Abstract
Myotonic dystrophy type 1 (DM1) is a microsatellite expansion disorder caused by the aberrant expansion of CTG repeats in the 3’ untranslated region of the DMPK gene. When transcribed, the toxic RNA CUG repeats sequester RNA binding proteins, which leads to disease symptoms. The expanded CUG repeats can adopt a double-stranded structure, and targeting this helix is a therapeutic strategy for DM1. In order to better understand the 5’CUG/3’GUC motif, and how it may interact with proteins and small molecules, we designed a short CUG helix attached to a GAAA tetraloop/receptor in order to facilitate crystal packing. Here we report the highest resolution structure (1.95 Å) to date of a GAAA tetraloop/receptor and the CUG helix it was used to crystallize. Within the CUG helix, we identify two different forms of non-canonical U-U pairs and reconfirm that CUG repeats are essentially A-form. An analysis of all non-canonical U-U pairs in the context of CUG repeats revealed six different classes of conformations that the non-canonical U-U pairs are able to adopt.
Microsatellite expansion diseases are the result of aberrant expansion of short repeating segments of DNA (between 1–10 bases) (1). These expansions can occur in either in coding regions where they result in abnormal proteins (2), or in noncoding regions of the genome where upon transcription, act through a toxic RNA gain-of-function mechanism (3, 4). Myotonic dystrophy (DM), which is the most common form of adult onset muscular dystrophy, affecting ~1 in 8000 individuals, is the result of microsatellite expansions (reviewed in (1, 3, 5)). DM type 1 (DM1) is caused by a CTG expansion in the 3’ untranslated region in the DMPK gene (6). When transcribed to RNA, these CUG repeats sequester RNA binding proteins within nuclear foci (7, 8). One family of sequestered proteins that co-localizes with expanded CUG repeats, the Muscleblind-like (MBNL) proteins, are regulators of alternative splicing, resulting in the mis-splicing of target transcripts, which leads to disease symptoms (9–14). A related disease, myotonic dystrophy type 2 (DM2), is very similar to DM1, except instead of a trinucleotide repeat, it is a tetranucleotide repeat (CCTG) in the first intron of the ZNF9 gene (15).
The expanded toxic CUG repeat RNA can form stable stem-loop structures (16), and a handful of CUG-repeat structures have been published demonstrating an essentially A-form structure, with the non-canonical U-U pairs forming only one hydrogen bond without distorting the backbone (17–19). However, two of these structures contained some degree of disorder, and all of the structures had very tight crystal packing, leaving little to no potential room for co-crystallization with ligands (either small molecules or short peptides). In order to overcome these issues, we decided to use an RNA crystallization motif in order to design specific intermolecular contacts and minimize close packing and introduce “free spaces” within the crystal.
We chose the GAAA tetraloop and its conserved 11-nucleotide receptor as the core for the crystal contacts. This tetraloop/receptor was initially identified as a tertiary motif in Group I and II self-splicing introns (20–22). The three adenosines in the loop flip outward and form specific hydrogen bonds with the 11-nucleotide receptor (22), making it an ideal candidate for engineering crystal contacts. This motif has been utilized to aid in the crystallization of larger RNA constructs, such as the hepatitis delta virus, among others (23, 24). Here, we utilized the tetraloop/receptor to crystallize a short RNA helix composed of CUG repeats allowing us to obtain crystals that diffracted to high resolution. Additionally, we were able to use the previously solved structure of the tetraloop/receptor as our search model, allowing us to quickly and easily solve the structure using molecular replacement.
MATERIALS and METHODS
Purification of RNA
RNA was purchased from Dharmacon and desalted per their instructions. RNA was then resuspended in 1x denaturing dye (25% w/w formamide, 1x TBE, 0.1% bromophenol blue, 0.1% xylene cyanol) and run on a denaturing 8% polyacrylamide (19:1) gel with 7 M urea. RNA was located using UV shadowing, excised and eluted overnight using the Elutrap Electroelution System (Whatman) in 1x TBE. RNA was then ethanol precipitated and resuspended in ddH2O and desalted using a Micro Bio-Spin column with Bio-Gel P2 Gel (Bio-Rad). RNA was brought to a final concentration of 0.5 mM in a solution of 15 mM NaCl, 5 mM Tris pH 7.5, and 5 mM MgCl2.
RNA crystallization
The RNAs were annealed by heating to 70 °C for 5 min and rapidly cooled to 4 °C. RNAs were screened for crystallization using the Mosquito nano-liter high throughput robot (TTP labtech) and the Natrix screen (Hampton Research) by the hanging drop vapor diffusion method at room temperature. Promising hits were screened in 4 µL hanging drops. The best crystals of trCUG-3 were grown from a mixture of 2 µL RNA solution and 2 µL well solution containing 4 mM MgSO4, 50 mM Tris pH 8.5, and 30% (w/v) 1,6-hexanediol. Crystals appeared in approximately one week.
Data collection
Crystals of at least 0.02 mm in the smallest dimension were mounted in rayon loops and flash frozen in liquid nitrogen. Experimental data were collected at Advanced Light Source BL 5.0.1 under a cryostream. The x-ray data were integrated, merged, and scaled using the HKL-2000 program suite (25), and converted to structure factors using the CCP4i GUI (26) for the CCP4 program suite (27). Data collection statistics are listed in Table 1.
Table 1.
Summary of data collection and refinement statistics
| Measurement | Value |
|---|---|
| Space group | R3 |
| Unit cell dimensions a, b, c Å | 70.34 70.34 68.24 |
| α, β, γ (°) | 90 90 120 |
| Resolution range, Å | 22.75 – 1.95 |
| Total number of reflections | 98787 |
| Number of unique reflections | 8621 |
| Average redundancy | 5.5 |
| % completeness | 99.3 (98.9) |
| I/σI | 37.0 (2.2) |
| Rmergea | 0.068 (0.60) |
| Average B-factors [atoms] | |
| nucleotides | 54.58 [744] |
| solvent | 60.53 [44] |
| Rfreeb | 0.266 |
| Rworkb | 0.208 |
Values in parentheses represent highest resolution shell
where I is the observed intensity and <I> is the average of intensities obtained from multiple observations of symmetry-related reflections.
where Fo and Fc are the observed and calculated structure amplitudes, respectively.
Structure determination
Structure was solved using the molecular replacement method by the program molrep (28), a part of the CCP4 program suite (27), with the tetraloop/receptor from PDB 1GID, the structure of group I ribozyme domain (22), as the search model. The 11-nucleotide receptor and the 4-nucleotide loop (plus the two adjacent base pairs) were successfully used as our search model. The model was manually rebuilt using Coot (29), and refined with refmac (30). Coot and PyMOL (31) were used to generate figures. Refinement statistics are listed in Table 1. Simulated annealing omit maps were created using CNS (32) by deleting all four uridines in the CUG portion of the structure while all other nucleotides remained fixed during annealing. The structure was deposited in the PDB (entry 4FNJ). Structural parameters were calculated using 3DNA (33).
RESULTS
Design of RNA constructs for crystallization
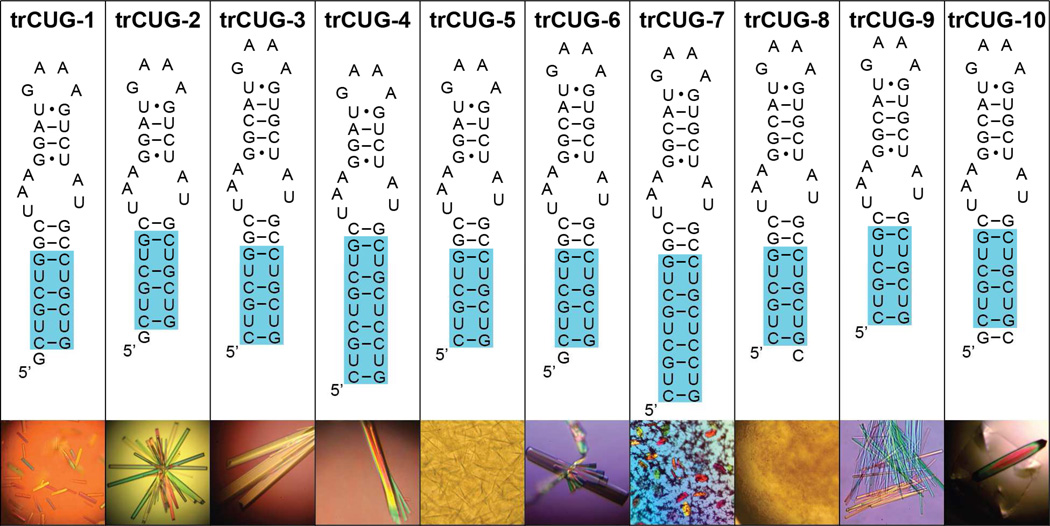
In order to overcome crystallographic disorder present in previous CUG repeat structures (17, 18), we designed ten different constructs utilizing the GAAA tetraloop/receptor to facilitate intermolecular contacts (Figure 1). Each construct contained either four or six CUG repeats, one or two stabilizing base pairs, the receptor, a short upper helix, and the GAAA tetraloop. The upper helix was kept short in order to favor intermolecular rather than intramolecular interactions. Only trCUG-8 failed to form any crystals, possibly due to the 3’ overhanging nucleotide. Of the remaining constructs, only trCUG-2, 3, 6, and 10 grew crystals of sufficient size to mount. Neither trCUG-2 nor trCUG-10 diffracted to better than 10Å. Both trCUG-3 and trCUG-6 diffracted to better than 3Å resolution; however, the 3’ overhanging G of trCUG-6 was disordered, rendering the models essentially the same.
Figure 1.
Sequence and predicted secondary structure of RNAs containing the CUG repeats and the tetraloop/receptor. Each RNA construct consists of four or six CUG repeats (highlighted in blue), the 11-nucleotide receptor, a short upper stem, and the GAAA tetraloop. Below each sequence is a representative picture of how well each RNA crystallized. The only sizable trCUG-6 crystal grew off a fiber.
Structure determination using tetraloop/receptor as a model for molecular replacement
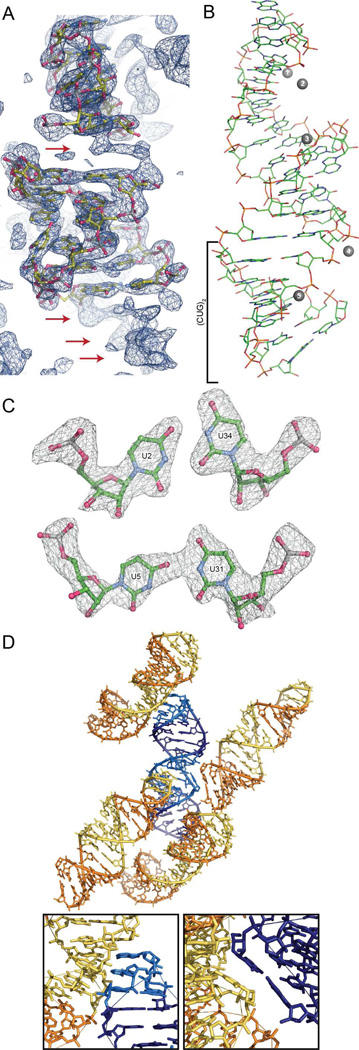
Since the sequence, and presumably the structure, of the tetraloop/receptor in trCUG-3 was the same as the tetraloop/receptor in the group I ribozyme domain structure determined by Cate et al. 22) (with the exception of the first base pair, which was reversed), this portion of their structure was successfully used for molecular replacement. The un-modeled base pairs were clearly visible in the maps and the nucleotides were easily modeled into the density (Figure 2A). Five peaks were observed in the maps that were unlikely to be waters due to geometric considerations. Based on previous structures of the tetraloop/receptor (22, 34, 35) and their proximity to the phosphate backbone, we modeled the peaks as magnesium ions (Figure 2B). The final model has 35 nucleotides, 44 water molecules, and five magnesium ions with an Rwork and Rfree of 0.208 and 0.266, respectively.
Figure 2.
Structure and crystal packing of trCUG-3. (A) Molecular replacement solution and 2Fo-Fc map (contoured at 1.5 σ) after one round of refinement. The density of the base pairs not yet modeled, both between the tetraloop/receptor and directly below the receptor, are clearly observable in the density (red arrows). (B) Tertiary structure of trCUG-3. The (CUG)2 region, containing four CUG repeats at the base of the hairpin, is denoted by a bracket. Magnesium ions are represented by numbered gray spheres. Waters are not shown. (C) Simulated annealing omit map (Fo-Fc) of both non-canonical U-U pairs contoured at 5 σ. (D) Crystal packing (or quaternary structure) of trCUG-3. Each RNA interacts with four other RNAs in the structure: at the loop, at the receptor, on the opposite side of the receptor and at the bottom of the helix. Tetraloop and receptor are denoted by lighter colors. Inset gives a closer view of each interaction.
Structure of trCUG-3
The 35-nucleotide trCUG-3 RNA crystallized in space group R 3. As designed, the RNA formed a hairpin with a GAAA tetraloop, a short five base pair stem, the tetraloop/receptor, and an eight base pair CUG-containing stem (Figure 2B). The upper stem begins and ends with a G•U wobble base pair. The lower stem contains two different non-canonical U-U pairs (Figure 2C), while the G-C base pairs formed Watson-Crick interactions. Each RNA makes specific crystal contacts with four other RNAs: two of these interactions were predicted tetraloop/receptor interactions, and the other two were the unanticipated stacking of the base of the helices against the opposite sides of the receptors of the symmetry mates (Figure 2D).
Within the CUG-containing stem, two different conformations of the non-canonical U-U pair were observed (Figure 2C). The lower U2-U34 non-canonical pair formed two hydrogen bonds, which resulted in a shorter C1’-C1’ distance (8.6 Å) as compared to the upper U5-U31 non-canonical pair, which formed one hydrogen bond and had a C1’-C1’ distance of 10.6 Å (Supplemental Table 1). Both non-canonical pairs were inclined towards the minor groove (λ1 < 54°), and as a result, do not appear to stack with the preceding C. The incline was more severe in the U5-U31 non-canonical pair, which is most likely a result of the single hydrogen bond. B-factors for the atoms that composed these pairs were not noticeably different from other atoms within the structure.
Examining the overall structure of the CUG portion of the lower helix (hereafter referred to as (CUG)2) revealed that it is primarily an A-form helix. The average helical rise of (CUG)2 is 2.39 Å (Supplemental Table 2), which is closer to an A-form conformation (2.83 Å) than a B-form conformation (3.29 Å) (36). The average helical twist is 35.9°, which is greater than the expected 32.5° for A-form, and not much less than the expected 36.5° for B-form. Base pairs in (CUG)2 are also steeply inclined (18.6°) as is expected for A-form helices. Local dimer step parameters, including roll, twist, slide, and most importantly zP and zP(h), also match well with average A-form values (Supplemental Table 2). Interestingly, the structure did not seem to be affected, beyond the first CU/UG step, by the crystal packing interaction that occurred at the base of the helix (Figure 2D). This base pair step was strongly tipped (20.6°), tilted (−11.3°), and rolled (19.4°) as compared to the other base pairs in the structure.
Within the structure, five suspected magnesium ions were identified. Two were near the GAAA tetraloop, two more flanked the 11-nucleotide receptor, and the final magnesium ion was near the center of the CUG portion of the lower helix (Figure 2B). Only the third and fourth magnesium ions were close enough to the RNA to be making direct contacts: the rest were making interactions via water molecules. None of the suspected magnesium ions appeared to be fully coordinated and the first and second magnesium ions near the loop interacted with four water molecules.
DISCUSSION
Comparison to other CUG helices
To date, four other crystal structures of CUG repeat RNA crystal structures have been published (17–19). The first, and longest, to be crystallized was a (CUG)6 duplex (17). Unfortunately, this construct crystallized with twofold translational disorder, complicating analysis. The next structure to be released was another duplex: G(CUG)2C (18). This structure contained three different duplexes within the same unit cell: the A+B duplex, the C+D duplex, and the E+E* duplex, which were related to one another by crystallographic symmetry. The authors of this publication also de-twinned the data from Mooers et al . The final two structures are different conformations of a third duplex: UUGGGC(CUG)3UCC (19). Using 3DNA, we compared the helical parameters of (CUG)2 to the (5’CUG/3’GUC)x portions of these six different helices observed in four separate crystal structures. (For simplicity, the de-twinned (CUG)6 data were used.)
Overall, all seven CUG helices appeared to be more similar to an A-form helix than a B-form helix, especially when parameters that demonstrate the greatest distinction between A-form and B-form are considered (roll, slide, inclination, x-displacement, zP, and zP(h)) (Supplemental Table 2). Although (CUG)2 from trCUG-3 has the greatest helical twist of all the published CUG helices (35.9°), it also has the smallest helical rise (2.39 Å). In general, it appears that the (CUG)2 model is consistent with existing models.
Non-canonical U-U pairs are dynamic
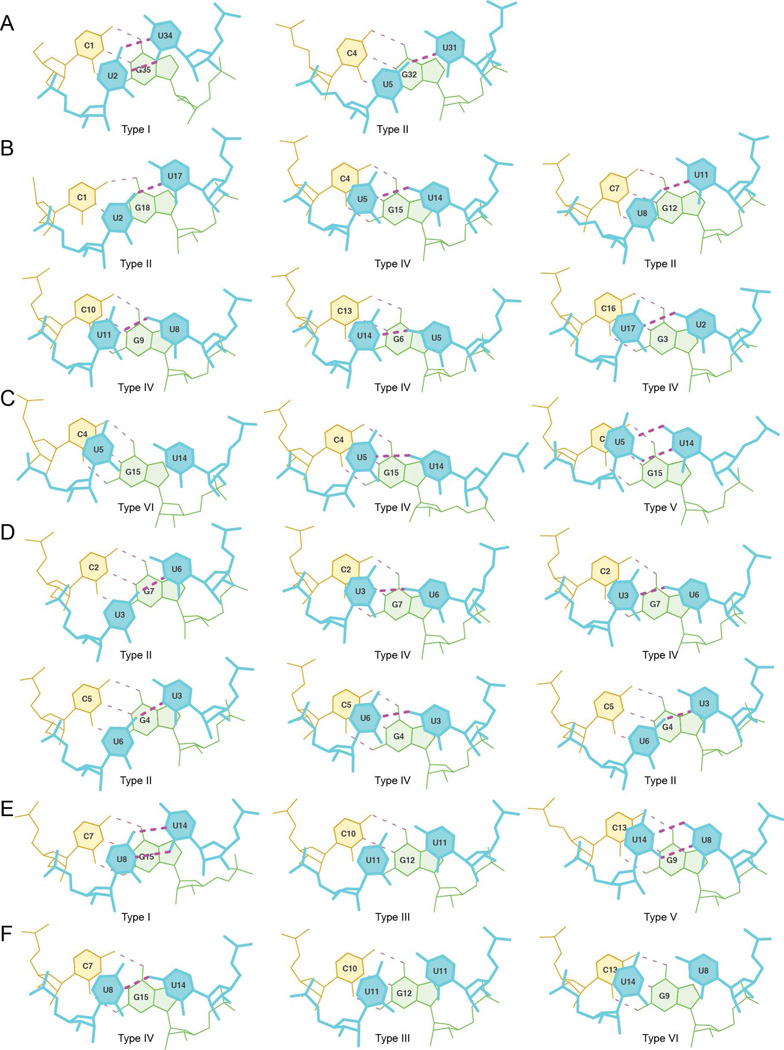
In addition to comparing helical parameters of other CUG structures, we also examined all the non-canonical U-U pairs, in the context of CUG repeats that were available (Figure 3, Supplemental Table 1). Non-canonical U-U pairs from all the CUG crystal structures (17–19) and an NMR structure (37) were compared. Examining these pairs revealed that the non-canonical U-U pair in the 5’CUG/3’GUC motif is dynamic, and at least six unique conformations have been captured in the structures. These six conformations were determined by considering the potential number of hydrogen bonds and direction of inclination (towards the major or minor groove, or not inclined). There is one other potential type of non-canonical U-U pair that has not been observed in the currently available data, which is a non-canonical pair with no hydrogen bonds inclined towards the minor groove. It is likely that this type of non-canonical pair occurs in the context of CUG repeats (as a counterpart to type VI, which also forms no hydrogen bonds, but is inclined toward the major groove) and has simply not been observed to date.
Figure 3.
Comparison of non-canonical U-U pairs in the context of CUG repeats. (A) Lower and upper non-canonical U-U pair of trCUG-3. The upper U5-U31 non-canonical pair forms the more typical (in the context of CUG repeats) one hydrogen bond, while the lower U2-U34 non-canonical pair forms two hydrogen bonds, shortening the C1’-C1’ distance. (B) Non-canonical U-U pairs of the de-twinned (CUG)6 structure. All non-canonical U-U pairs in this structure form one hydrogen bond, maintaining the C1’-C1’ distance typical of a Watson-Crick interaction. (C) Non-canonical U-U pairs from the NMR structure of CCGCUGCGG, forming zero, one, or two hydrogen bonds. (D) Non-canonical U-U pairs from G(CUG)2C crystal structure. The first column contains the non-canonical U-U pairs in duplex A+B, the second contains C+D and the third contains E+E*. Every non-canonical pair within the G(CUG)2C crystal structure forms one hydrogen bond. (E–F) Noncanonical U-U pairs from UUGGGC(CUG)3CUCC pdb entry 3SYW (E) and 3SZX (F).
Within the trCUG-3 structure, we found two of the six types: type I and II (Figure 3, Supplemental Table 1). The type I non-canonical pair forms two hydrogen bonds, shortening the C1’-C1’ distance, and is inclined towards the minor groove. One other type I non-canonical pair has been observed, in UUGGGC(CUG)3UCC (Figure 3E) (19), suggesting this conformation is not simply due to potential distortion of the helix of (CUG)2 due to the crystal packing at the base of the trCUG-3 helix. The other non-canonical pair we observed was a type II non-canonical pair, the second most frequent type of noncanonical U-U pair. The type II non-canonical pair is also inclined towards the minor groove but forms only one hydrogen bond. It was observed twice in (CUG)6 (Figure 3B), twice in the G(CUG)2C A+B duplex, and once in the E+E* duplex (Figure 3D). The most commonly observed non-canonical pair, type IV, is the counterpart to type II, and forms one hydrogen bond while being inclined toward the major groove. It was found four times in (CUG)6 (Figure 3B), in the NMR structures (Figure 3C), twice in the G(CUG)2C C+D duplex, once in the E+E* duplex (Figure 3D), and once in the UUGGGC(CUG)3UCC duplex (Figure 3F). Of course, in solution, it is probable that the non-canonical U-U pairs found in the 5’CUG/3’GUC motif sample all these conformations, and that these “types” of non-canonical U-U pairs are not discreet, but rather found in a continuum of different states. These data are consistent with the flexibility of the non-canonical U-U pairs observed in the NMR structure of the CCGCUGCGG duplex (Figure 3C), where the pairs have the ability to sample various conformations without disrupting the structure of the surrounding nucleotides (37).
Our findings confirm the observation by Kumar et al. that the U-U pairs found in the 5’CUG/3’GUC motif sample more conformations than the two forms of ‘stretched U-U wobble’ (a non-canonical U-U pair characterized by one hydrogen bond interaction between the uridines) identified by Kiliszek et al. However, our analysis suggests that the most common U-U interaction found in the 5’CUG/3’GUC motif is most likely of the ‘stretched U-U wobble’ variety. This is in contrast to Kumar et al. who conclude that the zero-hydrogen bond conformation is predominant. The difference between our analysis and Kumar’s is the use of the detwinned (CUG)6 data (18) versus the original twinned data modeled at half occupancy (17). In the original model, five of the six U-U pairs formed zero hydrogen bonds and the final pair was a ‘stretched U-U wobble’, while in the detwinned model, all the U-U pairs were ‘stretched U-U wobbles’. The higher proportion of ‘stretched U-U wobbles’ is consistent with the molecular dynamics simulations performed by Parkesh et al. in which they found 76.5% of non-canonical U-U pairs in the 5’CUG/3’GUC motif formed one hydrogen bond (37). Interestingly, our structure is the first crystal structure to contain a ‘stretched U-U wobble’ as well as a U-U pair that forms two hydrogen bonds.
Metal ions in the structure
Within the trCUG-3 crystal structure, we identified five potential magnesium ions. It is unsurprising that magnesium ions would be identified within the structure, especially the tetraloop/receptor region, since it has previously been shown that the tetraloop/receptor interaction is dependent on magnesium (38) or other divalent metal ions (39). Several other structures of the GAAA tetraloop/receptor have been published, so we compared our structure and metal ion sites with several previously published structures (Supplemental Figure 1). Comparing trCUG-3 and PDB 1GID (22), we observed that our first three magnesium ions near the cobalt hexamine ion found contacting the GAAA loop (Supplemental Figure 1B). Our fourth magnesium ion near the receptor has a nearby counterpart in 1GID. When compared to 1HR2 (34) (Supplemental Figure 1C), we observed a similar scenario: our first three magnesium ions were near one magnesium ion contacting the loop. The second, third, and fourth magnesium ions had counterparts in the 1HR2 structure. The additional ions observed in our structure may be due to the close proximity of our tetraloop and receptor as compared to either 1GID or 1HR2, or it could simply be that the magnesium ions are important for electrostatic screening, allowing close contact between RNA strands (40).
Engineered RNA contacts as a strategy for crystallization
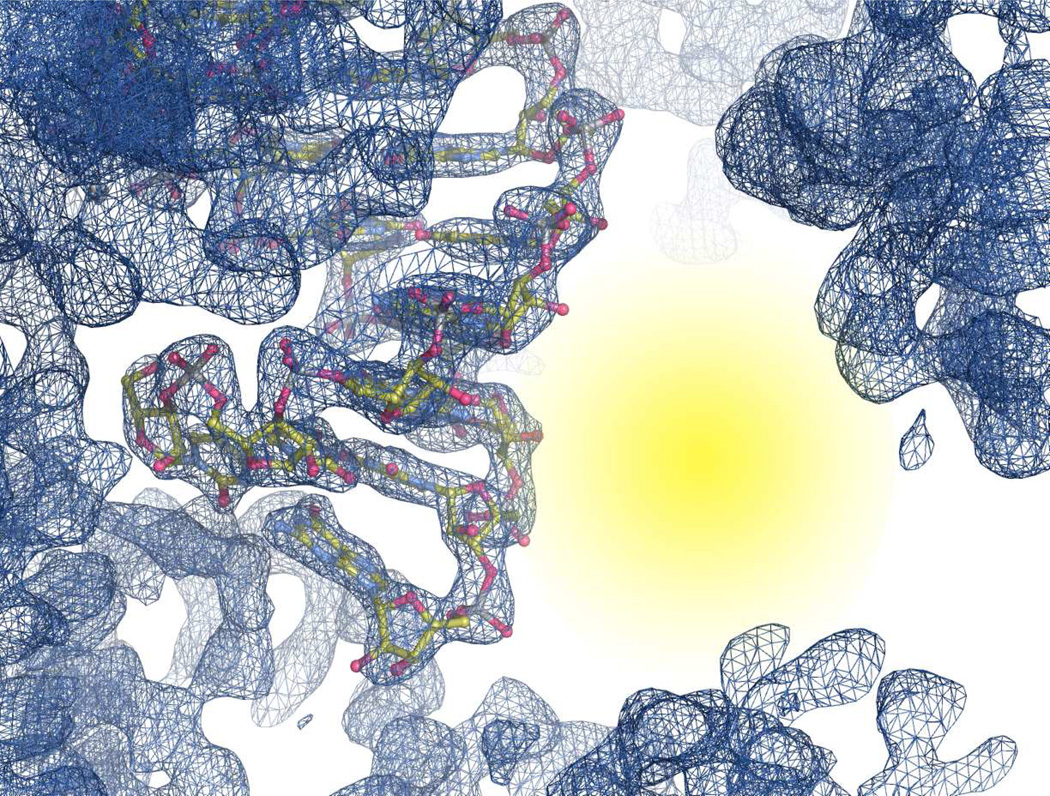
It is clear that utilizing the GAAA tetraloop/receptor was a successful strategy for crystallizing a short helix and avoids the pseudosymmetry problems associated with duplex RNA. Additionally, examining the crystal packing of this particular construct, we observed a large “open” space in the crystal lattice (Figure 4), which could potentially allow for co-crystallization of small molecules, short peptoids, and possibly small zinc-finger domains in complex with the CUG stem. This “open” space is unique to our tetraloop/receptor construct as compared to other CUG repeat containing crystal structures (17–19): these structures all crystalized in pseudo-infinite helices with very tight packing between helices, leaving little to no room for co-crystallization of an additional molecule.
Figure 4.
Structure and 2Fo-Fc map contoured at 1.5 σ near the CUG repeat stem. Yellow area highlights large open space near the minor groove of the (CUG)2 stem.
This GAAA tetraloop/receptor framework can also be utilized to crystallize other short repeat RNA stems, such as the CCUG repeats responsible for DM2 (8, 15), CAG repeats responsible for Huntington’s disease and several spinocerebellar ataxias, and CGG repeats responsible for fragile X-associated tremor ataxia syndrome, among many others (reviewed in (41)). We have already crystallized a CCUG helix utilizing the tetraloop/receptor construct, which diffracted to a resolution of better than 3 A (data not shown).
We have crystallized a CUG containing RNA construct that has captured two different orientations of the clearly mobile non-canonical U-U pair. By examining other crystal and NMR structures, it is clear that non-canonical U-U pairs, in the context of CUG repeats, can flex between many different conformations, from fully paired with one another, to only sharing a single hydrogen bond, to being too distant to interact with one another (Figure 3). Additionally, these non-canonical U-U pairs can go from being inclined towards the minor groove, through no inclination, all the way to being inclined towards the major groove. In our interpretation, this conformational heterogeneity suggests a weak interaction due to an apparent reduction in hydrogen bonding (averaged over all potential U-U conformations) and, more importantly, less efficient base stacking as compared to the stacking of the GC/GC steps within the helix (Supplemental Figure 2). The weak interaction between the uridines and neighboring bases may play an important role in MBNL proteins gaining access to single stranded CUG repeats in order to bind the Watson-Crick face of the GC dinucleotide as found in the MBNL1-RNA crystal structure (PDB 3D2S) (42). Interestingly, a recently published structure of CCG repeats shows that the non-canonical CC base pair appears to adopt fewer conformations than the U-U pair does (0–1 hydrogen bonds in comparison to 0–2 hydrogen bonds in the case of U-U) (43). Due to the arrangement of hydrogen bond donors/acceptors on the Watson-Crick face of cytosine, it is hard to envision the C-C pair adopting some of the conformations observed with the U-U pairs, but they do show similarly poor base stacking (43) and similar thermodynamic stability in the context of the CNG helix (44). Additionally, in the context of DM2, we predict that the C-U non-canonical pairs of the 5’CCUG/3’GUCC motif will have similarly weak/dynamic interactions as compared to the non-canonical U-U pairs found in the CUG helix, again allowing MBNL proteins to gain access to the Watson-Crick faces of the GC dinucleotide.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Nolen, J. Purcell, and members of the Berglund lab for helpful discussions and comments on the manuscript. Portions of this research were conducted at the Advanced Light Source, a national user facility operated by Lawrence Berkeley National Laboratory, on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Berkeley Center for Structural Biology is supported in part by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences.
Funding: These studies were supported by a grant (AR0599833) from NIAMS/NIH.
Footnotes
Supporting Information Available. Tables of non-canonical U-U pair structural parameters and helical parameters. A figure showing the comparison of the trCUG-3 tetraloop/receptor and previously crystallized tetraloop/receptors and a figure showing the stacking interactions of the three different base steps in the 5’CUG/3’GUC motif. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Cooper TA, Wan L, Dreyfuss G. RNA and Disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y-C, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 3.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 4.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated Disease. J Biol Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho DH, Tapscott SJ. Myotonic dystrophy: Emerging mechanisms for DM1 and DM2. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 8.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 9.Ho TH, Charlet-B N, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hino SI, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, Chihara K, Aoki Y, Nakamori M, Takahashi MP, Imaizumi K. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 11.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen S, Talukdar I, Liu Y, Tam J, Reddy S, Webster NJG. Muscleblind-like 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J Biol Chem. 2010;285:25426–25437. doi: 10.1074/jbc.M109.095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gates DP, Coonrod LA, Berglund JA. Autoregulated Splicing of muscleblind-like 1 (MBNL1) Pre-mRNA. J Biol Chem. 2011;286:34224–34233. doi: 10.1074/jbc.M111.236547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 16.Napierała M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable"slippery” hairpins. J Biol Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 17.Mooers BHM, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the"stretched U-U wobble”: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Park H, Fang P, Parkesh R, Guo M, Nettles KW, Disney MD. Myotonic dystrophy type 1 RNA crystal structures reveal heterogeneous 1 × 1 nucleotide UU internal loop conformations. Biochemistry. 2011;50:9928–9935. doi: 10.1021/bi2013068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 21.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 23.Ferre-D'Amare AR, Zhou K, Doudna JA. A general module for RNA crystallization. J. Mol. Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- 24.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 27.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.5.0.1. 2010 May [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 35.Davis JH, Foster TR, Tonelli M, Butcher SE. Role of metal ions in the tetraloop-receptor complex as analyzed by NMR. RNA. 2006;13:76–86. doi: 10.1261/rna.268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, Neidle S, Shakked Z, Sklenar H, Suzuki M, Tung CS, Westhof E, Wolberger C, Berman HM. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 37.Parkesh R, Fountain M, Disney MD. NMR spectroscopy and molecular dynamics simulation of RCCGCUGCGG) reveal a dynamic UU internal loop found in myotonic dystrophy type 1. Biochemistry. 2011;50:599–601. doi: 10.1021/bi101896j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 39.Downey CD, Fiore JL, Stoddard CD, Hodak JH, Nesbitt DJ, Pardi A. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45:3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Draper DE, Grilley D, Soto AM. Ions And RNA Folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 41.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystallographic characterization of CCG repeats. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broda M, Kierzek E, Gdaniec Z, Kulinski T, Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.