Abstract
Background
Exome sequencing is a recently implemented method to discover rare mutations for Mendelian disorders. Less is known about its feasibility to identify genes for complex traits. We used exome sequencing to search for rare variants responsible for a complex trait, low levels of serum high-density lipoprotein cholesterol (HDL-C).
Methods and Results
We conducted exome sequencing in a large French Canadian family with 75 subjects available for study of which 27 had HDL-C values < the 5th age-sex specific population percentile. We captured ~50 Mb of exonic and transcribed sequences of three closely related family members with HDL-C levels <5th age-sex percentiles and sequenced the captured DNA. Approximately 82,000 variants were detected in each individual of which 41 rare non-synonymous variants were shared by the sequenced affected individuals after filtering steps. Two rare non-synonymous variants in the ATP-binding cassette, sub-family A (ABC1), member 1 (ABCA1) and lipoprotein lipase (LPL) genes predicted to be damaging were investigated for co-segregation with the low HDL-C trait in the entire extended family. The carriers of either variant had low HDL-C levels and the individuals carrying both variants had the lowest HDL-C values. Interestingly, the ABCA1 variant exhibited a sex effect which was first functionally identified, and subsequently, statistically demonstrated using additional French Canadian families with ABCA1 mutations.
Conclusions
This complex combination of two rare variants causing low HDL-C in the extended family would not have been identified using traditional linkage analysis, emphasizing the need for exome sequencing of complex lipid traits in unexplained familial cases.
Keywords: genetics, HDL cholesterol, Exome sequencing, Rare variants
Introduction
Low HDL-C is the most common lipoprotein abnormality and established risk factor of coronary heart disease (CHD). Low HDL-C is caused by multiple genetic factors, common and rare, interacting with one another and with the environment and behavior. In the last two decades, significant effort has been devoted to the identification of low HDL-C susceptibility genes. This was initially done using the genome-wide linkage analysis.1–2 However, progress in identification of the actual disease genes was very slow despite the discovery of many linked intervals. More recently, genome-wide association studies (GWAS) have successfully identified multiple common variants associated with decreased levels of HDL-C.3
However, the sum of common variants identified so far through GWAS explains only a small fraction (10–15%) of the variance in the HDL-C levels.3 Hence, it has become evident that other types of DNA variants must contribute substantially to HDL-C levels as well. To identify new rare and low-frequency variants underlying low HDL-C, massive parallel sequencing technologies can be utilized. The whole-genome sequencing is the most complete approach, but it remains significantly more expensive than exome sequencing that only analyzes coding and transcribed regions which constitute less than 5% of the whole genome sequence.4 It is estimated that the protein coding regions of the human genome constitute about 85% of the disease-causing mutations.4
We used whole exome sequencing to search for rare variants conferring susceptibility to low HDL-C. We sequenced the exomes of closely related family members with low HDL-C from a large multigenerational French Canadian family with 75 subjects available for study and followed up the candidate variants by examining the co-occurrence patterns in the entire extended family.
Methods
Study samples
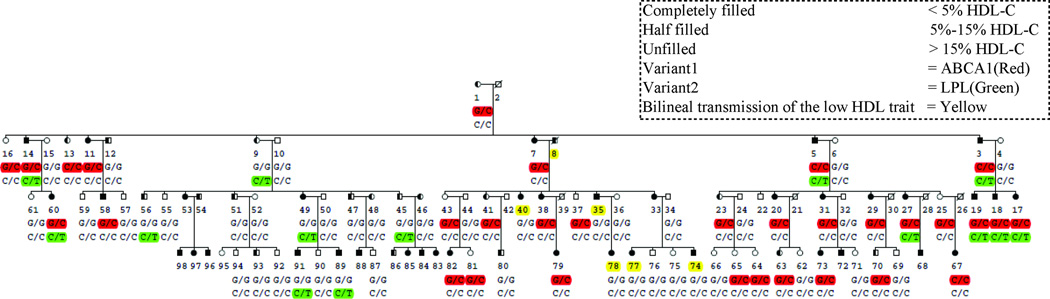
The study sample consists of a large multigenerational French Canadian family collected in the Cardiovascular Genetics Laboratory, McGill University Health Centre, Royal Victoria Hospital, Montreal, Canada, as described previously.5 There are 75 family members (35 males and 40 females) with both DNA and extensive demographic and clinical phenotype information available for study in this family. We selected three closely related family members with HDL-C levels ≤5th age-sex percentile from the uppermost generations (figure 1) for exome sequencing to focus on most severe cases and avoid genetic heterogeneity typical for complex lipid traits.
Figure 1.
ABCA1 and LPL variants co-segregate with low HDL-C in the multigenerational French Canadian low HDL-C family with 75 (35 males and 40 females) genotyped family members. All of the affected subjects who have HDL-C<the 5th age-sex specific population percentile comprise risk alleles for either one of the two variants or both, except in one separate branch the low HDL-C traits appears to be inherited from the affected spouse’s side (indicated in yellow). The subjects with both variants have a lower HDL-C than the subjects with only one variant. The subjects whose samples were exome sequenced are indicated by an arrow.
For a gene-sex interaction analysis, 10 additional French Canadian families with previously identified mutations in ABCA16–8 comprising 125 individuals were also included in the study. The affection status in all families was determined using the 5th age-sex specific population percentile of HDL-C.5 Family members were sampled (blood collection for lipoprotein analyses, DNA isolation for genetic studies and skin biopsy for culture of skin fibroblasts used in cellular cholesterol efflux assays) after a 12-h fast and discontinuation of lipid modifying medications for >4 weeks. Lipids and lipoproteins were measured using standardized techniques as described previously.6,9 The research protocol was approved by the Research Ethics Board of the McGill University Health Center, and all subjects gave informed consent.
Library construction and sequencing
Library construction was performed using 3 µg of genomic DNA and Agilent SureSelect All Exon Kit (50-Mb design) according to the manufacturer’s instructions. Further details of library construction and sequencing are given in the online-only Data Supplemental methods.
Data analysis
Exome sequencing
We converted the qseq files into a Sanger-formatted FASTQ files that were aligned to a reference sequence (hg19) using the default options of the Burroughs-Wheeler Aligner (BWA).10 Duplicates were removed and a pileup file was generated using SAMtools.11 The pileup file was used to run the quality control metrics including: a minimum read depth of 4, a maximum read depth of 600, a maximum of two SNPs per a window size of 10 bases, a minimum indel score of 25 for filtering nearby SNPs and Phred quality >40. The BED file supplied by Agilent was used to filter only those reads corresponding to the 50 Mb targets.
Annovar was used for functional annotation, dividing the variants into coding and non-coding variants.12 The coding variants were further divided into synonymous, nonsynonymous (missense), and stop gain or stop loss variants. The synonymous variants were subsequently discarded because they are less likely to be causal. The variants were filtered against the variants present in the HapMap,13 The 1000 Genomes Project14 and dbSNP13214 databases. Along with novel variants, we selected known rare variants with a minor allele frequency (MAF) <5%. These variants were classified into damaging and benign based on their predicted protein effect using PolyPhen15 and SIFT.16
Parametric linkage and association analysis
Two-point parametric linkage analysis was performed in the extended family using the ‘Location-Score’ option of the Mendel software17 as described in detail in the online-only Data Supplemental methods. Association analysis was performed using a measured genotype approach utilizing the ‘Polygenic-QTL’ option of Mendel,18 using continuous HDL-C levels with age and sex as covariates and allele counts of either the ABCA1 variant, LPL variant, both variants or none (i.e. null model). The heritability and variance explained were calculated as the percent change in total and genetic variance between the null model and the models including the genotypes as covariates. The LPL variant was further tested for association with log transformed TG values in a similar fashion.
Genotype by sex interaction
We included the extended family together with 10 additional families with previously identified mutations in ABCA16–8 in a gene-sex interaction analysis, comprising 200 individuals and 9 different mutations in ABCA1 (DelED1893, G616V, K776N, N1800H, Q2210H, R1851X, R2084X, R909X and S1731C). Genotype by sex interaction was tested by the SOLAR program19 using variance-component analysis for discrete traits. We compared models with and without the gene-sex interaction term while keeping the ABCA1 genotypes in both the null and interaction model. We assumed a dominant genetic inheritance, classifying carriers of a mutation as 1 and 0 otherwise, and a multiplicative interaction term, multiplying the genotype score by sex (men=1 and women=0). We also coded a sex-interaction term in which men and post-menopausal women (≥50 years) were coded as 1 and pre-menopausal women (<50 years) were coded as 0. Subjects with HDL-C levels <the age-sex specific 10th percentiles were classified as affected and subjects with HDL-C levels >the age-sex specific 20th percentiles as unaffected. P-values were generated by comparing the two models using a likelihood ratio statistic with one degree of freedom. Since the affection status is adjusted for gender, the inclusion of the main effect of sex in the model was no longer necessary. The binary HDL-C affection was tested because the variance of HDL-C levels in these ascertained families is reduced and thus limited for effective quantitative analysis.1
Cell culture
Human skin fibroblasts were obtained from 3.0-mm punch biopsies of the forearm of a healthy control subject and the affected proband homozygous for the ABCA1 S1731C variant. The fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 0.1% nonessential amino acids, penicillin (100 units/ml), streptomycin (100 µg/ml), and 10% fetal bovine serum.
Cellular cholesterol efflux assays
Cholesterol efflux was performed as described previously20–21 with minor modifications. Further details of cellular cholesterol efflux assay are given in the online-only Data Supplemental methods.
Results
Exome sequencing
To identify rare genetic variants underlying low HDL-C, we sequenced the entire exomes (~50 Mb) of 3 family members with HDL-C less than the 5th age-sex specific population percentile from a large multigenerational French Canadian family. The three sequenced family members were closely related, as one affected, his sibling and child were sequenced. Exome capture and sequencing were performed using the Agilent Sure Select in-solution method and Illumina Hiseq2000 platform as described in the Methods. We obtained an average of 90 million reads per person and successfully mapped ~90% of these reads to the reference sequence (Table 1). After quality control the mean coverage was 50X.
Table 1.
Summary of reads mapped to the human reference genome (hg19).
| Samples | L range* | H range† | Average |
|---|---|---|---|
| Total reads | 87816621 | 103315979 | 95566300 |
| Mapped reads | 80911740(82.45%) | 85185325(92.14%) | 83048533(87.29%) |
| Duplicate reads | 24513472(27.91%) | 34633913(33.52%) | 29573693(30.72%) |
| Unmapped reads | 6904881(7.86%) | 18130654(17.55%) | 12517768(12.70%) |
| Uniquely mapped reads | 50551412(48.93%) | 56398268(64.22%) | 53474840(56.58%) |
The lowest range limit of the total reads.
The highest range limit of the total reads.
Filtering of identified variants
On average 82,000 SNVs were detected in each individual. We focused on variants shared by all three exome sequenced subjects and filtered the variants based on their type, frequency, and functional predictions. Filtering for missense and stop gain or stop loss variants that were shared by all three affected individuals resulted in 3,428 non-synonymous variants and 31 stop gain or loss variants (Table 2). The transition/transversion (Ti/Tv) ratio of the coding variants was 3.4, whereas the Ti/Tv ratio of the non-coding was 2.5, in good agreement with the expected ratios.22
Table 2.
Number of variants shared by the 3 sequenced affected family members after a series of filtering steps.
| Variant Filter | Variants | Nonsynonymous | Stop gain or loss | Total |
|---|---|---|---|---|
| Shared by all 3* | Known† | 3389 | 30 | 3419 |
| Novel | 39 | 1 | 40 | |
| Total | 3428 | 31 | 3459 | |
| MAF<5% and new‡ | Known | 293 | 3 | 296 |
| Novel | 39 | 1 | 40 | |
| Total | 332 | 4 | 335 | |
| Damaging§ | Known | 52 | 3 | 55 |
| Novel | 24 | 1 | 25 | |
| total | 76 | 4 | 80 | |
| Functional# | Known | 18 | 3 | 21 |
| Novel | 19 | 1 | 20 | |
| Total | 37 | 4 | 41 |
Nonsynonymous and stop gain or loss variants shared by all three sequenced individuals.
Present in the HapMap, The 1000 Genomes Project and/or dbSNP132 databases.
Novel variants (not present in the HapMap, The 1000 Genomes Project and/or dbSNP132 databases) and known rare variants with a MAF<5%.
Novel and known rare (MAF<5%) variants predicted to be damaging either by PolyPhen and/or SIFT.
The novel and known rare (MAF<5%) variants located in genes functionally relevant to lipids; or in genes expressed in relevant tissue; and/or present within 500 kb of the 95 known lipid GWAS loci.9
The identified variants were further filtered against variants present in the HapMap13, 1000 Genomes Project14, and dbSNP13214 databases, resulting in 332 novel variants and known variants with MAF<5%. These variants were further filtered by selecting variants predicted to affect protein function using PolyPhen15 and SIFT16 and expressed in a relevant tissue including liver, adipose, and heart, resulting in 41 shared potentially functional variants that were either novel or known but relatively rare (Table 2). Among the shared variants there were two rare functional variants in the ABCA1 and LPL genes that are excellent susceptibility candidates as their key role in HDL-C metabolism is well established.23 We confirmed their presence by both Sanger sequencing and genotyping.
Rare missense variant in ABCA1
The ABCA1 (S1731C) variant is not present in dbSNP13214 or The 1000 Genomes Project14 data and is located in exon 38. This previously reported rare variant is changing a conserved amino acid from serine to cysteine and is known to result in decreased cholesterol efflux.7,24–26
In order to further determine the effect of the S1731C variant on cholesterol efflux, we used human fibroblasts from the affected proband homozygous for the S1731C variant, and compared these cells to a normal control. Assays were performed in 22OH/9CRA stimulated fibroblasts (to induce ABCA1 expression), and unstimulated cells, in the presence or absence of lipid free ApoA-I (Figure 2A). We observed a significant decrease (~40%) in apoA-I-mediated cellular cholesterol efflux in the proband, as compared to the control without the ABCA1 variant (P=1.23×10−4 using Student’s t-test and P=0.0495 using a non-parametric two-sample Wilcoxon rank sum test). These results are in agreement with previously documented findings.7,25,26 Low efflux levels were also observed in unstimulated cells, presumably due to basal levels of ABCA1 expression and the presence of other apoA-I binding sites at the cell surface. Also, as expected, background basal conditions of passive diffusion of cellular cholesterol were not affected by mutations at the ABCA1 gene locus.
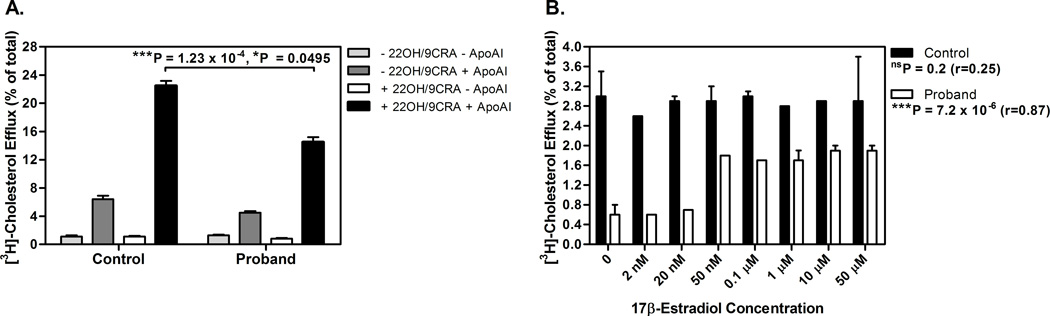
Figure 2.
A. Effect of the ABCA1 variant on cholesterol efflux in fibroblasts from a proband homozygous for S1731C and a healthy control. Fibroblasts were isolated by taking a biopsy from the forearm of the proband and a healthy individual, plated in 12-well plates and radiolabeled with [3H]-cholesterol for 48 hours. Cholesterol efflux was performed as described in Methods under background diffusion conditions (−22OH −ApoA-I), unstimulated (−22OH +ApoA-I) and stimulated (+22OH −ApoA-I, +22OH +ApoA-I) conditions, with or without ApoA-I. The proband had a significantly reduced ApoA-I-mediated efflux as compared to the control without the variant. Values represent the mean ±S.D. from triplicate wells. Results shown are a representative of three independent experiments. ***P=1.23×10−4 by Student's t-test; and *P=0.0495 using a non-parametric two-sample Wilcoxon rank sum test. 22OH indicates 22(R)-hydroxycholesterol; 9CRA, 9-cis-retinoic acid; and ApoA-I, apolipoprotein A-I.
B. Elevated concentrations of 17β-estradiol improve cholesterol efflux in the male proband with the ABCA1 S1731C variant. Fibroblasts from a male proband with the ABCA1 S1731C variant and a healthy male control were isolated by taking a biopsy from the forearm of plated in 24-well plates and radiolabeled with [3H]-cholesterol for 24 hours. Cholesterol efflux was performed as described in Methods, with addition of increasing concentrations (2 nM, 20 nM, 50 nM, 0.1µM, 1 µM, 10 µM and 50 µM) of 17β-estradiol while stimulating ABCA1 expression with 22OH/9CR for 17 hours. As in Figure 2A, experiments were done under four conditions (−22OH −ApoA-I, −22OH +ApoA, +22OH −ApoA-I, +22OH +ApoA-I). Efflux results were subsequently adjusted for background basal conditions of passive diffusion. The final stimulated ApoA-I mediated efflux condition is shown. Upon exposure to increasing estradiol concentrations (>20 nM), cholesterol efflux in the S1731C proband significantly increases. Of note, the overall [3H]-cholesterol efflux counts were lesser in magnitude than those observed in Figure 2A given the shorter labelling time period (24 hours). In addition, the difference in efflux between the control and proband was greater than in Figure 2A, given the different basal diffusion of the selected conditions (data not shown), which was now removed from the net ApoAI-mediated efflux. Values represent the mean ±S.D. from triplicate wells. Results shown are representative of three independent experiments. 22OH indicates 22(R)-hydroxycholesterol; 9CRA, 9-cis-retinoic acid; and ApoA-I, apolipoprotein A-I. ***P=7.2×10−6 (r=0.78) using a non-parametric Spearman trend test for the dose effect on efflux in the S1731C proband; and nsP=0.2 (r=0.25) using a non-parametric Spearman trend test for the dose effect on efflux in the wildtype control.
As the lipid levels of the ABCA1 S1731C variant carriers suggested a possible gene-gender effect (Tables 3–4), we further investigated whether exposure to 17β-estradiol steroid hormone endogenously expressed in females, possibly corrects the cholesterol efflux defect in fibroblasts from the S1731C male ABCA1 carrier during the 22OH/9CRA ABCA1 stimulation phase of 17 hours (Figure 2B). Interestingly, after adjusting for basal cholesterol diffusion, we observed that upon treatment with elevated doses of estradiol (>20 nM), efflux in the S1731C proband significantly increased (P=7.2×10−6, r=0.78 using a non-parametric Spearman trend test), while that in the wildtype control remained constant (P=0.2, r=0.25) (Figure 2B). Taken together, these results support a genotype-sex interaction effect, as hormonal regulation with 17β-estradiol partially restored the low efflux observed in the S1731C male proband but had no significant effect on the efflux of a wild-type control.
Table 3.
The lipid levels and other clinical characteristics of the three individuals that were exome sequenced.
| IND ID | ABCA1 | LPL | TC | TG | HDL-C | HDL % | BMI | LDL-C | AGE | SEX |
|---|---|---|---|---|---|---|---|---|---|---|
| Ind5 | C/C | C/T | 4.61 | 1.3 | 0.67 | <5 | 22.02 | 3.36 | 66 | Male |
| Ind14 | G/C | C/T | 4.6 | 6.1 | 0.62 | <5 | 25.81 | NA | 55 | Male |
| Ind19 | G/C | C/T | 2.43 | 2.4 | 0.62 | <5 | 27.22 | 0.7 | 21 | Male |
The lipid levels are shown in millimoles per liter.
Table 4.
The mean lipid levels and other clinical characteristics of individuals with different ABCA1 (S1731C) and LPL (P234L) genotypes.
| ABCA1/LPL | All Individuals |
TC | TG | HDL-C | AGE | BMI | LDL-C |
|---|---|---|---|---|---|---|---|
| CC/CC | 2 | 3.76(1.29) | 0.72(0.01) | 0.94(0.30) | 35(33.94) | 19.45(7.35) | 1.67(0.71) |
| CC/CT | 2 | 3.60(1.44) | 3.81(3.61) | 0.52(0.22) | 61(7.07) | 23.17(1.63) | 2.19(1.65) |
| GC/CC | 22 | 5.11(1.84) | 1.22(0.69) | 0.95(0.21) | 37(20.04) | 20.65(3.27) | 3.53(1.92) |
| GC/CT | 6 | 4.13(1.25) | 2.97(1.99) | 0.68(0.07) | 34.3(11.36) | 24.36(3.18) | 2.03(1.90) |
| GG/CC | 37 | 4.88(1.24) | 1.60(0.90) | 1.16(0.35) | 34.6(17.03) | 22.64(5.67) | 2.94(1.13) |
| GG/CT | 6 | 3.91(1.21) | 2.18(1.37) | 0.78(0.12) | 35(17.37) | 22.07(3.38) | 2.06(0.87) |
| Total | 75 | 4.74(1.47) | 1.68(1.21) | 1.01(0.33) | 36(17.86) | 22.12(4.75) | 2.95(1.51) |
| ABCA1/LPL | All Males | TC | TG | HDL-C | AGE | BMI | LDL-C |
| CC/CC | 0 | ||||||
| CC/CT | 2 | 3.59(1.44) | 3.81(3.61) | 0.52(0.22) | 61(7.07) | 23.17(1.63) | 2.19(1.65) |
| GC/CC | 3 | 4.59(1.59) | 0.94(0.45) | 0.89(0.21) | 30(17.44) | 22.08(3.03) | 3.28(1.56) |
| GC/CT | 3 | 3.59(1.09) | 4.43(1.84) | 0.65(0.06) | 35(17.78) | 25.28(2.25) | 0.35(0.49) |
| GG/CC | 22 | 4.73(1.35) | 1.45(0.89) | 1.06(0.26) | 34.3(17.44) | 22.88(3.62) | 2.97(1.31) |
| GG/CT | 4 | 4.02(1.54) | 2.62(1.51) | 0.73(0.08) | 26.2(11.67) | 23.39(3.30) | 2.04(1.08) |
| Total | 34 | 4.47(1.36) | 1.94(1.55) | 0.94(0.29) | 34.6(17.25) | 23.12(3.30) | 2.68(1.41) |
| ABCA1/LPL |
All Females |
TC | TG | HDL-C | AGE | BMI | LDL-C |
| CC/CC | 2 | 3.76(1.29) | 0.72(0.01) | 0.94(0.30) | 35(33.94) | 19.46(7.36) | 1.67(0.00) |
| CC/CT | 0 | ||||||
| GC/CC | 19 | 5.19(1.91) | 1.27(0.72 | 0.96(0.22) | 38.1(20.63) | 20.49(3.33) | 3.57(2.01) |
| GC/CT | 3 | 4.67(1.36) | 1.51(0.33) | 0.71(0.09) | 33.7(2.31) | 23.46(4.22) | 3.15(1.54) |
| GG/CC | 15 | 5.09(1.06) | 1.83(0.89) | 1.30(0.43) | 35.1(17.00) | 22.27(7.92) | 2.89(0.85) |
| GG/CT | 2 | 3.69(0.23) | 1.32(0.52) | 0.88(0.16) | 52.5(13.44) | 19.46(1.92) | 2.09(0.49) |
| Total | 41 | 4.97(1.53) | 1.47(0.78) | 1.06(0.36) | 37.2(18.47) | 21.29(5.58) | 3.17(1.57) |
The value in the parenthesis indicates standard deviation.
Rare missense variant in LPL
The identified LPL variant rs118204060 is present in the dbSNP13214 and The 1000 Genomes Project14 data with an unknown frequency. The rs118204060 located in exon 5 changes a conserved amino acid from proline to leucine (P234L). This variant was initially identified in familial chylomicronemia and was reported as P207L27–29 due to differences in genome builds. Upon sequence comparisons, we confirmed that they are indeed the same variant.
Investigation of the ABCA1 and LPL variants in the entire family
We examined the pedigree members for co-occurrence of non-synonymous ABCA1 and LPL variants with low HDL-C. By stratifying individuals by their HDL percentiles, we can see that all the affected family members with HDL-C<5th percentile carry a risk allele for either one or both of the variants (3 P234L, 11 S1731C, and 8 P234L/S1731C), except in one separate branch of the extended family in which the low HDL-C traits appears to be inherited from the affected spouse’s side (Figure 1). No family member with an HDL-C value greater than the 5th percentile had both LPL and ABCA1 variants. Furthermore no family member with HDL-C values greater than the 15th percentile had the LPL variant. Seven subjects (1 male and 6 females) had the ABCA1 variant with the HDL-C percentile of 22% for the male and with an average HDL-percentile of 35% for the six females (Figure 1). Two of the three exome sequenced subjects were heterozygous for both variants and one was homozygous for the ABCA1 variant and heterozygous for the LPL variant. There were four homozygous subjects for the ABCA1 variant two of which were also heterozygous for the LPL variant, whereas the LPL variant was heterozygous in all 14 family members it was observed (Figure 1). Thus, a heterozygous, milder form of LPL deficiency exists in this family. Accordingly, the LPL variant P234L is also associated with elevated levels of triglycerides (1.65±0.27 mmol/l, P=6.14×10−3). In addition, we observed that the subjects with both variants have a lower HDL-C than the subjects with only one variant, and that the subjects heterozygous or homozygous for the ABCA1 variant do not differ in the HDL-C levels (Table 4).
Explained variance and heritability
We estimated that the effect of the ABCA1 and LPL variants on continuous HDL-C measurements in the extended family is −0.17±0.08 mmol/l (P=0.025) and −0.27±0.09 mmol/l (P=0.006), respectively. Together these two variants explain 60% of the genetic variance in this family and 26% of the total (genetic + environment) variance in this family, which amounts to 46% of the heritability explained as assessed in a measured genotype analysis.18 We also repeated the analysis while excluding the three affected subjects that were exome sequenced to reduce the potential for ascertainment bias. In this analysis, the effect sizes of ABCA1 and LPL remained the same (−0.18±0.08 and −0.27±0.10 mmol/l, respectively) and the additive and total variance explained were 50% and 24%, respectively, with 34% of the heritably explained. Importantly, if the subfamily with the bilineal introduction of the low HDL-C trait through the affected spouse is excluded from these analyses, virtually all of the additive variance of HDL-C and virtually all of the heritability of HDL-C is explained by the ABCA1 and LPL variants, suggesting that the ABCA1 and LPL variants can explain the low HDL-C in the ‘non-bilineal’ part of the extended family.
Genome-wide linkage analyses
To further investigate that we did not miss a major susceptibility variant, we performed a whole-genome two-point linkage analysis for low HDL-C using a dominant mode of inheritance. We first estimated using the SLINK simulation program30 that under the assumption of homogeneity the maximum lod score this family can provide is 4.34. However, none of the actual 553 microsatellite markers reached this lod score, most probably due to the existence of multiple low HDL-C variants in the family (i.e. heterogeneity). In more detail, no lod scores >3 were observed anywhere in the genome. The only lod score >2.0 was observed on chromosome 21 for marker D21S1255. However, we noticed that this signal on chr 21seems to arise from the bilineal branch (Figure 1) since the signal diminishes to lod score of 0.6 when we excluded this subfamily from the analysis and increases to 2.5 when we analyzed this bilineal branch of the family alone. Hence the genome-wide linkage data suggest that there might be another susceptibility variant on chr 21 that accounts for the low HDL-C in the bilineal subfamily branch of the extended family. However, since we did not sequence any family members from this branch, none of the 3,459 filtered-out variants would be good candidates. Importantly, we observed lod scores >1 near the LPL and ABCA1 genes (lod scores of 1.62 and 1.28 10.3 Mb and 5.8 Mb from LPL and ABCA1, respectively). Without the bilineal branch these lod scores increased to 2.14 and 1.45, respectively.
Genotype by sex interaction
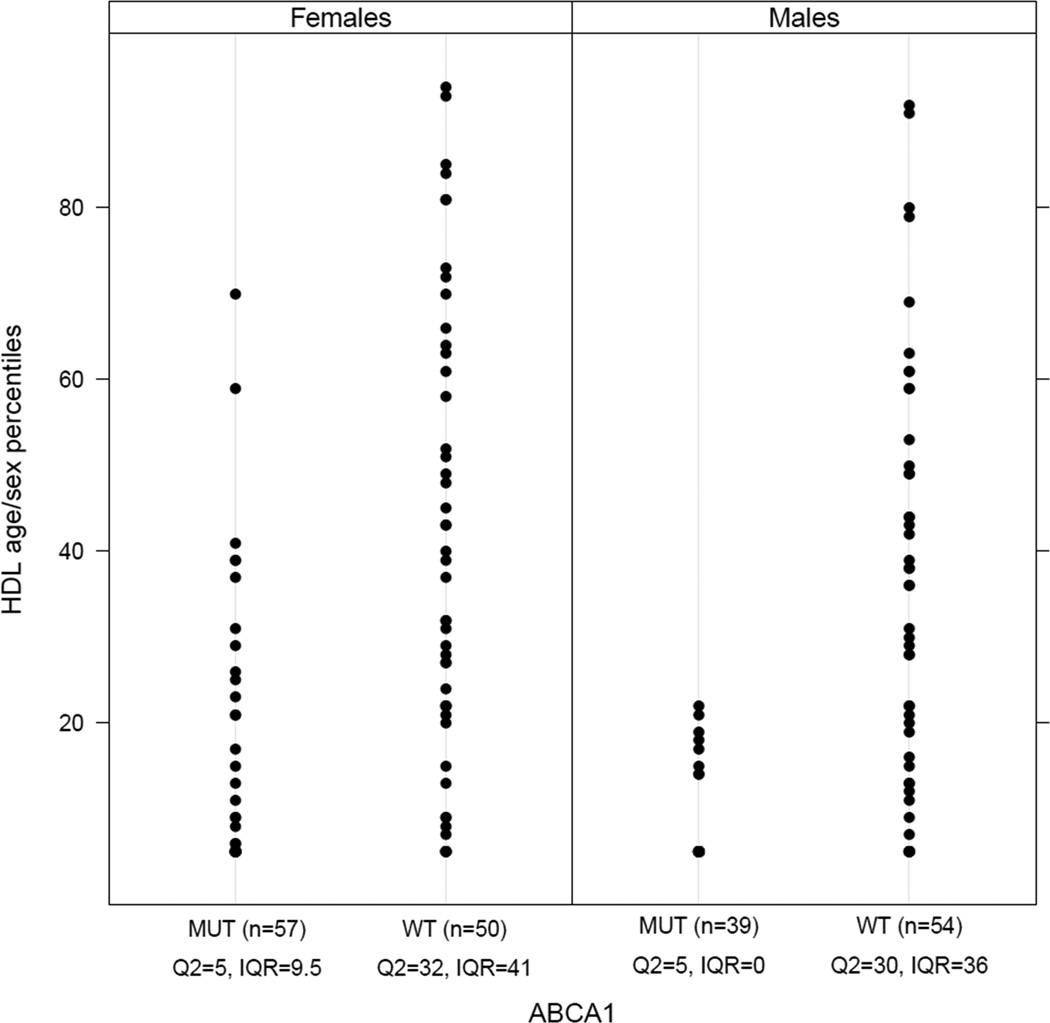
The effect of the ABCA1 S1731C variant on low HDL-C levels appears more profound in the males than in females in the extended family (Table 4). Furthermore, our efflux study also suggested a gene x sex interaction (Figure 2B). Although the frequency of the S1731C variant may be individually too rare for testing genetic interactions (as large sample sizes are necessary), rare variants with large phenotypic effects are collectively common in low HDL-C families.26 We hypothesized that the apparent sex effect may not be restricted to the S1731C allele, but rather it may generally extend to ABCA1 alleles with major phenotypic effects. Thus to further investigate this intriguing relationship between ABCA1 and sex, we examined the collective effect of multiple rare variants in ABCA1 by sex on HDL-C affection. All in all, 10 additional low HDL-C French Canadian families with known mutations in ABCA16–8 were included in the sex interaction analysis using the SOLAR program19, comprising to a total of 93 males and 107 females. The percentage of mutation carriers was 42% and 53% in males and females, respectively (Figure 3). The S1731C variant was present in 3 of these additional low HDL-C pedigrees7 and together with the exome sequenced family the association signal for the main effect of S1731C on low HDL-C status resulted in a p-value of 0.008. In all 11 families, we observed, as expected, a highly significant main effect for ABCA1 genotypes (P=1×10−09), as well as a significant ABCA1 genotype x sex interaction on the qualitative HDL-C affection (P=0.03) (Figure 3). Furthermore, the interaction effect appeared to be more pronounced when comparing pre-menopausal women (age<50 years) to men and post-menopausal women (P=0.003).
Figure 3.
Sex-dependent effect of ABCA1 variants. Figure 3 shows the age-sex specific population HDL-C percentiles by ABCA1 genotypes and sex in 200 French Canadian family members from 11 French Canadian families with different ABCA1 mutations (DelED1893, G616V, K776N, N1800H, Q2210H, R1851X, R2084X, R909X and S1731C). MUT indicates carriers of a mutation in the ABCA1 gene and WT stands for wild-type genotype (i.e. non-carriers). Q2 stands for the median quartile and IQR for interquartile range (Q3-Q1) of HDL-C percentiles. The distributions of the age-sex percentiles are similar between genders in the WT genotype group whereas in the MUT genotype group the distribution is more restricted to the lower tail in males than in females. This difference describes the significant result of the genotype-sex interaction analysis. It should be noted that the displayed age-sex percentiles are not adjusted for relatedness whereas the family relation was taken into account in the genotype-sex interaction analysis performed using SOLAR.
Discussion
By using exome sequencing we identified two functional rare variants in the ABCA1 and LPL genes, co-segregating with low HDL-C and explaining a major proportion of the HDL-C variance and heritability in an extended family. We also observed a sex effect for ABCA1 variants, male carriers exhibiting significantly lower HDL-C levels than females. Furthermore, none of the unaffected family members had the LPL variant or both variants. Our study exemplifies how utilization of exome sequencing was critical to reveal the complex combination of two variants of which one is less severe in females. Traditional linkage analysis was unable to elucidate this type of complex pattern of variants in this extended family,31 suggesting that many such combinations have been missed in previous linkage analyses of complex traits.
ABCA1 and LPL are major players of lipid metabolism. The ABCA1 is a key protein involved in reverse cholesterol transport (RCT) that transports cellular cholesterol to lipid-poor acceptor apolipoproteins, such as apolipoproteinA-I.32,33 As a result, the apolipoprotein is released with the extracted phospholipid and cholesterol, forming nascent HDL particles. Mutations disrupting the normal function of ABCA1 result in little or no circulating HDL.34 Previous studies have shown that cell lines with the identified ABCA1 variant S1731C exhibit low levels of protein expression,25 and that cells transfected with the S1731C allele express abundant ABCA1 mRNA but fail to generate significant amounts of ABCA1 protein.25 Furthermore, the cholesterol efflux of S1731C has been shown to be reduced to 12.3–68.0% of the wildtype.7,25,26 Here, we observed a ~40% cholesterol efflux reduction in the proband homozygous for the S1731C variant as compared to a normal control, in line with the earlier findings.7,25,26 In our previous paper,7 we showed that three heterozygous subjects with the S1731C variant have cholesterol efflux values of 63%, 66%, and 68% of the wildtype. Thus, about the same 40% decrease is observed in the heterozygous subjects as in the homozygous subject with the S173C variant, in line with their similar HDL-C values (Table 4). Although the phenotype data suggest that the S1731C variant does not have a gene dose effect, this conclusion warrants additional functional studies in future, as there are only 4 homozygotes in the family two of which are also heterozygous for the LPL variant, and furthermore the variant has a large range (12.3–68.0% of the wildtype) in its effect on the cholesterol efflux.7,25,26
The main function of LPL is to hydrolyze triglycerides in order to deliver fatty acids to the tissue. LPL also hydrolyzes very-low-density lipoproteins (VLDL). Sequence variation in LPL has been reported to be associated with the risk of CHD, TGs and HDL-C.3 An efficient LPL function is associated with lower TG and low-density lipoproteins (LDL) and higher HDL. Regarding the identified P207L variant, individuals with this mutation have reduced HDL particles compared with the control subjects.35 Previous studies have also shown that missense mutations in exon 5 of the LPL gene where the P207L variant resides are the most common cause of LPL deficiency.36,37 Importantly, Ma et al. reported that upon site-directed in vitro mutagenesis this variant produces a catalytically inactive lipoprotein lipase protein which is the cause of the lipoprotein lipase deficiency in the patients.29 Taking together these previous data, along with our PolyPhen15 and SIFT16 predictions, it is highly likely that both identified variants S1731C and P207L affect protein function.
The two identified variants, S1731C and P207L have been reported previously in French Canadian dyslipidemic individuals but not in normal controls.7,24–29 The S1731C ABCA1 variant was present in three French Canadian dyslipidemic families with low HDL-C levels7 but not in 528 chromosomes from French Canadian subjects with normal HDL-C levels.24 It was also absent in 108 French Canadian subjects with high HDL-C.26 The P207L LPL variant was previously observed in 37 unrelated French Canadian patients with lipoprotein lipase deficiency with 54 mutant alleles present in that study sample.29 In the same study, the variant was also genotyped in 34 unrelated patients with LPL deficiency from ancestries other than French Canadian. Only one German patient was found to be heterozygous for the risk allele. Furthermore, 11 out of 180 French Canadian hyperlipidemic cases were heterozygous for the P207L variant, whereas none of the 170 normolipidemic controls had the P207L variant.29
It is important to study the effect of sex on lipid traits to better understand the sex-specific differences in incidence of dyslipidemia and cardiovascular disease. The results of an earlier study demonstrated that ABCA1 has a sex-specific effect as elevated levels of ABCA1 were observed in females,38 which is in line with the higher HDL-C levels and the lower risk of females for coronary artery disease. In this study, we observed that functional mutations in ABCA1 affecting the cholesterol efflux7,25,26 have a larger effect on HDL-C levels in male than female carriers of these variants. It is possible that the observed genotype-sex interaction results from the previously observed gender differences in ABCA1 expression levels,38 because if males have lower baseline levels of ABCA1, the effect of the mutations could be even more profound in males. These interesting sex-specific mechanisms of ABCA1 may involve hormonal regulation of ABCA1, a hypothesis supported by our efflux experiment (Figure 2B), demonstrating that exposure of fibroblasts of a male proband with the ABCA1 S1731C variant to increasing concentrations of 17β-estradiol led to a significantly increased efflux in the male proband with the ABCA1 variant. These intriguing findings warrant further investigation in future studies.
Our results demonstrate that two relatively rare functional ABCA1 and LPL variants contribute to the risk of low HDL-C in a unique combination involving a sex-effect in an extended family. We first identified a set of variants by filtering the variants shared by the exome sequenced affected family members for variant type, frequency, and functional predictions. Because filtering has limitations caused by heterogeneity of complex traits,4 we then utilized the extended family structure for statistical analysis exploring how much of the trait variance and heritability the two key ABCA1 and LPL variants explain. Thus, our study highlights the fact that the filtering strategy used in exome studies of Mendelian disorders4 is not directly applicable for complex disorders and that new methodologies that incorporate multiple susceptibility variants within a family are warranted. As the two variants explain a major part of the variance in HDL-C and are shown to be functional,7,25,26,29 they represent the key underlying HDL-C variants in this family, though other rare and common variants are likely to explain the remaining portion of the variance. As both ABCA1 and LPL are known to affect HDL-C, our study did not reveal a novel HDL gene. However, our study does highlight the importance of exome sequencing of dyslipidemic families, because traditional linkage or haplotype analysis cannot detect complex segregation of several functional rare variants due to the inherent parametric restrictions of linkage analysis. This type of underlying biological complexity must have contributed to the low lod scores and weak success of linkage analysis in gene identification of complex lipid traits. In this study, we demonstrate for how family-based exome sequencing can successfully identify multiple rare variants to be followed up utilizing the effective co-segregation information available in extended dyslipidemic families. To the best of our knowledge, our study is the first described example of two functional rare variants conferring the susceptibility to low HDL-C in an extended family.
Supplementary Material
Acknowledgments
We thank the family members who participated in this study. We also thank Cindy Montes and UCLA core facilities for laboratory technical assistance.
Funding Sources: This research was supported by the grants HL095056 and HL-28481 from the National Institutes of Health (PP, JSS); by grants MOP 97752 from the Canadian Institutes of Health Research (CIHR) and from the Heart and Stroke Foundation of Canada (JG); and by the American Heart Association grant 11POST7380028 (MVPLR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures: None.
References
- 1.Pajukanta P, Allayee H, Krass KL, Kuraishy A, Soro A, Lilja HE, et al. Combined analysis of genome scans of Dutch and Finnish families reveals a susceptibility locus for high-density lipoprotein cholesterol on chromosome 16q. Am J Hum Genet. 2003;72:903–917. doi: 10.1086/374177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soro A, Pajukanta P, Lilja HE, Ylitalo K, Hiekkalinna T, Perola M, et al. Genome scans provide evidence for low-HDL-C loci on chromosomes 8q23, 16q24.1-24.2, and 20q13.11 in Finnish families. Am J Hum Genet. 2002;70:1333–1340. doi: 10.1086/339988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dastani Z, Quiogue L, Plaisier C, Engert JC, Marcil M, Genest J, et al. Evidence for a gene influencing high-density lipoprotein cholesterol on chromosome 4q31.21. Arterioscler Thromb Vasc Biol. 2006;26:392–397. doi: 10.1161/01.ATV.0000198243.83781.a3. [DOI] [PubMed] [Google Scholar]
- 6.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 7.Alrasadi K, Ruel IL, Marcil M, Genest J. Functional mutations of the ABCA1 gene in subjects of French-Canadian descent with HDL deficiency. Atherosclerosis. 2006;188:281–291. doi: 10.1016/j.atherosclerosis.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Marcil M, Brooks-Wilson A, Clee SM, Roomp K, Zhang LH, Yu L, et al. Mutations in the ABC1 gene in familial HDL deficiency with defective cholesterol efflux. Lancet. 1999;354:1341–1346. doi: 10.1016/s0140-6736(99)07026-9. [DOI] [PubMed] [Google Scholar]
- 9.Marcil M, Yu L, Krimbou L, Boucher B, Oram JF, Cohn JS, et al. Cellular cholesterol transport and efflux in fibroblasts are abnormal in subjects with familial HDL deficiency. Arterioscler Thromb Vasc Biol. 1999;19:159–169. doi: 10.1161/01.atv.19.1.159. [DOI] [PubMed] [Google Scholar]
- 10.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The 1000 Genomes Project Consortium. A map of human genome variation from populationscale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, et al. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69:504. [Google Scholar]
- 18.Lange K, Sinsheimer JS, Sobel E. Association testing with Mendel. Genet Epidemiol. 2005;29:36–50. doi: 10.1002/gepi.20073. [DOI] [PubMed] [Google Scholar]
- 19.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcil M, Bissonnette R, Vincent J, Krimbou L, Genest J. Cellular phospholipid and cholesterol efflux in high-density lipoprotein deficiency. Circulation. 2003;107:1366–1371. doi: 10.1161/01.cir.0000056764.53152.f9. [DOI] [PubMed] [Google Scholar]
- 21.Denis M, Haidar B, Marcil M, Bouvier M, Krimbou L, Genest J., Jr Molecular and cellular physiology of apolipoprotein A-I lipidation by the ATP-binding cassette transporter A1 (ABCA1) J Biol Chem. 2004;279:7384–7394. doi: 10.1074/jbc.M306963200. [DOI] [PubMed] [Google Scholar]
- 22.Clark MJ, Chen R, Lam HY, Karczewski KJ, Chen R, Euskirchen G, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissglas-Volkov D, Pajukanta P. Genetic causes of high and low serum HDL-cholesterol. J Lipid Res. 2010;51:2032–2057. doi: 10.1194/jlr.R004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clee SM, Zwinderman AH, Engert JC, Zwarts KY, Molhuizen HO, Roomp K, et al. Common genetic variation in ABCA1 is associated with altered lipoprotein levels and a modified risk for coronary artery disease. Circulation. 2001;103:1198–1205. doi: 10.1161/01.cir.103.9.1198. [DOI] [PubMed] [Google Scholar]
- 25.Brunham LR, Singaraja RR, Pape TD, Kejariwal A, Thomas PD, Hayden MR. Accurate prediction of the functional significance of single nucleotide polymorphisms and mutations in the ABCA1 gene. PLoS Genet. 2005;1:e83. doi: 10.1371/journal.pgen.0010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 27.Brisson D, Ledoux K, Bossé Y, St-Pierre J, Julien P, Perron P, et al. Effect of apolipoprotein E, peroxisome proliferator-activated receptor alpha and lipoprotein lipase gene mutations on the ability of fenofibrate to improve lipid profiles and reach clinical guideline targets among hypertriglyceridemic patients. Pharmacogenetics. 2002;12:313–320. doi: 10.1097/00008571-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Normand T, Bergeron J, Fernandez-Margallo T, Bharucha A, Ven Murthy MR, Julien P, et al. Geographic distribution and genealogy of mutation 207 of the lipoprotein lipase gene in the French Canadian population of Québec. Hum Genet. 1992;89:671–675. doi: 10.1007/BF00221960. [DOI] [PubMed] [Google Scholar]
- 29.Ma Y, Henderson HE, Murthy V, Roederer G, Monsalve MV, Clarke LA, et al. A mutation in the human lipoprotein lipase gene as the most common cause of familial chylomicronemia in French Canadians. N Engl J Med. 1991;324:1761–1766. doi: 10.1056/NEJM199106203242502. [DOI] [PubMed] [Google Scholar]
- 30.Weeks DE, Ott J, Lathrop GM. SLINK: a general simulation program for linkage analysis. Am J Hum Genet. 1990;47:204. [Google Scholar]
- 31.Dastani Z, Pajukanta P, Marcil M, Rudzicz N, Ruel I, Bailey SD, et al. Fine mapping and association studies of a high-density lipoprotein cholesterol linkage region on chromosome 16 in French-Canadian subjects. Eur J Hum Genet. 2010;18:342–347. doi: 10.1038/ejhg.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oram JF, Lawn RM. ABCA1. The gatekeeper for eliminating excess tissue cholesterol. J Lipid Res. 2001;42:1173–1179. [PubMed] [Google Scholar]
- 33.Marcil M, Bissonnette R, Vincent J, Krimbou L, Genest J. Cellular phospholipid and cholesterol efflux in high-density lipoprotein deficiency. Circulation. 2003;107:1366–1371. doi: 10.1161/01.cir.0000056764.53152.f9. [DOI] [PubMed] [Google Scholar]
- 34.Albrecht C, Baynes K, Sardini A, Schepelmann S, Eden ER, Davies SW, et al. Two novel missense mutations in ABCA1 result in altered trafficking and cause severe autosomal recessive HDL deficiency. Biochim Biophys Acta. 2004;1689:47–57. doi: 10.1016/j.bbadis.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Ruel IL, Gaudet D, Perron P, Pascot A, Després JP, Bergeron J, et al. Determinants of HDL particle size in patients with the null (P207L) or defective (D9N) mutation in the lipoprotein lipase gene: the Québec LipD Study. Atherosclerosis. 2002;162:269–276. doi: 10.1016/s0021-9150(01)00716-x. [DOI] [PubMed] [Google Scholar]
- 36.Peterson J, Ayyobi AF, Ma Y, Henderson H, Reina M, Deeb SS, et al. Structural and functional consequences of missense mutations in exon 5 of the lipoprotein lipase gene. J Lipid Res. 2002;43:398–406. [PubMed] [Google Scholar]
- 37.Garenc C, Couillard C, Laflamme N, Cadelis F, Gagné C, Couture P, et al. Effect of the APOC3 Sst I SNP on fasting triglyceride levels in men heterozygous for the LPL P207L deficiency. Eur J Hum Genet. 2005;13:1159–1165. doi: 10.1038/sj.ejhg.5201469. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Klein K, Sugathan A, Nassery N, Dombkowski A, Zanger UM, et al. Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease Transcriptional profiling of human liver identifies sexbiased genes associated with polygenic dyslipidemia and coronary artery disease. PLoS One. 2011;6:e23506. doi: 10.1371/journal.pone.0023506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.