Abstract
One of the major challenges in the development of targeted nanoparticles (NPs) for cancer therapy is to discover targeting ligands that allow for differential binding and uptake by the target cancer cells. Using prostate cancer (PCa) as a model disease, we developed a cell-uptake selection strategy to isolate PCa-specific internalizing 2'-Omethyl RNA aptamers (Apts) for NP incorporation. Twelve cycles of selection and counter-selection were done to obtain a panel of internalizing Apts, which can distinguish PCa cells from non-prostate and normal prostate cells. After Apt characterization, size minimization, and conjugation of the Apts with fluorescently-labeled polymeric NPs, the NP-Apt bioconjugates exhibit PCa specificity and enhancement in cellular uptake when compared to non-targeted NPs lacking the internalizing Apts. Furthermore, when docetaxel, a chemotherapeutic agent used for the treatment of PCa, was encapsulated within the NP-Apt, a significant improvement in cytotoxicity was achieved in targeted PCa cells. Rather than isolating high-affinity Apts as reported in previous selection processes, our selection strategy was designed to enrich cancer-cell specific internalizing Apts. A similar cell-uptake selection strategy may be used to develop specific internalizing ligands for a myriad of other diseases and can potentially facilitate delivering various molecules, including drugs and siRNAs, into cells.
Keywords: nanoparticles, internalization, in vitro selection, aptamer, targeted cancer therapy
Targeted nanoparticle (NP) therapeutics have shown great potential for cancer therapy, as they provide enhanced efficacy and reduced side effects.1–3 These features are mainly due to the improved accumulation of NPs in tumors, and active intracellular delivery of NPs into cancer cells. Indeed, intracellular delivery of NP therapeutics results in higher drug concentration inside the cells, and thus is more efficacious than non-internalized nanotherapeutics.4–6 In addition, intracellular NP delivery is particularly important for the development of nucleic acid-based therapeutics (e.g., genes and siRNAs), as these macromolecules cannot readily cross the cell membrane.7
For intracellular delivery of NPs, one strategy is to modify their biophysiochemical properties, such as surface topography and charge, allowing for rapid NP internalization.8 This strategy has the limitation of non-specificity whereby NP uptake occurs indiscriminately. The other strategy is to incorporate NPs with targeting ligands, which enhance cellular uptake via receptor-mediated endocytosis and provide cell-targeting specificity.1 Most targeted NPs under pre-clinical and clinical development utilize ligands that are isolated from well-characterized cancer antigens. However, only limited number of antigens have been characterized for cancer cell recognition,9 and some of these characterized antigens cannot mediate the internalization of their assoaciated ligands. Therefore, a robust targeted internalizing NP delivery platform needs to be established where development can be achieved without pre-characterization of target antigens.
Recently, aptamers (Apts) have emerged as a promising class of ligands for targeted NP delivery.3, 10, 11 Apts are single-stranded RNA or DNA oligonucleotides that fold into three-dimensional conformations with high binding affinity and specificity. They have shown low immunogenicity. The relatively small size of Apts allows for more efficient penetration into biological compartments.12 Moreover, Apts can be manipulated and produced by a chemical synthesis process, which is less prone to batch-to-batch variability than other biologic products.13 Because of these favorable features, we used Apts as model ligands to develop a targeted internalizing NP-Apt platform.
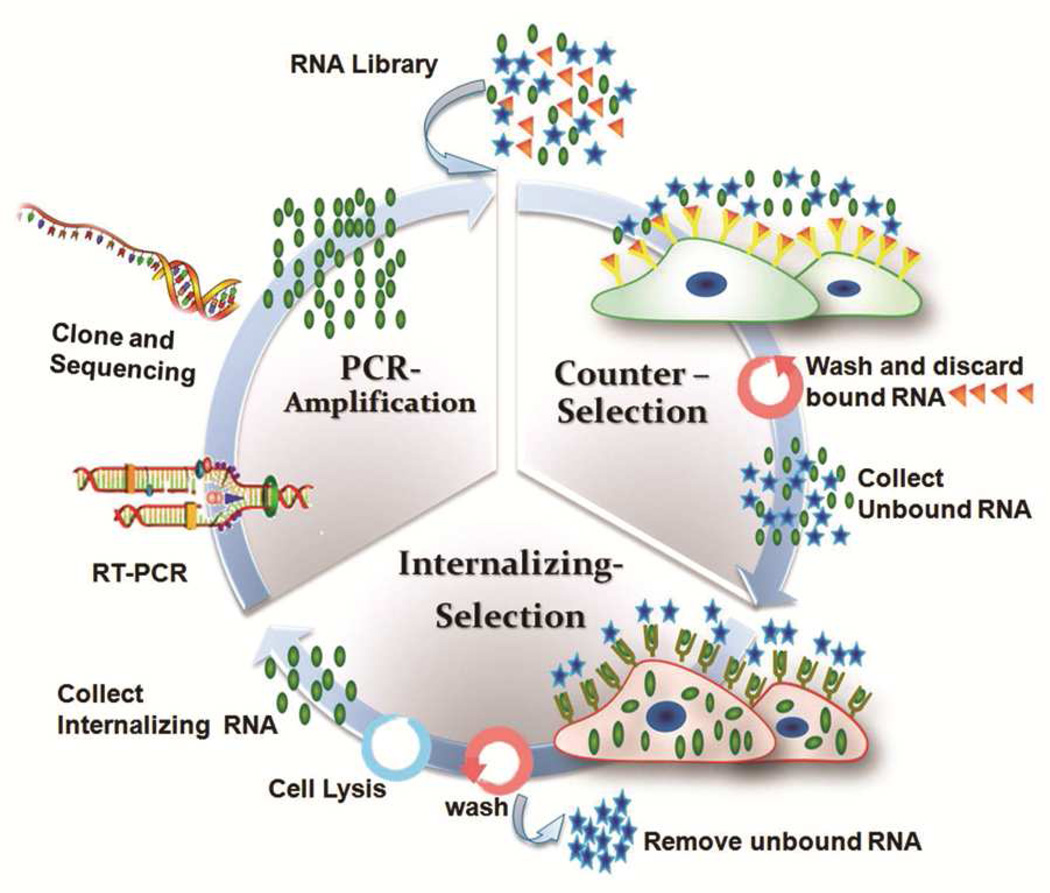
To achieve this goal, we designed a unique selection strategy to enrich internalizing Apts for NP incorporation: First, we chose to isolate Apts directly against live cancer cells, and thus the evolved Apts can recognize cancer cells without pre-characterization of the targeted cancer antigens. Using this strategy, a single selection process potentially generates Apts that can target multiple antigens on cancer cells, which in turn yields a diverse candidate pool of Apts facilitating multi-antigen targeting. Second, stringent counter selections were used to remove Apt candidates that interacted with non-target cells, contributing to the target-cell-specificity of the evolved Apts. Most importantly, the selection was specially designed to enrich internalizing Apts rather than highest affinity Apts as reported in previous SELEX (Systematic Evolution of Ligands by EXponential enrichment) processes,14–18 which may evolve Apts that have bound to cells without internalizing. For example, Shangguan et al. systematically developed “Cell-SELEX” strategy wherein the selection was performed at 4°C to enrich Apts that specifically bound to target cells.17, 19 Among the more than 30 isolated Apts, only one Apt was reported to have the internalization feature.20 Some of other isolated Apts bind to target cells at 4°C, whereas lose their binding capabilities at 37°C, which could hinder their applications as drug delivery vehicles.21 Towards the specific goal, we performed the selection at physiological temperature (37°C), where cells and their membrane receptors are biologically active and continue to function in endocytosis. Additionally, we selectively collected internalizing Apts after removing non-internalized membrane-bound Apts. Moreover, the isolated RNA Apts were introduced with 2' O-methyl (OMe) modification during the selection process, which facilitates the resistance of nuclease degradation inside the intra-cellular environments.22 Characterized by the cellular uptake of the Apts, we termed the process “cell-uptake selection” (Figure 1).
Figure 1.
Schematic protocol of cell-uptake selection for evolving disease-specific internalizing Apts. The 2'OMe-RNA pools, transcribed from initial DNA library, were incubated with prostate normal cells (counter-selection). After washing, the unbound RNAs were presented to prostate cancer cells for binding and cell uptake. After washing or trypsin treatment, and cell lysis, those internalizing RNAs were extracted (cell-uptake selection). The collected RNAs, after reverse transcription, were amplified by PCR. The PCR products were transcribed into 2'-OMe modified RNAs for the next round of selection, or cloned and sequenced for Apt identification in the last-round selection.
As the proof-of-concept demonstration of cell-uptake selection, we isolated herein cell-specific internalizing 2'-OMe RNA Apts against prostate cancer (PCa) cells. The selected PCa-specific internalizing Apts were further characterized and conjugated to drug-encapsulated NPs for targeted PCa therapy.
RESULTS AND DISCUSSION
To demonstrate the robust and predictable features of our selection strategy, we performed two distinct but identical selections against PC3 and LNCaP cells. They represent two distinct PCa epithelial cell lines that differ in their androgen responsiveness: androgen-responsive (LNCaP) and androgen-independent (PC3). RWPE-1 (prostate normal epithelial cell line), BPH-1 (prostate benign hyperplastic epithelial cell line), and PrEC (prostate normal epithelial cell line) have differential surface antigen expression as compared with LNCaP or PC3,23 and serve as model counter-selection cell lines to prevent the collection of RNAs that could bind to common surface antigens present on non-cancer cells. The starting RNA Apt candidate library was composed of 77 base long degradation-resistant RNA oligonucleotides incorporating 2'-OMe modified ATP, CTP, and UTP.24 The partly 2'-OMe-modified oligonucleotides were initially incubated with counter-selection cell lines (RWPE-1, BPH-1, and PrEC) consecutively and the RNA sequences remaining in the supernatant were continually collected. The collected RNAs were incubated with the target cells (either PC3 or LNCaP) at 37°C to allow for binding and cellular uptake. The cells were then extensively washed (rounds 1–12) and either lysed to collect the internalized RNAs (rounds 1–6), or treated with trypsin to remove the majority of membrane-bound RNAs prior to cell lysis and collection of internalized RNAs (rounds 7–12). The stringency of the selection was slowly increased by diminishing both the number of PC3 and LNCaP cells and the incubation time during the selection (rounds 1–12), and further increased by complicating the RNA pools through mutagenic PCR (round 7).25 The progress of selection, measured by the number of PCR cycles needed to amplify the chosen material for the next round, is shown in Figure S1. As rounds of selection progressed, the needed PCR-cycle number steadily decreased from the 3rd round, but did not decrease from 10th up to 12th round, thus indicating the saturation of Apt candidate enrichment.
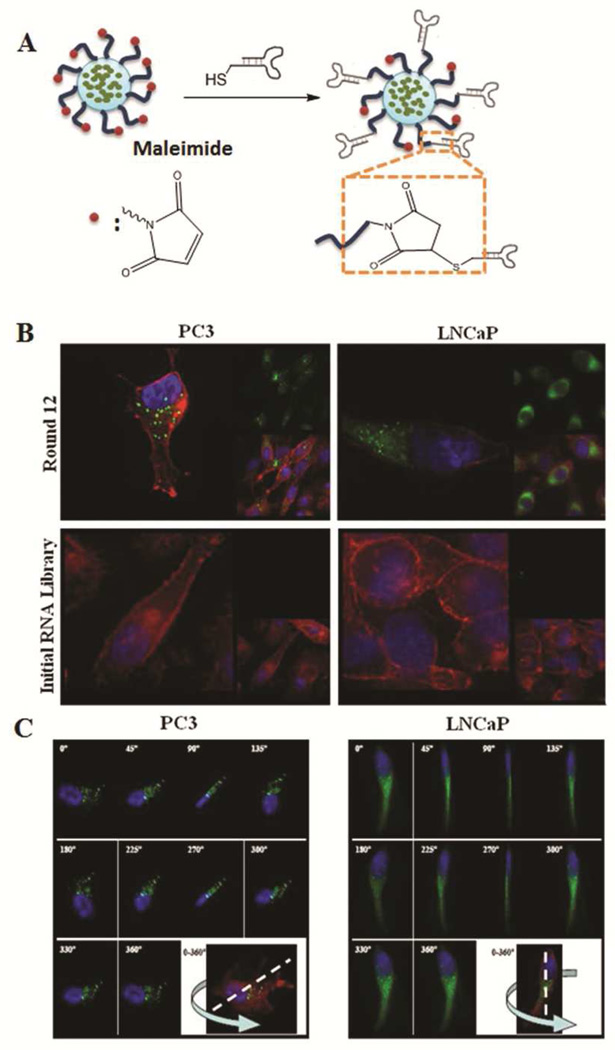
Prior to identification of specific sequences in the round-12 RNA pool, we first confirmed that the enriched RNA pools (round 12 LNCaP and round 12 PC3), which represent many distinct Apt candidates, could be internalized and transported with NPs into target cancer cells. As a model NP platform, we used the hybrid lipid-polymer NP that has been designed and systematically investigated by our group.26–28 The hybrid NP consists of (i) a poly(D,L-lactide-co-glycolide) (PLGA) hydrophobic core for drug encapsulation, (ii) a lipid monolayer, and (iii) a poly(ethylene glycol) (PEG) shell. PEG was conjugated to 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) at one end for interspersing into the lipid monolayer, and was functionalized with maleimide group at the other end for targeting ligand modification. This hybrid NP is prepared in a single-step process via nanoprecipitation and self-assembly, and the yielded NP has the size of 50–100 nm and ζ potential of −10 to −20 mV, providing favourable physiochemical properties for drug delivery application. The conjugation of NP to RNA pool relies on maleimide-thoil chemistry (Figure 2A). Briefly, the vicinal hydroxyl groups in the unmodified 5'-end GTP of RNA pool were oxidized into aldehyde groups by periodate. These aldehyde groups further reacted with free amine group of cystamine to introduce thiol groups. The resulting thiolated RNA pools were then incubated with maleimide-functionalized NPs encapsulating NBD (22-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-23,24-bisnor-5-cholen-3β-ol) to form NP (NBD)-RNA pool bioconjugates. As demonstrated in Figure 2B, the presence of the selected RNA round 12 LNCaP or PC3 pools greatly facilitated the uptake of the green fluorescent NPs into the target LNCaP or PC3 cells, separately. By contrast, control NPs similarly conjugated with the initial random library were not taken up by target cells at detectable levels. Figure 2C represents a panel of images across the Z-axis of a single cell with 3-D image deconvolution demonstrating the intracellular source of fluorescent signal, consistent with NP uptake within LNCaP or PC3 cells. The cell-uptake selection was shown to have successfully enriched a pool of Apt candidates that are specifically internalized by the cancer cells.
Figure 2.
Demonstration of the internalization of selected round 12th RNAs-NP conjugates. A) RNAs and NP were conjugated by using maleimide-thiol chemistry. B) Cellular uptake of selected RNAs-NP conjugates. In all the images, the nucleus is in blue (DAPI), cytoskeleton is in red (Rhodamine Phalloidin), and NP is in green (NBD dye). upper left: NP- PC3 Round12th RNAs conjugates in PC3 cells; lower left: NP-initial RNA library conjugates in PC3 cells; upper right: NP-LNCaP Round 12th RNAs conjugates in LNCaP cells; lower right: NP-initial RNA library conjugates in LNCaP cells. C) Three dimensional reconstruction of cell images confirm the NP-Round 12 RNAs conjugates are inside the PC3 cells (left) and LNCaP cells (right).
We next separately cloned and sequenced the enriched PC3- and LNCaP-round 12 pools by using high-throughput genome sequencing methods. The sequences were sorted into putative families by aligning consensus motifs, and termed XEO1, XEO2, …, etc. XEO2, XEO9 and their homologues represented 12% and 10% of the selected 68 sequences in the PC312th round pool, separately. XEO6 and its homologues represented 14% of the selected 65 sequences in the LNCaP 12th pool. These three abundant sequences, along with their truncated forms (XEO2mini and XEO6mini, described in supporting information, Figure S2), were considered as the best internalizing Apt candidates for further characterization (Table 1).
Table 1.
Sequences of selected internalizing Apts.
| Aptamer Source | Size | Sequences | |
|---|---|---|---|
| PC3 Round 12 |
LNCaP Round 12 |
||
| XE02 | 77 | 5’-GG GAG AGG AGA GAA ACG UUC UCG CUG ACU GAC CUG GCG AGG AUU GAC GCU GAU GGA UCG UUA CGA CUA GCA UCG AUG-3’ |
|
| XEO2mini | 34 | 5’-CAC GAC GCU GAU GGA UCG UUA CGA CUA GCA UCG C-3’ |
|
| XEO6 | 77 | 5’-GG GAG AGG AGA GAA ACG UUC UCG GGC GCG AGA CGA UCC GCU AUG AUG GCU GUG GGA UCG UUA CGA CUA GCA UCG AUG-3’ |
|
| XEO6mini | 50 | 5’-CGG GCG CGA GAC GAU CCG CUA UGA UGG CUG UGG GAU CGU UAC GAC UAG CA-3’ |
|
| XEO9 | 77 | 5’-GG GAG AGG AGA GAA ACG UUC UCG UUU GUG AAU ACG CGC GUU GUC CCU UGA GUG GGA UCG UUA CGA CUA GCA UCG AUG-3’ |
|
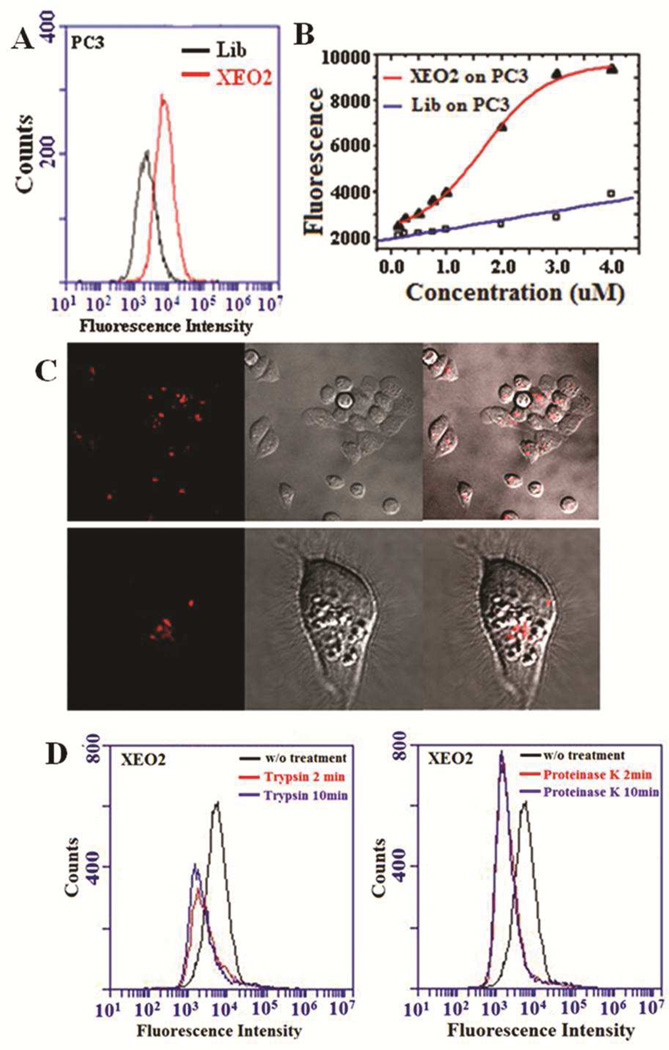
We proceeded to characterize the internalization of the selected Apts. Because specific sequences had been identified, the synthesis, modification and labelling of Apts were directly performed by RNA synthesizers. This solid-phase chemical synthesis process is straightforward and accessible to be scaled up. Cy3-labeled Apts were incubated with target cells (PC3 or LNCaP) at 37°C for 2 h to allow for cellular uptake. Cells were then treated with trypsin to remove the external binding fluorescence signal that could interfere with the detection of the intracellular Apts,20, 29 followed by flow cytometry analysis. Cells were incubated with similarly synthesized Cy3-labeled initial RNA random library as a control and trypsinized to determine non-specific background uptake. Figure 3 shows the representative results from one of the selected Apts (XEO2). Compared with the initial library, the XEO2 profile showed a clear right shift in cytometric analysis, suggesting uptake by PC3 cells (Figure 3A). We further evaluated uptake of Cy3-labeled XEO2 during 2h incubation with various concentrations. The internalization of the selected XEO2 Apt was enhanced in a concentration-dependent fashion and reached a plateau in target PC3 cells (Figure 3B). By comparison, uptake of the initial library showed only a slight linear increase. The difference in the cellular uptake profiles indicates that, unlike the non-specific cellular uptake shown by random sequences, receptor-mediated endocytosis might participate in the specific and efficient cellular uptake of the selected XEO2 Apt.30–32 Confocal images further confirmed the cellular internalization of Cy3-labeled XEO2 (Figure 3C).
Figure 3.
Internalization of Apt XEO2. A) Representative flow cytometric profiles showing XEO2 internalization signals in PC3 cells. The black curve represents the background uptake of unselected initial library. B) Uptake efficiency of XEO2 by PC3 Cy3-labeled XEO2 was incubated with target cells at different concentrations. Fluorescence signals from inside cells were determined by flow cytometry. C) Representative confocal images showing the distributions of Cy3-labeled XEO2 inside PC3 cells. Left: fluorescence image; middle: widefield image; right: overlay of fluorescence and widefield images. D) Effects of trypsin (left) and proteinase K (right) treatment on the binding of XEO2. The PC3 cells were pre-treated with trypsin or proteinase K for 2 or 10 min before incubation with the Apts.
Besides XEO2, the other selected sequences also exhibited cellular uptake into target cancer cells (Table 2, additional examples were shown in Figure S3, S4, S6). Using R value as the criteria (Table 2) to measure internalization capacity, we quantitatively compared selected Apts with a well-studied A10 Apt that bind to prostate-specific membrane antigen (PSMA). A10 gets taken up into PSMA-expressed cells such as LNCaP, but not PC3 cells that do not express PSMA antigens. As shown in Figure S7, the R value of A10 in LNCaP cells was 1.45 (1<R<1.5, ++). As such, the internalization capacity of XEO2, XEO6, XEO6mini (XEO6 truncated form), and XEO9 (R>2, ++++, as summarized in Table 2) was higher than that of A10 (1<R<1.5, ++) in LNCaP cells, indicating the robust feature of “cell-uptake selection” strategy. In addition, our strategy allows, for the first time, to discover a group of new internalizing Apts XEO2, XEO2mini, and XEO9, which can get taken up into PC3 cells with high internalization capacity (R>2, ++++, as summarized in Table 2). To the best of our knowledge, no cancer antigens and targeting Apt ligands have currently been identified for PC3 cells.33 Our strategy has the advantage for enabling the design and engineering of ligand-targeted NPs without prior knowledge of target antigens.
Table 2.
Cellular uptake of selected aptamers by different cell lines.
| Cell lines | Cell Source | XEO2 | XEO2mini | XEO6 | XEO6mini | XEO9 |
|---|---|---|---|---|---|---|
| PC3 | Prostate carcinoma (androgen- independent) | ++++ | ++++ | + | + | ++++ |
| LNCaP | Prostate carcinoma (androgen-dependent) | ++++ | + | ++++ | ++++ | ++++ |
| RWPE-1 | Prostate normal epithelial | ++ | ---- | ---- | ---- | ++ |
| BPH | Prostate benign hyperplastic epithelial | ++ | + | ---- | ---- | ++ |
| HeLa | Cervical carcinoma | ---- | ---- | ---- | ---- | ---- |
| SKBR3 | Breast carcinoma | + | + | + | + | + |
| A375 | Melanoma | ---- | ---- | ---- | ---- | ---- |
| U373MG | Brain glioblastoma-astrocytoma | ---- | ---- | ---- | ---- | ---- |
| T98G | Brain glioblastoma | ---- | ---- | ---- | ---- | ---- |
| U87MG | Brain glioblastoma-astrocytoma | ---- | ---- | ---- | ---- | ---- |
| A549 | Lung carcinoma | ---- | ---- | ---- | ---- | ---- |
| SKOV3 | Ovary adenocarcinoma | ---- | ---- | ---- | ---- | ---- |
Note: The internalization capacity of selected aptamers in different cell lines was evaluated by R value as the following threshold. The mean fluorescence of selected sequence (MF sequence) in the FACS analysis was normalized to the mean fluorescence of initial library (MF lib) in the same experiment condition. R= (MF sequence −MF lib) / MF lib.
R < 0.5;
0.5≤R≤1;
1 < R < 1.5;
1.5 ≤ R < 2;
R > 2.
To ascertain whether these Apts were binding to cell-surface membrane proteins, cells were pre-treated with proteases, including trypsin and proteinase-K, before incubation with Cy3-labeled Apts. For example, although XEO2 showed the binding affinity of 117 nM with PC3 cells (Figure S5), it lost the binding characteristics against target cells after protease treatments (Figure 3D), indicating that its target molecules are most likely membrane proteins. Protease treatment assays similarly showed these selected Apts likely bound to membrane proteins (Figure S8). Further characterization of the protein could lead to the discovery of novel PCa biomarkers.
Taken together, multiple internalizing Apts targeting the same cancer cells were generated from a single selection process. Using multiple Apts for development of NP-Apt conjugates may be most clinically useful, whereas conventional single antigen-targeted NP platforms may be confounded by the heterogeneous pattern of intra- and inter-tumoral antigen expression.34, 35 Such a group of internalizing Apts isolated from our designed selection can collectively interact with multiple antigens on cancer cells, and potentially be utilized to develop a multi-antigen targeted NP platform to address this limitation.
We subsequently assessed the cell-type specificity of selected internalizing Apts. As illustrated in Table 2, Apts XEO2 and XEO9 showed specific uptake into both LNCaP and PC3 cells. Apts XEO6 and XEO6mini showed specific uptake only into LNCaP cells. Apt XEO2mini showed specific uptake only into PC3 cells. All these five sequences showed much less favourable uptake into other cell lines, including BPH, RWPE-1, HeLa, SKBR3, A375, U373MG, T98G, U-87MG, A549, and SKOV-3. The slightly uptake into some of these cells lines may be due to the fact that some biomarkers, which are expressed in prostate cancer cells, are also expressed in non-prostate cancer cells albeit at a relatively lower expression level. For example, PSMA over-expressed in PCa cells, is also expressed at various degrees in normal prostate and other normal tissues, including whole brain, kidney, liver and small intestine,36 and is similarly over-expressed on the neovasculature of most non-prostate solid tumors.37, 38 The XEO2mini, XEO6, and XEO6mini had the most specific internalization profiles among the selected Apts, and thus may be promising for targeted delivery applications.
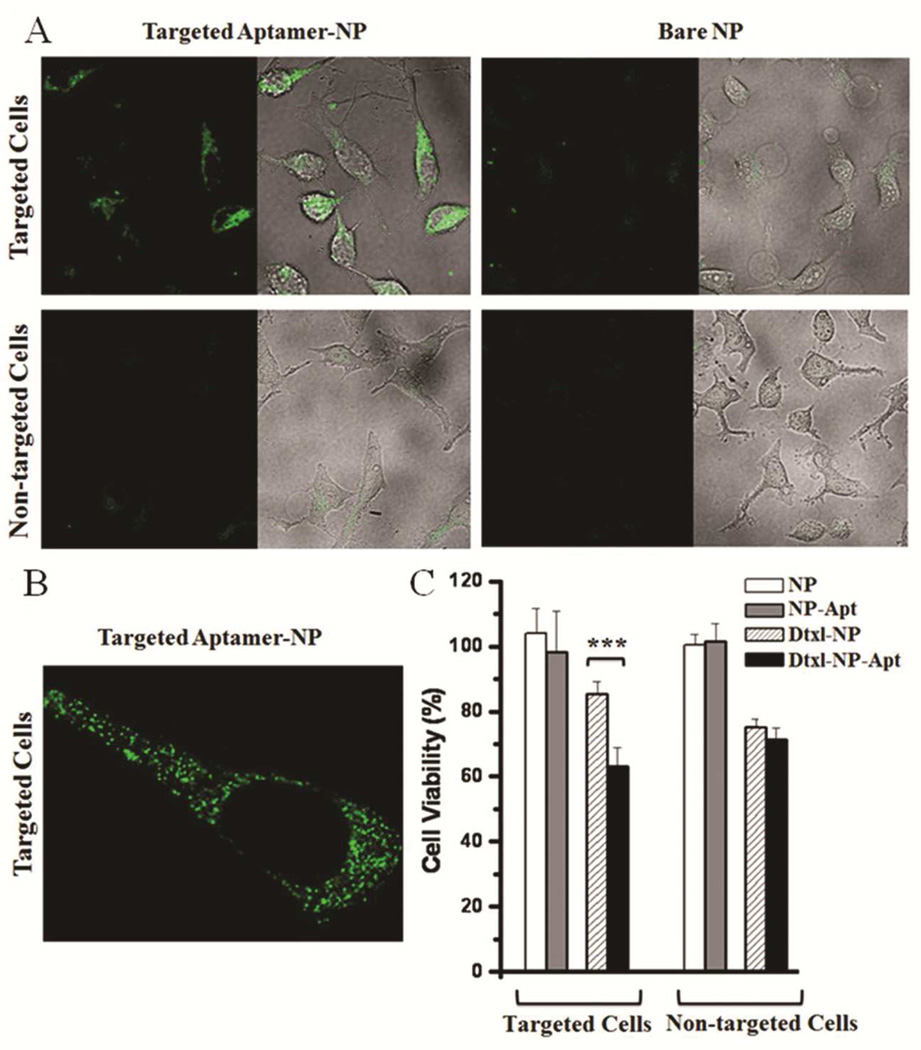
To investigate the feasibility of using the selected internalizing Apts for NP incorporation into potential applications, we used the XEO2mini as a representative Apt to develop a model system of NP-Apt bioconjugates. The conjugation of Apt XEO2mini and NP was achieved by using maleimide-thiol chemistry — the Apt was modified by solid-phase synthesis with a thiol group at its 5' end, and the NP was pre-functionalized with maleimide. We previously have demonstrated the optimal density of A10 Apt on the NP surface for in vitro and in vivo efficacy.39 With the determined optimal density of one Apt per 1180 nm2 of NP surface area,39 we anticipate our NPs with a diameter of 80 nm have approximately the density of 17 Apts per NP. We visualized the cellular uptake of the NP-Apt XEO2mini (NP-Apt) by encapsulating NBD inside the NPs; though for clinical applications, small molecule drugs, siRNAs or other therapeutics may be encapsulated. PC3 and HeLa cells were employed as model target and non-target cell lines, respectively. As shown in Figure 4A, the cellular uptake of NP(NBD)-Apt was significantly enhanced in the target cells compared with that of the non-conjugated NP(NBD). The differential uptake of the NP(NBD)-Apt was not observed in the non-target cells. The background NBD signal represented nonspecific cellular uptake of NPs and any free NBD released from the NPs during incubation. The high magnification imaging (Figure 4B) shows the cellular uptake and cytoplasmic distribution of the NP(NBD)-Apt inside the target cells. In addition, flow cytometry analysis was performed to confirm specific cellular uptake of the targeted NP-Apt (Figure S9).
Figure 4.
A) Representative confocal images showing specific uptake of NP-XEO2mini conjugates in different cells. NBD cholesterol is encapsulated in the NP. In each image, left panel: fluorescent image; right panel: overlay of fluorescence and optical image. Targeted NP-XEO2mini (left) and bare NP (right) incubated with target PC3 cells (top) and non-target HeLa cells (bottom). B) Cellular distributions of NP-XEO2mini (NBD) in PC3 cells by high magnification confocal imaging. C) Cytotoxicity study of Dtxl-encapsulated NP-Apt conjugates (Dtxl-NP-Apt), Dtxl-encapsulated NP without Apt (Dtxl-NP), NP-Apt conjugates without Dtxl (NP-Apt) and control NP without Dtxl (NP). ***, P<0.001 by two sample student’s t-test.
With the model system of the XEO2mini-conjugated NPs, we next investigated its potential efficacy for drug delivery by encapsulating docetaxel (Dtxl) inside the NPs. A control experiment was first performed by incubating the cells with Apt XEO2mini or NPs without drug in both non-conjugated and Apt-conjugated forms. No obvious cytotoxicity was found in either target or non-target cell lines (Figure 4C and Figure S10), confirming the non-cytotoxicity of NPs and Apt XEO2mini. After loading with Dtxl, we observed the differential cytotoxicity of Dtxl-NP in non-target and target cells, which may be due to the differences in the non-specific uptake of NPs and in the IC50 of Dtxl between two cell lines.40–42 To exclude these intrinsic factors, we compared the cytotoxic effects of Dtxl-NP-Apt and Dtxl-NP in the same cell line, and thus each line is its own control. As shown in Figure 4C, the Dtxl-NP-Apt (71.45% ± 3.60) showed similar cytotoxicity to the Dtxl-NP (75.33% 2.21) in non-target cells (mean ± SD, n=5, P>0.05). In contrast, the Dtxl-NP-Apt (63.10% ± 5.81) was significantly more cytotoxic than the Dtxl-NP (85.47% ± 3.65) in target cells (mean ± SD, n=5, P<0.001). The significant increase in cellular cytotoxicity is presumably through Apt-targeted intracellular delivery and release of Dtxl in target cells. Previously, we had developed Dtxl-encapsulated and A10 Apt-targeted NP that bound to extracellular domain of the PSMA protein on the surface of PCa cells, and explored the efficacy of this system in vitro and in vivo.3 In that study, we showed an enhancement in the cytotoxicity of A10-conjugated Dtxl-NP-Apt (42% ± 2) compared with Dtxl-NP lacking the A10 Apt (61% ± 5).3 Our newly-developed internalizing NP-Apt system showed at least equivalent or more favorable enhancement in therapeutic efficacy than A10 Apt-targeted NP delivery system, demonstrating the potential of this system for targeted cancer therapy. More importantly, unlike the A10 targeted NPs which recognized the well characterized PSMA protein, the current platform allows us to develop equally efficacious or better targeted NPs even when the target antigen is unknown.
CONCLUSION
In summary, we have developed a targeted NP platform for cancer therapy by incorporating Apts isolated from a novel cell-uptake selection process. The selection was uniquely designed to enrich cancer cell-specific internalizing Apts rather than highest affinity Apts as reported in previous selection processes. After modifying NPs with these selected Apts, the NP-Apt bioconjugates demonstrated enhanced therapeutic efficacy in target cancer cells. Further engineering of NPs with a diverse pool of Apts would facilitate the development of multi-ligand targeted NP platforms. In this platform, detailed knowledge of the target antigens on the cell surface is not needed, simplifying the process of NP development. Further characterization of the target antigens may lead to the discovery of important PCa biomarkers. This internalizing NP-Apt platform can be similarly applied in a wide variety of other oncologic diseases, and can potentially facilitate the delivery of various molecules, including drugs and siRNAs, into target cells.
METHODS
Cell Lines
LNCaP, PC3, SKBR3, HeLa, RWPE-1, A375, U373MG, T98G, U-87MG, A549, and SKOV-3 were from ATCC (Manassas). BPH-1 was from Vanderbilt University Medical Center (Nashville). PrEC was from Cambrex (Hopkinton). Cells were grown according to the manufacturer’s specifications. All cell lines were used within three to ten passages from their acquisition. The internal authentication has been performed by monitoring growth rate and tracking the changes in morphology.
RNA Library and Primers
The DNA library (~ 9 × 1014) 5'-CATCGATGCTAGTCGTAACGATCC-30N-CGAGAACGTTTCTCTCCTCTCCCTATAGTGAGTCGTATTA-3' (Operon) was amplified by PCR (5 min at 95°C, followed by cycles of 0.5 min at 95°C, 0.5 min at 65°C, and 1 min at 72°C, followed by 2 min at 72°C), with Reverse-Primer 5'-CATCGATGCTAGTCGTAACGATCC-3' and Forward-Primer 5'-TAATACGACTCACTATAGGGAGAGGAGAGAAACGTT CTCG-3'. The resultant dsDNA was precipitated and separated by gel filtration. Partly 2'-O-Methyl-modified RNAs were obtained by overnight incubation at 37 °C of the reaction mixture: 200 nM template, 200 mM HEPES, 40 mM DTT, 10% PEG8000, 0.01% Triton X-100, 2 mM spermidine, 1.0 mM each of GTP, 2'-O-methyl ATP, CTP and UTP (Trilink), 5.5 mM MgCl21.5 mM MnCl210 U/ml inorganic pyrophosphatase (Sigma-Aldrich), and 200 nM T7 RNA polymerase.24 The resultant transcripts were precipitated in 3M LiCl at −80 °C, followed by ethanol precipitation.
Cell-uptake selection
The RNA library (1.5 nmol) was briefly denatured at 90 °C in 20 ml of selection buffer (EBSS with 1 mM MgCl2), cooled slowly and then warmed up to 37 °C before consecutive incubations with three counter-selection cell-lines (RWPE-1, BPH-1, and PrEC). After each incubation (60 min for the first 5 rounds, 45 min afterwards), the unbound RNAs were collected for the next incubation. After consecutive incubations with these three counter-selection cells, the pool was exposed to the positive-selection cells, LNCaP or PC3, with varied incubation time: 60 min for rounds 1–2, 45 min for rounds 3–5 and 30 min for rounds 6–12. The cells were then extensively washed (rounds 1–12) and either lysed to collect the internalized RNAs (rounds 1–6), or treated with trypsin to remove the membrane-bound RNAs prior to cell lysis for the collection of internalized RNAs (rounds 7–12). The internalized RNAs were then extracted using Trizol Reagent (Invitrogen). Selected RNAs were treated with RQ1 DNase (Promega), before reverse-transcription and PCR amplification. To monitor the enrichment of internalizing Apt candidates, a semi-quantitiative PCR method was used to quantify the numbers of PCR cycles carried out to obtain the same amount of PCR products. Briefly, the collected reverse-transcribed DNAs of each round were equivalently separated into several reaction tubes and simultaneously run through different numbers of PCR cycles. The products were loaded in parallel on the agarose gel and quantified by the intensity of specific bands. The experiments were repeated three times to determine the number of cycles necessary to achieve a given amount. The PCR reaction was then repeated by running the desired number of PCR cycles. Subsequently, the PCR products were purified, transcribed into modified RNA, treated with DNase and precipitated with LiCl, followed by ethanol precipitation before starting the next cycle. During the selection, the number of PC3 and LNCaP cells exposed to the RNA library progressively decreased, starting with 1x107 and diminishing by 1–2x106 cells every other round until reaching 1x106 for round 12. After 7 rounds of selection, the material was amplified with 14 cycles of mutagenic PCR, (template DNA=25 µg/µL; MgCl2=7 mM; Tris= 10 mM; KCl=50 mM; primers=2 M; dCTP & dTTP =1 mM; dGTP & dATP=0.2 mM; enzyme=0.05 U/L; and MnCl2=0.5 mM; annealing 3 min) to introduce occasional mutations (roughly 0.79% mutations per position; 0.24% mutations per sequence). After 12 rounds of selection, sequences were cloned into the pCR-4 TOPO plasmid, using the TOPO-TA Cloning Kit (Invitrogen).
Identification of selected pools
To identify the internalization of selected RNA pools, NBD-encapsulated NPs with maleimide groups were prepared using the combination of self-assembly and nanoprecipitation method as previously described.26 The RNA pool was then oxidized to form aldehyde derivatives and cystamine was incubated with RNA for 2 h to functionalize the RNA with a free thiol reacting group. Subsequently, sodium sulphite (2X) was added to remove the excess oxidant and cystamine. NPs were further incubated with RNAs for 12 h at room temperature with gentle stirring to form NP-RNA bioconjugates. For confocal imaging, cells were incubated with NP (NBD)-RNAs in selection buffer for 1h, washed, fixed with 4% formaldehyde, followed by 0.1% Triton-X100, stained with rhodamine-phalloidin, and mounted with DAPI. Cells were visualized with 1.4 NA oil-immersion 25x or 60x objectives, and individual images were taken along their z-axis at 0.1-µm intervals with a confocal microscope (Carl Zeiss).
Measurement of Apt binding affinity
The binding affinity of XEO2 Apt was determined by incubating PC3 cells (5 ×105) at 37 °C for 30 min in the dark with varying concentrations of Cy3-labeled Apt in a 500-µl volume of binding buffer. Cells were then washed twice with 700 µl of the binding buffer with 0.1% sodium azide, suspended in 400 µl of binding buffer with 0.1% sodium azide, and subjected to flow-cytometric analysis within 30 min. The Cy3-labeled unselected RNA library was used as a negative control to determine nonspecific binding. All of the experiments for binding assay were repeated two times. The mean fluorescence intensity of target cells labeled by Apts was used to calculate for specific binding by subtracting the mean fluorescence intensity of nonspecific binding from unselected library RNAs. The equilibrium dissociation constants (Kd) of the Apt–cell interaction were obtained by fitting the dependence of fluorescence intensity of specific binding on the concentration of the Apts to the equation Y = B max X / (Kd +X), using SigmaPlot (Jandel, San Rafael, CA).
Internalization characterization of selected Apts
All the specific sequence candidates and initial libraries were synthesized by a solid-phase process and were directly conjugated with Cy3 at the 5' end (Thermo Sci.), followed by purification using reverse-phase RNA enzyme-free HPLC. For flow cytometry analysis, Cy3-labelled Apts were heated at 95 °C for 5 min, then slowly cooled down to room temperature for 2 h. Cells (105) were then incubated with a serial concentration of Cy3-labelled Apts (125 nM-4 µM for upake efficacy analysis, and 3 µM for cell-specific analysis) in 500 µl binding buffer [4.5 g/l glucose, 1 mM MgCl2, 0.1 mg/ml yeast tRNA, and 1 mg/ml BSA in EBSS] at 37 °C for 2 h. After washing with 700 µl binding buffer (with 0.1 % NaN3), cells were incubated with pre-warmed trypsin (500 µl, 0.25%) / EDTA (0.53 mM) at 37 °C for 10 min. Subsequently, FBS (50 µl) was added, and cells were centrifuged. The cell pellets were washed with binding buffer (700 µl, with 0.1 % NaN3) once again and suspended in 300 µl binding buffer (with 0.1 % NaN3). The fluorescence was determined with a FACScan cytometer (Accuri C6 Cytometers) by counting 20000 events (note: only living cells were counted). For confocal imaging, cells (104) were washed and incubated with Cy3 labeled Apts (200 nM) in binding buffer at 37°C for 2 h. After extensive washing with cold binding buffer three times, the cells were fixed and kept in dark before imaging.
Proteinase Treatment for Cells
Cell monolayers were detached by nonenzymatic cell dissociation solution (Invitrogen Corporation, Carlsbad, CA), filtered with a 40-µm cell strainer (Becton, Dickinson and Company, Franklin Lakes, NJ). PC3 or LNCaP cells (2×105) were incubated with 500µl 0.25% trypsin / 0.53 mM EDTA in HBSS or 0.1 mg/ml proteinase K in PBS at 37°C for 2 and 10 min. FBS was then immediately added to quench the proteinase digestion. After washing with 700 µl binding buffer, the treated cells were incubated with Cy3-labelled Apt (1 µM) in a 500-µl volume of binding buffer at 37°C for 30 min. Cells were then washed twice with 700 µl pre-warmed binding buffer (with 0.1% NaN3), and suspended in 300 µl binding buffer. The cell suspension was transferred into FACS tube with 40-µm cell strainer cap (Becton, Dickinson and Company, Franklin Lakes, NJ), and subjected to flow cytometric analysis within 30 min. The Cy3-labeled Apt under the same condition, but without proteinase treatment, was applied in showing cell-specific binding profile.
Cellular uptake and cytotoxicity study of Apt XEO2mini-NP bioconjugates
NPs were firstly prepared via nanoprecipitation and self-assembly.26 For Apt conjugation, the disulfide-terminated XEO2mini synthesized by IDT were reduced with 5mM of TCEP (Neutral pH, Thermo Scientific) in PBS (PH=7.4) for 30 min at room temperature. Free TCEP was removed by G-25 Sephadex column (Roche Diagnostics). Apt was added into the prepared NPs and incubated for 2 h with gentle stirring, followed by washing with Amicon tubes. To identify the specific cellular uptake of XEO2mini-NP bioconjugates, PC3 or HeLa cells (105) were incubated with pre-warmed binding buffer for 30 min, and then further incubated with 0.5 mg/ml NP-Apt (NBD) or NP (NBD) at 37°C for 30 min. For cytotoxicity studies, the PC3 and HeLa cells (8×103) were seeded in 96-well plates to allow growth for 24 h. On the day of the experiment, the cells were washed once and incubated with pre-warmed binding buffer for 30 min. With the addition of Dtxl-NP-Apt, Dtxl-NP (100 µg/ml, Dtxl with 5% weight ratio of PLGA), and XEO2mini Apt (2.5 µM) the cells were further incubated in binding buffer for 30 min. The cells were then washed twice, and fresh media were added for further growth for 48 h. Cell viability was measured by MTT cell proliferation assay kits (Invitrogen).
Supplementary Material
Acknowledgment
Etgar Levy-Nissenbaum dedicates this work in memory of Emilia Levy. We thank Jack W. Szostak, Brian Haines and Noam Shomron for helpful discussions and suggestions. We thank Rosa Larralde-Ridaura for helping with the sequence identification. This research was supported by National Institutes of Health Grants CA151884 and EB003647, the David Koch—Prostate Cancer Foundation Award in Nanotherapeutics, and the USA Department of Defence Prostate Cancer Research Program PC 051156.
Footnotes
Supporting Information Available:
Figures for selection progress identified by the number of PCR cycles, predicted secondary structures of Apts XEO2 and XEO6, XEO2mini internalization by flow cytometry and confocal analysis, binding curve of Apt XEO2; internalization profiles of XEO2 and A10 in LNCaP cells, effects of proteinase K or trypsin treatment on XEO9 and XEO2mini binding profile, targeted delivery of NP-XEO2mini (NBD) by flow cytometry analysis, and cytotoxicity study of XEO2mini Apt in PC3 cells and HeLa cells. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Davis ME, Chen ZG, Shin DM. Nanoparticle Therapeutics: An Emerging Treatment Modality for Cancer. Nat. Rev. Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 2.Langer R. Drug Delivery and Targeting: Review. Nature. 1998;392:5–10. [PubMed] [Google Scholar]
- 3.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted Nanoparticle-Aptamer Bioconjugates for Cancer Chemotherapy in Vivo. Proc. Natl. Acad. Sci. U S A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugano M, Egilmez NK, Yokota SJ, Chen FA, Harding J, Huang SK, Bankert RB. Antibody Targeting of Doxorubicin-Loaded Liposomes Suppresses the Growth and Metastatic Spread of Established Human Lung Tumor Xenografts in Severe Combined Immunodeficient Mice. Cancer Res. 2000;60:6942–6949. [PubMed] [Google Scholar]
- 5.Sapra P, Allen TM. Internalizing Antibodies Are Necessary for Improved Therapeutic Efficacy of Antibody-Targeted Liposomal Drugs. Cancer Res. 2002;62:7190–7194. [PubMed] [Google Scholar]
- 6.Park JW, Hong K, Kirpotin DB, Colbern G, Shalaby R, Baselga J, Shao Y, Nielsen UB, Marks JD, Moore D. Anti-Her2 Immunoliposomes: Enhanced Efficacy Attributable to Targeted Delivery. Clin. Cancer Res. 2002;8:1172–1181. [PubMed] [Google Scholar]
- 7.Whitehead KA, Langer R, Anderson DG. Knocking Down Barriers: Advances in Sirna Delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petros RA, DeSimone JM. Strategies in the Design of Nanoparticles for Therapeutic Applications. Nat. Rev. Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 9.Sawyers CL. The Cancer Biomarker Problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 10.Cao Z, Tong R, Mishra A, Xu W, Wong GC, Cheng J, Lu Y. Reversible Cell-Specific Drug Delivery with Aptamer-Functionalized Liposomes. Angew. Chem. Int. Ed. Engl. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 11.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-Aptamer Bioconjugates: A New Approach for Targeting Prostate Cancer Cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 12.Bouchard PR, Hutabarat RM, Thompson KM. Discovery and Development of Therapeutic Aptamers. Annu. Rev. Pharmacol. Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 13.Keefe AD, Pai S, Ellington A. Aptamers as Therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. In Vitro Selection of Rna Molecules That Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: Rna Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A Tenascin-C Aptamer Identified by Tumor Cell Selex: Systematic Evolution of Ligands by Exponential Enrichment. Proc. Natl. Acad. Sci. U S A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raddatz MS, Dolf A, Endl E, Knolle P, Famulok M, Mayer G. Enrichment of Cell-Targeting and Population-Specific Aptamers by Fluorescence-Activated Cell Sorting. Angew. Chem. Int. Ed. Engl. 2008;47:5190–5193. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 19.Sefah K, Shangguan D, Xiong X, O'Donoghue MB, Tan W. Development of DNA Aptamers Using Cell-Selex. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 20.Xiao Z, Shangguan D, Cao Z, Fang X, Tan W. Cell-Specific Internalization Study of an Aptamer from Whole Cell Selection. Chemistry. 2008;14:1769–1775. doi: 10.1002/chem.200701330. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA Aptamer-Micelle as an Efficient Detection/Delivery Vehicle toward Cancer Cells. Proc. Natl. Acad. Sci. U S A. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic Characterization of Ribonuclease-Resistant 2'-Modified Hammerhead Ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 23.Liu B, Conrad F, Cooperberg MR, Kirpotin DB, Marks JD. Mapping Tumor Epitope Space by Direct Selection of Single-Chain Fv Antibody Libraries on Prostate Cancer Cells. Cancer Res. 2004;64:704–710. doi: 10.1158/0008-5472.can-03-2732. [DOI] [PubMed] [Google Scholar]
- 24.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C. Direct in Vitro Selection of a 2 -O-Methyl Aptamer to Vegf. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Cadwell RC, Joyce GF. Randomization of Genes by Pcr Mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Chan JM, Gu FX, Rhee JW, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. Self-Assembled Lipid--Polymer Hybrid Nanoparticles: A Robust Drug Delivery Platform. ACS Nano. 2008;2:1696–1702. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan JM, Zhang L, Yuet KP, Liao G, Rhee JW, Langer R, Farokhzad OC. Plga-Lecithin-Peg Core-Shell Nanoparticles for Controlled Drug Delivery. Biomaterials. 2009;30:1627–1634. doi: 10.1016/j.biomaterials.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Chan JM, Zhang L, Tong R, Ghosh D, Gao W, Liao G, Yuet KP, Gray D, Rhee JW, Cheng J, et al. Spatiotemporal Controlled Delivery of Nanoparticles to Injured Vasculature. Proc. Natl. Acad. Sci. U S A. 2010;107:2213–2218. doi: 10.1073/pnas.0914585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CCN, Castro JE, Motta M, Cottam HB, Kyburz D, Kipps TJ, Corr M, Carson DA. Selection of Oligonucleotide Aptamers with Enhanced Uptake and Activation of Human Leukemia B Cells. Hum. Gene Ther. 2003;14:849–860. doi: 10.1089/104303403765701141. [DOI] [PubMed] [Google Scholar]
- 30.Perera RM, Zoncu R, Johns TG, Pypaert M, Lee FT, Mellman I, Old LJ, Toomre DK, Scott AM. Internalization, Intracellular Trafficking, and Biodistribution of Monoclonal Antibody 806: A Novel Anti-Epidermal Growth Factor Receptor Antibody. Neoplasia. 2007;9:1099–1110. doi: 10.1593/neo.07721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen HM, Cahill CM, McPherson PS, Beaudet A. Receptor-Mediated Internalization of [3h]-Neurotensin in Synaptosomal Preparations from Rat Neostriatum. Neuropharmacology. 2002;42:1089–1098. doi: 10.1016/s0028-3908(02)00054-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu CC, Castro JE, Motta M, Cottam HB, Kyburz D, Kipps TJ, Corr M, Carson DA. Selection of Oligonucleotide Aptamers with Enhanced Uptake and Activation of Human Leukemia B Cells. Hum. Gene Ther. 2003;14:849–860. doi: 10.1089/104303403765701141. [DOI] [PubMed] [Google Scholar]
- 33.Larkin SE, Zeidan B, Taylor MG, Bickers B, Al-Ruwaili J, Aukim-Hastie C, Townsend PA. Proteomics in Prostate Cancer Biomarker Discovery. Expert Rev. Proteomics. 2010;7:93–102. doi: 10.1586/epr.09.89. [DOI] [PubMed] [Google Scholar]
- 34.Heppner GH. Tumor Heterogeneity. Cancer Res. 1984;44:2259–2265. [PubMed] [Google Scholar]
- 35.Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative Splicing and Biological Heterogeneity in Prostate Cancer. Nature Reviews Urology. 2009;6:454–460. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 36.O'Keefe DS, Bacich DJ, Heston WD. Comparative Analysis of Prostate-Specific Membrane Antigen (Psma) Versus a Prostate-Specific Membrane Antigen-Like Gene. Prostate. 2004;58:200–210. doi: 10.1002/pros.10319. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Moy P, Kim S, Xia Y, Rajasekaran A, Navarro V, Knudsen B, Bander NH. Monoclonal Antibodies to the Extracellular Domain of Prostate-Specific Membrane Antigen Also React with Tumor Vascular Endothelium. Cancer Res. 1997;57:3629–3634. [PubMed] [Google Scholar]
- 38.Chang SS, Reuter VE, Heston WDW, Bander NH, Grauer LS, Gaudin PB. Five Different Anti-Prostate-Specific Membrane Antigen (Psma) Antibodies Confirm Psma Expression in Tumor-Associated Neovasculature. Cancer Res. 1999;59:3192–3198. [PubMed] [Google Scholar]
- 39.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Precise Engineering of Targeted Nanoparticles by Using Self-Assembled Biointegrated Block Copolymers. Proc. Natl. Acad. Sci. U S A. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Istomin YP, Zhavrid EA, Alexandrova EN, Sergeyeva OP, Petrovich SV. Dose Enhancement Effect of Anticaner Drugs Associated with Increased Temperature in Vitro. Exp. Oncol. 2008;30:56–59. [PubMed] [Google Scholar]
- 41.Fizazi K, Sikes CR, Kim J, Yang J, Martinez LA, Olive MC, Logothetis CJ, Navone NM. High Efficacy of Docetaxel with and without Androgen Deprivation and Estramustine in Preclinical Models of Advanced Prostate Cancer. Anticancer Res. 2004;24:2897–2903. [PubMed] [Google Scholar]
- 42.Takara K, Sakaeda T, Yagami T, Kobayashi H, Ohmoto N, Horinouchi M, Nishiguchi K, Okumura K. Cytotoxic Effects of 27 Anticancer Drugs in Hela and Mdr1-Overexpressing Derivative Cell Lines. Biol. Pharm. Bull. 2002;25:771–778. doi: 10.1248/bpb.25.771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.