transporter turnover [65], [66]. However, unlike Npr1, the Hal4 and Hal5 kinases do not seem to function in response to nitrogen starvation or rapamycin treatment [65]. Nonetheless, it would be of interest to identify which kinases, in addition to Rim15, participate in the condition-specific turnover of Hxt3 and Hxt7.
Rsp5 is an essential E3 ubiquitin ligase responsible for the ubiquitylation and subsequent endocytosis of nutrient transporters [24], [33], [34], [52]. An assortment of arrestin-like proteins can function as adaptors for recruiting Rsp5 to nutrient transporters targeted for degradation [54], [55]. Here we show that Hxt3 endocytosis in response to glucose starvation is dependent on Rsp5 and Art8, confirming Rsp5 and the arrestin-like adaptors as major players needed for the endocytosis of Hxt3. The glucose-induced degradation of Hxt7 is known to be dependent on Rsp5 [24], [56], and here we confirm that the rapamycin-induced degradation is as well. Interestingly, Art8 was not needed for the endocytosis of Hxt7, suggesting that specificity exists between the Art adaptors and its specific hexose transporter target proteins. Importantly, very little, if any, turnover of Hxt3 occur in the rsp5-1 ts mutant, confirming Rsp5 as an essential E3 ubiquitin ligase in Hxt3 turnover. We could not use the rsp5-1ts allele in the turnover of Hxt7 as the strain could not tolerate rapamycin treatment during the turnover experiment (data not shown). We therefore used the less dominant rsp5-3 ts allele to study Hxt7 turnover. Hxt7 was clearly stabilized in the plasma membrane, but some internalization was observed. These observations implicate two E3 ubiquitin ligases, Rsp5 and the Vid30c, in the nutrient-mediated endocytosis and degradation of Hxt3 and Hxt7. Nutrient transporters have been shown to be direct targets for Rsp5-mediated ubiquitylation, suggesting a more indirect function for the Vid30c where it does not directly ubiquitylate the target Hxt. It is therefore of great importance to identify the precise role the Vid30c in Hxt turnover. Investigation into the signaling surrounding Hxt turnover has implicated the Ras/cAMP/PKA pathway and Rim15 in both unique turnover events. The strikingly similar retention of Hxt3 and Hxt7 in the plasma membranes of the vid28Δvid30Δ double mutant and cells expressing Ras2Val19 could serve as the foundation to investigate a potential functional link between the Vid30c and the Ras/cAMP/PKA.
Supporting Information
Deletion of components of the Vid30c causes a slight increase in HXT3 transcription. BY4742 (WT), vid30Δ, gid2Δ, vid28Δ, gid8Δ, gid9Δ (A) and vid28Δvid30Δ (B) expressing HXT3-GFP were cultured in glucose media as described in the methods (time 0). Following a switch to ethanol media, samples were collected at the indicated times and analyzed by northern blot analysis. Membranes were probed for GFP and ACT1 as a loading control.
(TIF)
The turnover of Hxt7 is dependent on Rsp5. BY4742 (WT) and rsp5-3 (A) and BY4742 (WT), art3 Δ, art4 Δ, art6 Δ and art8 Δ (B) expressing HXT7-GFP were cultured in raffinose media as described in the methods, time 0. After treated with rapamycin, samples were collected at the indicated times and analyzed by fluorescence microscopy (Top) and Western analysis with anti-GFP antibodies (Bottom). Identical blots were also probed with anti-ADH antibody as a loading control. (C) BY4742 (WT), art3Δ, art4Δ, art6Δ and art8Δ expressing HXT3-GFP were cultured in glucose media as described in the methods, time 0. Following a switch to ethanol media, samples were collected at the indicated times and analyzed by fluorescence microscopy.
(TIF)
Yeast strains used in this study.
(DOCX)
Acknowledgments
We thank Mark Longtine, Enzo Martegani, and Christopher Walkey for kindly supplying plasmids.
Funding Statement
This research was funded by: NSERC (Discovery Grant #262276-08), OMAFRA (SR9231), and Genome Canada/Genome British Columbia (151WIN) grants awarded to GvdM, and an Ontario Graduate Scholarship to CS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, et al. (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cameron S, Levin L, Zoller M, Wigler M (1988) cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae . Cell 53: 555–566. [DOI] [PubMed] [Google Scholar]
- 3. Smith A, Ward MP, Garrett S (1998) Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J 17: 3556–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beck T, Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- 5. Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev 13: 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, et al. (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kamada Y, Sekito T, Ohsumi Y (2004) Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol 279: 73–84. [DOI] [PubMed] [Google Scholar]
- 8. Powers T, Dilova I, Chen CY, Wedaman K (2004) Yeast TOR signaling: a mechanism for metabolic regulation. Curr Top Microbiol Immunol 279: 39–51. [DOI] [PubMed] [Google Scholar]
- 9. Powers T, Walter P (1999) Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae . Mol Biol Cell 10: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, et al. (2005) PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol 55: 862–880. [DOI] [PubMed] [Google Scholar]
- 11. Petkova MI, Pujol-Carrion N, Arroyo J, Garcia-Cantalejo J, Angeles de la Torre-Ruiz M (2010) Mtl1 is required to activate general stress response through Tor1 and Ras2 inhibition under conditions of glucose starvation and oxidative stress. J Biol Chem 285: 19521–19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C (2005) A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J 24: 4115–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmelzle T, Beck T, Martin DE, Hall MN (2004) Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol 24: 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, et al. (2003) TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell 12: 1607–1613. [DOI] [PubMed] [Google Scholar]
- 15. Hixson CS, Krebs EG (1980) Characterization of a cyclic AMP-binding protein from bakers’ yeast. Identification as a regulatory subunit of cyclic AMP-dependent protein kinase. J Biol Chem 255: 2137–2145. [PubMed] [Google Scholar]
- 16. Haney SA, Broach JR (1994) Cdc25p, the guanine nucleotide exchange factor for the Ras proteins of Saccharomyces cerevisiae, promotes exchange by stabilizing Ras in a nucleotide-free state. J Biol Chem 269: 16541–16548. [PubMed] [Google Scholar]
- 17. Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, et al. (1987) The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell 48: 789–799. [DOI] [PubMed] [Google Scholar]
- 18. Matsumoto K, Uno I, Oshima Y, Ishikawa T (1982) Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A 79: 2355–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka K, Lin BK, Wood DR, Tamanoi F (1991) IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A 88: 468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, et al. (1990) S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60: 803–807. [DOI] [PubMed] [Google Scholar]
- 21. Nikawa J, Cameron S, Toda T, Ferguson KM, Wigler M (1987) Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae . Genes Dev 1: 931–937. [DOI] [PubMed] [Google Scholar]
- 22. Wilson RB, Tatchell K (1988) SRA5 encodes the low-Km cyclic AMP phosphodiesterase of Saccharomyces cerevisiae . Mol Cell Biol 8: 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cameroni E, Hulo N, Roosen J, Winderickx J, De Virgilio C (2004) The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle 3: 462–468. [PubMed] [Google Scholar]
- 24. Krampe S, Boles E (2002) Starvation-induced degradation of yeast hexose transporter Hxt7p is dependent on endocytosis, autophagy and the terminal sequences of the permease. FEBS Lett 513: 193–196. [DOI] [PubMed] [Google Scholar]
- 25. Snowdon C, Hlynialuk C, van der Merwe G (2008) Components of the Vid30c are needed for the rapamycin-induced degradation of the high-affinity hexose transporter Hxt7p in Saccharomyces cerevisiae . FEMS Yeast Res 8: 204–216. [DOI] [PubMed] [Google Scholar]
- 26. Roberts GG, Hudson AP (2006) Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol Genet Genomics 276: 170–186. [DOI] [PubMed] [Google Scholar]
- 27. Snowdon C, Schierholtz R, Poliszczuk P, Hughes S, van der Merwe G (2009) ETP1/YHL010c is a novel gene needed for the adaptation of Saccharomyces cerevisiae to ethanol. FEMS Yeast Res 9: 372–380. [DOI] [PubMed] [Google Scholar]
- 28. Pitre S, Dehne F, Chan A, Cheetham J, Duong A, et al. (2006) PIPE: a protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs. BMC Bioinformatics 7: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun B, Pfirrmann T, Menssen R, Hofmann K, Scheel H, et al. (2011) Gid9, a second RING finger protein contributes to the ubiquitin ligase activity of the Gid complex required for catabolite degradation. FEBS Lett 585: 3856–3861. [DOI] [PubMed] [Google Scholar]
- 30. Santt O, Pfirrmann T, Braun B, Juretschke J, Kimmig P, et al. (2008) The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell 19: 3323–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL (2004) Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem 279: 49138–49150. [DOI] [PubMed] [Google Scholar]
- 32. Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, et al. (2003) Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell 14: 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hatakeyama R, Kamiya M, Takahara T, Maeda T (2010) Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol Cell Biol 30: 5598–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Springael JY, De Craene JO, Andre B (1999) The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2 domain is competent for ubiquitination but not for subsequent endocytosis of the Gap1 permease. Biochem Biophys Res Commun 257: 561–566. [DOI] [PubMed] [Google Scholar]
- 35. Leon S, Haguenauer-Tsapis R (2009) Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp Cell Res 315: 1574–1583. [DOI] [PubMed] [Google Scholar]
- 36. Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, et al (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- 37. Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- 38. Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J (2005) Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast 22: 1061–1068. [DOI] [PubMed] [Google Scholar]
- 39. Cruz MC, Goldstein AL, Blankenship J, Del Poeta M, Perfect JR, et al. (2001) Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob Agents Chemother 45: 3162–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson LC, Gibbs JB, Marshall MS, Sigal IS, Tatchell K (1987) CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae . Science 235: 1218–1221. [DOI] [PubMed] [Google Scholar]
- 41. Onodera J, Ohsumi Y (2004) Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae . J Biol Chem 279: 16071–16076. [DOI] [PubMed] [Google Scholar]
- 42. Hedbacker K, Carlson M (2008) SNF1/AMPK pathways in yeast. Front Biosci 13: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benanti JA, Cheung SK, Brady MC, Toczyski DP (2007) A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol 9: 1184–1191. [DOI] [PubMed] [Google Scholar]
- 44. Mbonyi K, Thevelein JM (1988) The high-affinity glucose uptake system is not required for induction of the RAS-mediated cAMP signal by glucose in cells of the yeast Saccharomyces cerevisiae . Biochim Biophys Acta 971: 223–226. [DOI] [PubMed] [Google Scholar]
- 45. Crechet JB, Poullet P, Camonis J, Jacquet M, Parmeggiani A (1990) Different kinetic properties of the two mutants, RAS2Ile152 and RAS2Val19, that suppress the CDC25 requirement in RAS/adenylate cyclase pathway in Saccharomyces cerevisiae . J Biol Chem 265: 1563–1568. [PubMed] [Google Scholar]
- 46. Toda T, Cameron S, Sass P, Zoller M, Wigler M (1987) Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50: 277–287. [DOI] [PubMed] [Google Scholar]
- 47. Schmidt A, Beck T, Koller A, Kunz J, Hall MN (1998) The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J 17: 6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. MacGurn JA, Hsu PC, Smolka MB, Emr SD (2011) TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147: 1104–1117. [DOI] [PubMed] [Google Scholar]
- 49. Martin DE, Soulard A, Hall MN (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979. [DOI] [PubMed] [Google Scholar]
- 50. Ramachandran V, Herman PK (2011) Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae . Genetics 187: 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reinders A, Burckert N, Boller T, Wiemken A, De Virgilio C (1998) Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev 12: 2943–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R (1996) Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem 271: 10946–10952. [DOI] [PubMed] [Google Scholar]
- 53. Springael JY, Andre B (1998) Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae . Mol Biol Cell 9: 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725. [DOI] [PubMed] [Google Scholar]
- 55. Nikko E, Sullivan JA, Pelham HR (2008) Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep 9: 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krampe S, Stamm O, Hollenberg CP, Boles E (1998) Catabolite inactivation of the high-affinity hexose transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett 441: 343–347. [DOI] [PubMed] [Google Scholar]
- 57. Hoffman M, Chiang HL (1996) Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae . Genetics 143: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van der Merwe GK, Cooper TG, van Vuuren HJ (2001) Ammonia regulates VID30 expression and Vid30p function shifts nitrogen metabolism toward glutamate formation especially when Saccharomyces cerevisiae is grown in low concentrations of ammonia. J Biol Chem 276: 28659–28666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Griffioen G, Thevelein JM (2002) Molecular mechanisms controlling the localisation of protein kinase A. Curr Genet. 41: 199–207. [DOI] [PubMed] [Google Scholar]
- 60. Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81. [DOI] [PubMed] [Google Scholar]
- 61. Santangelo GM (2006) Glucose signaling in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 70: 253–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103: 253–262. [DOI] [PubMed] [Google Scholar]
- 63. Dennis PB, Fumagalli S, Thomas G (1999) Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev 9: 49–54. [DOI] [PubMed] [Google Scholar]
- 64. Vidan S, Mitchell AP (1997) Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol 17: 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Perez-Valle J, Jenkins H, Merchan S, Montiel V, Ramos J, et al. (2007) Key role for intracellular K+ and protein kinases Sat4/Hal4 and Hal5 in the plasma membrane stabilization of yeast nutrient transporters. Mol Cell Biol 27: 5725–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Perez-Valle J, Rothe J, Primo C, Martinez Pastor M, Arino J, et al. (2010) Hal4 and Hal5 protein kinases are required for general control of carbon and nitrogen uptake and metabolism. Eukaryot Cell 9: 1881–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deletion of components of the Vid30c causes a slight increase in HXT3 transcription. BY4742 (WT), vid30Δ, gid2Δ, vid28Δ, gid8Δ, gid9Δ (A) and vid28Δvid30Δ (B) expressing HXT3-GFP were cultured in glucose media as described in the methods (time 0). Following a switch to ethanol media, samples were collected at the indicated times and analyzed by northern blot analysis. Membranes were probed for GFP and ACT1 as a loading control.
(TIF)
The turnover of Hxt7 is dependent on Rsp5. BY4742 (WT) and rsp5-3 (A) and BY4742 (WT), art3 Δ, art4 Δ, art6 Δ and art8 Δ (B) expressing HXT7-GFP were cultured in raffinose media as described in the methods, time 0. After treated with rapamycin, samples were collected at the indicated times and analyzed by fluorescence microscopy (Top) and Western analysis with anti-GFP antibodies (Bottom). Identical blots were also probed with anti-ADH antibody as a loading control. (C) BY4742 (WT), art3Δ, art4Δ, art6Δ and art8Δ expressing HXT3-GFP were cultured in glucose media as described in the methods, time 0. Following a switch to ethanol media, samples were collected at the indicated times and analyzed by fluorescence microscopy.
(TIF)
Yeast strains used in this study.
(DOCX)