Abstract
Terminase enzymes are responsible for the excision of a single genome from a concatemeric precursor (genome maturation) and concomitant packaging of DNA into the capsid shell. Here, we demonstrate that lambda terminase can be purified as a homogenous “protomer” species and we present a kinetic analysis of the genome maturation and packaging activities of the protomeric enzyme. The protomer assembles into a distinct maturation complex at the cos sequence of a concatemer. This complex rapidly nicks the duplex to form the mature left end of the viral genome, which is followed by procapsid binding, activation of the packaging ATPase, and translocation of the duplex into the capsid interior by the terminase motor complex. Genome packaging by the protomer shows high fidelity with only the mature left end of the duplex inserted into the capsid shell. In sum, the data show that the terminase protomer exhibits catalytic activity commensurate that expected of a bone fide genome maturation and packaging complex in vivo and that both catalytically-competent complexes are composed of four terminase protomers assembled into a ring-like structure that encircles duplex DNA. This work provides mechanistic insight into the coordinated catalytic activities of terminase enzymes in virus assembly that are generalizable to all of the dsDNA viruses.
Keywords: Virus assembly, DNA packaging, terminase motor, viral ATPase
The developmental pathway for the double-stranded DNA (dsDNA) viruses represents an ordered assembly process in which many aspects are conserved across adenovirus, herpesvirus, and bacteriophage groups (1-3). A key step in virus assembly involves the packaging of a viral genome into a preformed procapsid shell (4-8). The preferred packaging substrate is generally a linear concatemer of genomes that is the product of viral DNA replication (immature DNA) (9, 10). Genome packaging requires excision of a single genome from the concatemer (genome maturation) and concomitant translocation of duplex into the capsid shell powered by ATP hydrolysis (DNA packaging). The terminase enzymes perform both of these functions and the motors package DNA to near liquid crystalline density generating up to 50 atmospheres of pressure within the capsid shell (4, 7, 8). The reactions are strongly conserved among all of these viruses, both prokaryotic and eukaryotic, and terminase enzymes may provide a novel target for antiviral therapeutics. Bacteriophage lambda is prototypical of these viruses and it has been extensively characterized by genetic, biochemical, and biophysical approaches. Our lab has harnessed the lambda system as a model with which to interrogate the mechanistic features of the packaging process.
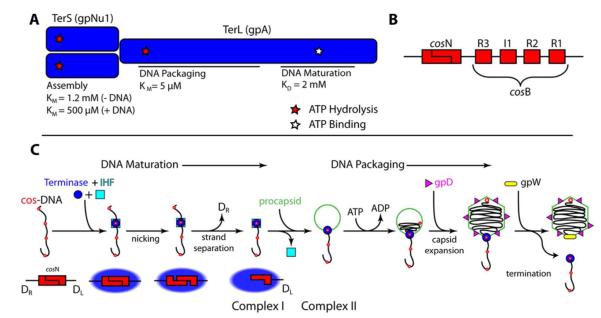
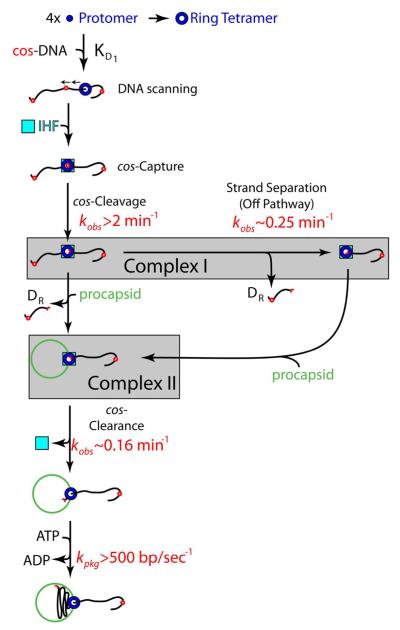
Lambda terminase is composed of two gene products - gpA, the large terminase subunit (TerL) and gpNu1, the small terminase subunit (TerS), in a TerL1•TerS2 heterotrimer complex (Figure 1A) (11, 12). This “protomer” possesses several catalytic activities related to genome maturation and packaging (13, 14) and current models describing virion assembly are summarized as follows (see Figure 1C):
Multiple terminase protomers assemble at a cohesive end site (cos), which represents the junction between successive genomes in the concatemer (Figure 1B). The assembly process is mediated by the terminase TerS subunit and Escherichia coli integration host factor (IHF), which cooperatively bind and bend the duplex at the cosB sub-site (15). This provides a duplex architecture to which the protomer binds with high affinity; however, the stoichiometry of IHF and terminase protomers bound in this maturation complex remains unclear.
The endonuclease activity of TerL introduces symmetric nicks into the duplex at the cosN sub-site (Figure 1B). Subsequent separation of the nicked duplex, catalyzed by the so-called helicase activity of TerL, affords the mature, 12-base single-stranded mature left end of the genome (DL) tightly bound by the enzyme. This stable intermediate is commonly referred to as Complex I (Figure 1C) (14).
The maturation complex next binds to the portal ring of an empty procapsid to yield the packaging motor complex (Complex II, Figure 1C); the structural features of the packaging motor also remain unclear. Whatever the case, assembly of the motor triggers terminase release from the cos site and translocation of DNA into the procapsid, powered by ATP hydrolysis (cos-clearance).
DNA packaging triggers expansion of the procapsid shell to the mature capsid conformation, which then binds the gpD decoration protein (16, 17). This provides structural integrity to the shell so that it can withstand the internal force generated by the tightly packaged DNA (17, 18). Upon reaching the downstream cos site in the concatemer, the terminase motor engages the terminal cos sequence (cos-capture). This terminates translocation activity and again activates the end maturation activities of the enzyme. Duplex nicking and strand separation afford the mature DR end of the packaged genome and the gpW adaptor protein replaces terminase at the portal vertex. Subsequent addition of the gpFII protein and a pre-assembled tail affords an infectious virus, while the terminase•DL-DNA complex (regenerated Complex I) binds a second procapsid to initiate a second round of processive packaging (14, 19).
Figure 1. Terminase Has Multiple Catalytic Activities Required for Maturation and Packaging the Viral Genome.
Panel A. The terminase protomer is composed of one TerL and two TerS subunits. The TerL subunit provides all of the catalytic activities of the enzyme in two functional domains as indicated in the figure (maturation and packaging domains). The TerS subunits are required for site-specific assembly of the maturation complex at cos. Three ATP binding sites have been identified in the protomer – the assembly site, the packaging ATPase site, and DNA maturation site - and are indicated with stars. Experimentally determined binding constants are presented below each indicated site. Panel B. The cos sequence of the lambda genome is multi-partite. The terminase TerL subunit introduces symmetric nicks within the cosN sub-site to generate the 12 base “sticky” ends of the mature lambda genome. Cooperative assembly of the TerS subunit and IHF at the cosB sub-site mediates specific assembly of terminase at cos. IHF binds to the I1 consensus sequence while TerS binds to the three “R” elements. Panel C. Current model for genome maturation and packaging by lambda terminase. Concatemeric (immature) DNA is presented as a line with multiple cos sites depicted as red dots. Lower graphic shows details of terminase-catalyzed duplex nicking and strand-separation reactions. Details are provided in the text.
The enzymology of lambda terminase has been extensively studied and this work has yielded significant insight into the mechanism of genome maturation and packaging. Most recent studies have utilized highly purified terminase preparations; however, we recently demonstrated that the purified enzyme exists as a mixture of the 5.1 S protomer in slow equilibrium with a heterogeneous, “13.3 S species”† (11, 12). We refer to this purified preparation as the terminase “mix”. Undoubtedly, the structural heterogeneity of the terminase mix is responsible, at least in part, for the complex behavior previously observed in kinetic and biophysical interrogation of the enzyme, and this has complicated mechanistic interpretation of the data. Here, we show that a homogenous preparation of the lambda terminase protomer may be isolated in high yield and we have defined conditions for the long-term storage of the enzyme. We further describe a kinetic interrogation of the Genome maturation and packaging activities of the protomer and contrast this with the pure, but heterogeneous terminase mix used in published studies. While many activities are similar, important differences between the protomer and the terminase mix are observed which are relevant to the function of the enzyme. In sum, the present studies suggest that the terminase protomer is the biologically relevant species during a productive viral infection in vivo and this work provides mechanistic insight into the coordinated activities of terminase enzymes in virus assembly.
EXPERIMENTAL PROCEDURES
Materials and Protein Constructs
Tryptone, yeast extract, agar, and ampicillin were purchased from Fisher Scientific. Terrific broth was purchased from Difco. All nucleoside triphosphates were purchased from Sigma-Aldrich. Chromatography media was purchased from GE Healthcare Life Sciences. Mature lambda DNA was purchased from Invitrogen. All other materials were of the highest quality available. The plasmid pCT-λ, a 12 kb plasmid that contains a unique wild-type cos sequence, was purified by published procedure (20). Cell lysis utilized a Thermo Scientific IEC “French” laboratory press. All protein purifications utilized the Amersham Biosciences AÄKTApurifier core 10 System from GE Healthcare. Full-length, native sequence Escherichia coli Integration Host Factor (IHF) was purified from HN880 cells as previously described (21).
Expression and Purification of Lambda Terminase
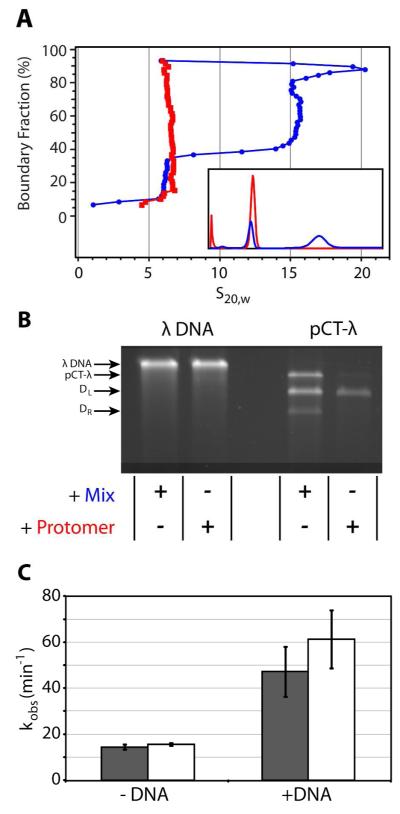
The terminase enzyme used in this study was expressed from OR1265[pQH101] cells as previously described (22). This vector expresses full-length, native sequence gpNu1 and full-length, native sequence gpA with six histidines directly appended to the C-terminal Glu residue of the large gpA subunit. Expression and purification of the terminase “mix” was as previously described (22), with modification to optimize the yield of the protomer. Briefly, purified terminase eluted from the HisTrap FF column was dialyzed overnight at 4°C against buffer Q (20 mM Phosphate buffer, pH 6.8, containing 100 mM NaCl, 1 mM EDTA, 7 mM β-ME, and 10% glycerol (v/v)). The dialysate was loaded onto a 1 mL HiTrap Q column and bound proteins were eluted with a 20-column volume gradient to buffer Q containing 1 M NaCl. The terminase containing fractions (~300 mM NaCl) were pooled and aliquots were stored at −80 C. These samples contained both the homogenous protomer and the heterogeneous “assembled” species (Figure 2A) and are referred to as the “Terminase Mix”. We have previously demonstrated that the self-association behavior and the catalytic activities of the H6-terminase mix are indistinguishable from those of the native, untagged enzyme (22).
Figure 2. Purification and Packaging Activity of the Lambda Terminase Protomer.
Panel A. Sedimentation velocity data for the terminase mix (blue) and isolated protomer (red) were analyzed using the van Holde-Weischet method as described in Experimental Procedures. The mix clearly contains both the protomer and assembled species while the isolated protomer is homogenous. Inset – c(s) analysis of the same data using Sedfit. Panel B. Fidelity of DNA Packaging by the Mix and by the Isolated Terminase Protomer. The DNase protection assay was performed as described in Experimental Procedures using 100 nM terminase protomer or terminase mix and 5 nM mature lambda DNA (λ-DNA) or pCT-λ, as indicated. The position of mature λ DNA, uncleaved pCT-λ, and the DL and DR products resulting from cos-cleavage of pCT-λ are indicated at left. There is no difference between the protomer and the mix when packaging a mature lambda genome (left). The mix packages full-length pCT-λ and both nuclease products in a linked maturation/packaging reaction. In contrast, the protomer packages the matured DL genome end with high fidelity. Panel C. The observed rate of ATP hydrolysis by the terminase protomer was determined in the presence of 50 μM (grey bars) and 1 mM (white bars) ATP. Duplex DNA (pCT-λ) was added to the reaction mixture as indicated. Each bar represents the average of at least three separate experiments with standard deviations indicated.
To isolate the terminase protomer, a one mL aliquot of purified terminase mix was applied to a HiPrep S-300 HR gel filtration column (120 mL) equilibrated and developed with buffer Q. The terminase protomer eluted at ~ 65 mL and the protomer containing fractions were pooled, aliquoted, and stored at −80°C. The TerL1•TerS2 protomer concentration was determined spectrally (ε280 = 15 000 M−1 cm−1).
Sedimentation Velocity Analytical Ultracentrifugation Analysis
Sedimentation velocity (SV) experiments were performed using terminase that had been purified as described above. All experiments were performed in buffer Q containing 350 mM NaCl or 100 mM NaCl for the terminase mix and the protomer, respectively. Data were collected using a Beckman XL-A analytical ultracentrifuge (Beckman Instruments, Inc., Fullerton, CA) using 12 mm Epon charcoal two sector centerpieces at 42,000 rpm. Absorbance data were collected at 280 nm, using a spacing of 0.001 cm, with four averages in the continuous scan mode; scans were collected every 15 minutes. Samples were run at 7°C. The raw data were analyzed using both the UltraScan comprehensive data analysis suite (23, 24) and the SedFit/SedPhat data analysis packages (25, 26).
Terminase Activity Assays and Kinetic Analysis
The cos-cleavage endonuclease and strand separation assays were performed by published procedure (21). The single-turnover reaction time courses were fit to an exponential rate equation in the form below:
where Products represent the fraction of substrate converted to product at time t and A∞ is the extent of the reaction at infinite time. Ai represents the fraction of the rate associated with the “ith” phase of the reaction when fitted for multiple rates and ki is the associated observed rate constant. The kinetic data were fit to both single (n=1) and double (n=2) exponential rate equations. A double exponential time course was deemed appropriate only if (i) the quality of the fit (φ2) improved by an order of magnitude and if (ii) the derived rate constants differed by at least 10-fold. The Microsoft Excel solver function was used with error minimization to achieve the reported fits.
The synchronous DNA packaging assay was performed as described previously (27, 28), with modification as described in Supporting Information. The ATPase assay was conducted as described above for the DNA packaging reaction except that [α-32P]ATP (1000 – 3000 cpm/pmol) was added to the reaction mixture and the reaction was allowed to proceed at 37°C. ATP hydrolysis was quantified by thin layer chromatography as previously described (13, 29).
RESULTS
Isolation of the Homogenous Terminase Protomer
The goal of this study is to characterize the biochemical properties and catalytic activity of the terminase protomer. This requires an efficient purification protocol that affords a homogenous enzyme preparation in high yield. We previously demonstrated that purified lambda terminase is composed of a homogenous TerL1•TerS2 heterotrimer complex (the protomer) and a heterogeneous higher-order assembly of approximately four protomers (the assembled species) (11). The assembled species slowly dissociates ultimately yielding ~ 30% homogenous protomer after two weeks of incubation in Tris buffer, pH 8, containing 150 mM NaCl (12). We further demonstrated that the association state of the enzyme is influenced by NaCl concentration and by temperature (11); however, preparative isolation of the protomer was time consuming and inefficient. In an effort to purify homogenous protomer in high yield, the effect of salt and pH on the association state of the enzyme was investigated in greater detail and the purification protocol was optimized. Lowering the pH to 6.8 and decreasing the NaCl concentration to 100 mM results in rapid dissociation of the assembled species to afford ~ 50% homogenous protomer after only two hours incubation time (data not shown). Further decreases in pH or salt hasten the process, but result in significant loss of protein due to aggregation (not shown). The optimized purification protocol described in Experimental Procedures affords 4 mg protomer per liter of cells that is > 95% pure as determined by SDS-PAGE and that is > 95% homogenous as determined by sedimentation velocity analytical ultracentrifugation analysis (Figure 2A). The purified protomer can be stored at −80 C for over 8 months without evidence of self-association or loss of catalytic activity.
Fidelity of Genome Packaging by the Terminase Protomer
Genome maturation in vivo affords the DR and DL genome ends, but only the latter is utilized as a packaging substrate (see Figure 1C) (14). In contrast, in vitro packaging assays indicate that both DR and DL ends, as well as the uncleaved substrate can be packaged into the capsid depending on experimental conditions (27). These published studies used the purified “terminase mix”, which contains both protomer and heterogeneous assembled species as described above. We proposed that the protomer represents the biologically relevant species during a productive viral infection (11, 12) and here we directly compare the fidelity of DNA packaging by the terminase protomer and the terminase mix. As shown previously, both the protomer and the terminase mix efficiently package a full-length lambda genome in the presence of IHF (Figure 2B) (11). While the data demonstrate that both preparations exhibit high processivity (the entire genome is packaged), this experiment does not interrogate fidelity because packaging could start at either genome end. We next examined packaging coupled to Genome maturation using pCT-λ, a 12 kb linearized plasmid that contains an intact cos sequence (20). In this assay, the terminase mix efficiently matures the duplex at cos and then packages the matured DL end; however, the mix also packages the DR end and in addition the un-cleaved, full-length substrate (Figure 2B). In other words, the terminase mix packages DNA non-discriminately. In contrast, the protomer also matures the duplex at cos but shows a strong preference for packaging only the DL containing duplex with little to no packaging of either the matured DR end or the substrate duplex observed (Figure 2B). Hence, coupled maturation and packaging with the terminase protomer establishes fidelity in the packaging reaction.
ATP Binding and Hydrolysis by the Terminase Protomer
DNA packaging is fueled by a high-affinity ATPase catalytic site in the N-terminus of the TerL subunit (the packaging ATPase site) (14). In addition, lambda terminase possesses a second ATP binding site in the C-terminus of TerL that regulates the genome maturation activities of the enzyme and a third ATP binding site in the TerS subunit that regulates terminase binding to DNA (the maturation and assembly ATP binding sites, respectively; Figure 1A). Kinetic and mutagenesis studies of the terminase mix identified two catalytically active ATPase sites in the enzyme - the high affinity packaging ATPase in TerL and a low affinity site in TerS (28-33); the genome maturation site is catalytically silent (34). In most proteins where nucleotides regulate biological activity, NTP and NDP‡ stabilize alternate conformations of the protein and catalytic turnover at the site is extremely slow. For instance, G-proteins involved in signal transduction pathways are activated by GTP but are inactive with GDP bound; slow hydrolysis of the bound GTP (kcat ~ 10−2 – 10−3 min−1) acts as a “switch mechanism” that allows regulation of biological activity (35). Within this context, ATP hydrolysis by the assembly site in the terminase mix is unusual. We hypothesized that the observed low-affinity ATPase activity is aberrant and the result of improper protomer associations in the assembled species contained in the terminase mix. To directly test this hypothesis, we performed a kinetic analysis of ATP hydrolysis by the isolated protomer to define catalytically active ATPase sites in the enzyme.
We have historically used 50 μM ATP to isolate ATP hydrolysis to the high-affinity packaging ATPase site (Km= 5 μM) (13, 30, 32). The nucleotide concentration is then increased to 1 mM to interrogate additional ATP hydrolysis by the maturation site in the terminase mix (Km~ 500 μM) (13, 29, 30, 32). Consistently, increased ATP hydrolysis by the terminase mix is observed at elevated ATP concentrations (data not show); however, when a similar approach is taken with the terminase protomer, no significant increase in ATP hydrolysis is observed with increasing concentration of ATP (Figure 2C). We interpret these data to indicate that the low affinity ATP binding site in TerS is catalytically silent in the terminase protomer, even in the presence of DNA; this is exactly what is expected of a regulatory nucleotide-binding site. In sum, the highly purified terminase protomer possesses (i) an ATP binding site in TerS that regulates DNA binding activity and (ii) an ATP binding site in the C-terminus of TerL that regulates genome maturation activity; catalytic turnover at these sites is undetectable. In addition, the motor possesses a high-affinity packaging ATPase catalytic site in the N-terminus of TerL that fuels DNA translocation into the capsid (see Figure 1A).
Kinetic Analysis of Genome Maturation by the Terminase Protomer
As depicted in Figure 1C, maturation of the genome end by lambda terminase in vitro involves nicking of the duplex at cosN and ejection of the DR end to afford Complex I, a stable intermediate in the absence of procapsids (t1/2~ 8 hours) (36). The reaction time course thus represents a single-catalytic turnover by the enzyme and the data are analyzed according to an exponential increase in product formation. Previous studies in our lab have demonstrated multiple exponential phases in the time course for the cos-cleavage reaction (20, 37). We suggested that terminase assembly at cos is the rate-limiting step in genome maturation; however, this interpretation is complicated by the presence of structural heterogeneity in the purified preparation, as discussed previously (37) and above.
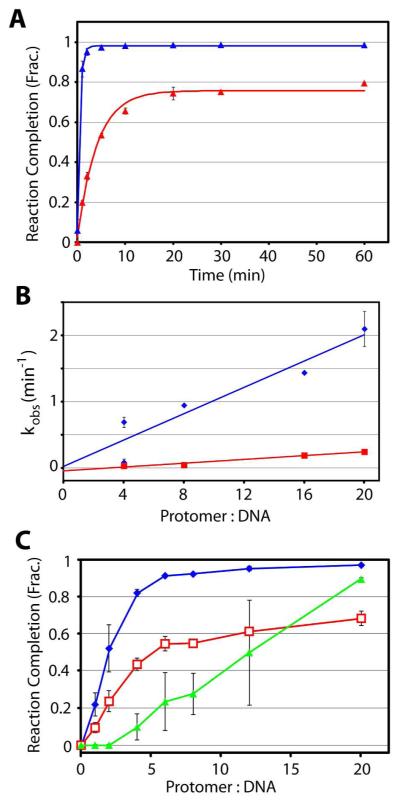
Here, we describe a detailed kinetic analysis of the cos-cleavage and subsequent strand separation reactions by the isolated terminase protomer using pCT-λ at a concentration of 5 nM. At low protomer concentrations (20 nM), the cos-cleavage reaction time course is poorly described by a single-exponential function and a double-exponential rate equation is required to adequately describe the data (Figure S1, dashed line and solid line, respectively). This analysis affords rate constants for the fast and slow phases of the reaction that are similar to those previously obtained in our lab (Table 1) (20, 37). Presumably, the slow rate constant reflects slow assembly of a catalytically-competent nuclease complex at cos under limiting protomer concentrations. In contrast, at protomer concentrations greater than 20 nM (4-fold excess enzyme) the slow phase disappears and the reaction time course is well described by a single-exponential rate equation (blue data, Figure 3A). Interestingly, the observed rate constant is linearly dependent on protomer concentration (Figure 3B, Table 1). The catalytically competent maturation complex is composed of multiple protomers (vide infra) and these data indicate that terminase assembly is not the rate-limiting step in the kinetic time course under these conditions. If this were the case, a higher-order concentration dependence would be observed. Notwithstanding, these data indicate that the rate of cos-cleavage, and by extension the rate of protomer assembly at cos, is fast under conditions that mimic those observed during a productive virus infection in vivo (~ 100 nM terminase) (38, 39).
Table 1.
Kinetic Analysis of the Genome Maturation Reactions.
| [Protomer] |
cos-Cleavage Reaction Rate |
Strand Separation Reaction Rate |
Cleavage:Separation Reaction Extent |
|---|---|---|---|
| 100 nM | kobs= 2.1 min−1 | kobs= 0.25 min−1 | 1.4 |
| 80 nM | kobs= 1.4 min−1 | kobs= 0.19 min−1 | 1.4 |
| 40 nM | kobs= 0.95 min−1 | kobs= 0.045 min−1 | 1.4 |
| 20 nM |
kfast= 0.69 min−1 (60% total amplitude) kslow= 0.086 |
kobs= 0.033 min−1 | 1.4 |
Figure 3. Genome End Maturation in Vitro is Limited by a Slow Strand Separation Rate.
Panel A. Time courses for the cos cleavage (blue) and strand separation (red) reactions in the presence of 100 nM terminase protomer. Each data point represents the average of at least three separate experiments with standard errors indicated (in some cases the error is smaller than the data point and is obscured). The solid line is the best fit of the data to a single exponential time course; derived kinetic parameters are reported in Table 1. Panel B. The observed rates of the cos-cleavage (blue) and strand-separation (red) reactions as a function of terminase protomer concentration. The data points at 4- and 20-fold excess terminase represent the average of three separate experiments with error bars indicated; the data points for 8- and 16-fold excess terminase are the result of a single experiment. Analysis of the data affords bi(molecular rate constants kon= (2.8 ± 0.5) × 105 M−1 sec−1) and kon= (0.43 ± 0.05) × 105 M−1 sec−1) for cos-cleavage and strand-separation, respectively. Panel C. Terminase Stoichiometry in the Maturation and Packaging Complexes. The extent of the cos-cleavage (blue), strand separation (red), and DNA packaging (green) reactions are shown as a function of increasing protomer concentration.
In contrast to the cos-cleavage reaction, the kinetic time course for the strand separation reaction is well described by a single-exponential rate equation at all protomer concentrations examined. Two observations are of interest; (i) the observed rate of strand separation is an order of magnitude slower than duplex nicking and (ii) the extent of the strand-separation reaction is significantly less than that observed for cos-cleavage under all reaction conditions (Figures 3A, 3B, and Table 1). These observations have implications on the nature of Complex I in the packaging pathway in vivo and are discussed further below.
Stoichiometry of the Maturation and Packaging Motor Complexes
Early models hypothesized that a terminase dimer assembles at the cosN sub-site to introduce symmetric nicks into the duplex (see Figure 1) (40, 41). This model is based on the symmetric nature of cosN (42), the identification of a putative bZIP protein dimerization motif in the TerL subunit (43), and in analogy to the “orthodox” type II restriction endonuclease enzymes (44). In contrast, it has been presumed that the packaging motor is composed of an oligomeric terminase ring, in analogy to the translocating ring helicases (45-47). These models predict that additional terminase protomers are recruited to the maturation complex to complete a higher-order packaging motor complex. We have rigorously demonstrated that the protomer can assemble into a stable tetrameric ring complex in solution ([TerL1•TerS2]4) and proposed a simplified model in which the ring tetramer is responsible for both the maturation and packaging reactions (11, 12). As a first step to address this hypothesis, we examined the extent of genome maturation as a function of protomer concentration; several features of this analysis are of interest. (i) The extent of strand separation is always less than the extent cos-cleavage at all protomer concentrations and (ii) the extents of both reactions reach an apparent maximum at around 4 - 6 protomers per DNA molecule (Figure 3C). We interpret these data to indicate that there are no major differences in the protomer stoichiometry between the two catalytic complexes during nicking and separation reactions. The data further place an upper limit on the number of protomers assembled into the maturation complex. This analysis presumes that the catalytic activity of the preparation is 100%, which is rarely the case. If we assume a more reasonable ~ 90% active enzyme preparation, this places an upper limit of four to five protomers in the maturation complex.
We next examined the effect of protomer concentration on the DNA packaging activity of terminase and the data suggest a more complex relationship. In contrast to the end maturation reactions, little to no packaging activity is detectable below a protomer:DNA stoichiometry of four (Figure 3C). The extent of the reaction increases with further increase in protomer concentration until essentially 100% of the input DNA has been packaged in the presence of 20-fold excess protomer. It is noteworthy that under these conditions, the extent of packaging (~100%) exceeds the extent of strand separation by the enzyme (~50%; Figure 3C). Control studies indicate that procapsids affect neither the rate nor the extent of the strand separation by the enzyme (data not shown). This observation indicates that the packaging motor can utilize the nicked, annealed duplex bound by terminase in addition to the strand-separated duplex in Complex I (see Figure 1).
Motor Assembly and Regulation of the Packaging ATPase Site
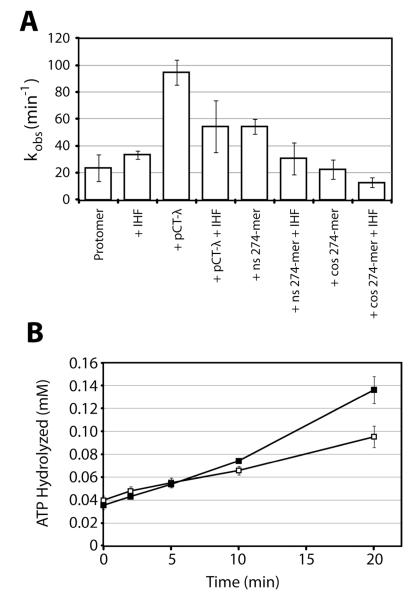
The protomer possesses a weak basal ATPase activity while the isolated ring tetramer efficiently hydrolyzes ATP (11). Figure 4A demonstrates that ATP hydrolysis by the protomer is also stimulated by duplex DNA and at biologically relevant enzyme concentrations (100 nM). Interestingly, addition of IHF to the reaction mixture attenuates DNA-stimulated ATPase activity. The substrate used in this experiment (pCT-λ) is a 12 kb duplex that contains the cos-sequence (200 bp) and thus the I1 recognition element (Figure 1B) (20). Based on these and published observations, we hypothesized that non-specific DNA promotes ring tetramer assembly on the duplex and thus stimulates ATP hydrolysis; in contrast site-specific, cooperative assembly of IHF and the protomer at a cos site engender a maturation complex in which the packaging ATPase is down-regulated (Figure 1A) (21, 37). We reasoned that the vast excess of non-specific DNA in pCT-λ (12 kb) relative to the 200 bp cos-sequence precludes complete sequestration of all the terminase complexes and incomplete abrogation of the packaging ATPase activity. To directly test this hypothesis, we utilized short (274 bp) duplexes that contained the entire cos sequence (cos-274) or that was of random sequence (NS-274). Consistent with our hypothesis, the short non-specific duplex stimulates ATP hydrolysis, though not as strongly as longer pCT-λ duplex (Figure 4A). In contrast and as predicted, basal ATP hydrolysis by the protomer is unaffected by the cos-274 duplex and may in fact be attenuated in the presence of both cos-274 and IHF.
Figure 4. Activation of ATPase Activity in the Packaging Motor Complex.
Panel A. The ATPase activity assay was conducted as described in Experimental Procedures using 200 nM terminase protomer, 1 mM [α-32P]ATP, plus IHF and/or DNA as indicated. The observed rate is reported as an average of at least three separate experiments with standard deviation indicated. Panel B. The ATPase/cos-cleavage reaction was conducted as described in Experimental Procedures using 200 nM protomer and allowed to proceed for five minutes. Buffer (open squares) or 40 nM procapsids (closed squares) was then added to initiate the DNA packaging reaction. Aliquots were removed at the indicated times and ATP hydrolysis quantified by TLC assay. Each data point represents the average of at least three separate experiments with standard deviation indicated. The addition of procapsids increases ATPase activity with procapsids.
The next step in the packaging pathway is binding of procapsids to Complex I (Figure 1). This triggers “cos-clearance”, which includes release of the motor from the cos site and activation of the packaging ATPase to power translocation. We next examined the effect of procapsids on the ATPase activity of Complex I. The data presented in Figure 4B shows that the protomer possesses modest ATPase activity during maturation of the genome ends and in Complex I, but that ATP hydrolysis is stimulated upon subsequent addition of procapsids. Given that turnover at the maturation and assembly ATP binding sites is not observed (vide supra), this increase in activity must reflect ATP hydrolysis by the packaging ATPase in TerL.
DISCUSSION
The packaging of a viral genome into a pre-formed procapsid shell is a highly conserved process in the complex double-stranded DNA viruses. Terminase enzymes catalyze both the genome maturation and DNA packaging reactions and they are essential to virus assembly and infectivity. Given their central role, terminases may serve as specific targets for anti-viral therapeutics. Unfortunately, purification of these enzymes in a soluble, well-behaved, and homogenous state has been problematic in all viral systems and this has been a major impediment to their biochemical characterization. The isolation of lambda terminase in a soluble, homogenous, and highly active form has allowed us to interrogate the catalytic properties of the enzyme and to probe the packaging pathway in detail, without the ambiguity associated with a heterogeneous enzyme preparation.
What is the Nature of “Complex I”?
Complex I was first described as a stable terminase•DNA intermediate isolated from infected cells in the absence of procapsids (48). This intermediate could be chased into a packaging complex (Complex II) with the addition of procapsids. We previously demonstrated that the product of the maturation reaction in vitro is composed of terminase tightly bound to the mature DL end of the genome but that has ejected the DR end (36). We proposed that this accurately reflected Complex I isolated in vivo and current models incorporate this concept, as depicted in Figure 1C; however, the data presented here clearly demonstrate that separation of the nicked strands is quite slow and in fact un-necessary for duplex utilization by the packaging motor. Based on these data, we propose that while ejection of the DR end from the maturation complex occurs in vitro, this reaction does not accurately reflect the natural packaging pathway in vivo. Rather, terminase protomers assemble into a maturation complex at cos and rapidly nick the duplex strands. Separation of the nicked, annealed strands is slow and procapsids quickly capture this intermediate. Either procapsid binding or activation of the packaging motor triggers ejection of the DR end, which leads to cos-clearance and translocation of DNA into the capsid shell (Scheme 1). This revised model is harmonious with published data and further addresses a conundrum that has been present in the literature for over 30 years; although cos-cleavage and strand separation reactions are quite efficient in vitro, matured ends are not observed in the absence of procapsids in vivo. We suggest that while the reaction can easily be observed in vitro, strand separation from the maturation complex is sufficiently slow as to preclude the generation of free, mature ends within the cell. For the remainder of this work we use the terms genome maturation complex and packaging motor complex rather than the vague complex I and II terminology.
Scheme 1.
Kinetic Model for Maturation and Packaging by the λ Terminase Protomer.
Stoichiometry of the Genome Maturation and Packaging Motor Complexes
We have proposed that the terminase ring tetramer observed in solution is representative of the complex utilized for both genome maturation and packaging activities (12) and the data presented here are consistent with this hypothesis. With respect to duplex nicking, this model is analogous to the Type IIE and IIF restriction endonuclease enzymes where the catalytically competent enzyme complex is a tetramer within which two subunits each bind to one copy of the recognition sequence (44, 49). This interaction induces looping of the DNA between the sites and nicking of the duplex at either one or both of the recognition elements. We propose that the terminase ring tetramer adopts a similar strategy, bending and wrapping DNA at the cos site and introducing site-specific nicks into the duplex at cosN using two symmetrically disposed TerL subunits. The other two subunits similarly bind DNA but are catalytically silent. This model is consistent with the observation that terminase binding to cos, especially in the presence of IHF, bends and wraps the duplex occluding over 250 bp of DNA (50).
The maturation complex next binds to the procapsid to complete the packaging motor complex. On the surface our data suggest that additional protomers must be recruited to assemble a functional motor. It is feasible that additional subunits are recruited to the putative ring tetramer to engender a complex of higher-order stoichiometry; however, we disfavor this model based on the stability of the ring once formed (11, 12). Another possibility is that a second terminase ring could be recruited to power translocation, as is observed in the eukaryotic MCM and viral SV40 helicases where two hexameric rings form the active motor complex (51, 52). We suggest a third possibility. Strong cooperative interactions between subunits are presumed, if not directly demonstrated, in all known biological motors. Preliminary data in our lab indicate that this is true of the terminase packaging motor as well and that incorporation of a single defective protomer into the motor has profound effects on DNA packaging activity (Andrews and Catalano, in preparation). If we assume that our purified protomer preparation is ~ 90% active (vide supra), the defective subunits could significantly alter the observed dependence of protomer concentration on packaging activity. Unfortunately, the present data do not allow discrimination between these possibilities and biochemical, biophysical, and structural studies are currently underway to provide a detailed description of the packaging motor complex. Presently, we prefer a simple and unified model where both the maturation complex and the packaging motor complex are composed of a single terminase ring tetramer encircling viral DNA.
The Terminase Protomer Establishes Fidelity in Genome Packaging
In vitro packaging using the purified, but heterogeneous terminase mix exhibits low packaging fidelity and varying amounts of the DR strand and even the uncleaved substrate are packaged with reasonable efficiency (27, 28). This is similarly observed in other viral systems such as bacteriophage T4 and SPP1 where in vitro packaging using purified terminase is essentially non-specific (53-55). In contrast, packaging by the lambda terminase protomer exhibits high fidelity and only the matured DL end is packaged into the capsid shell, as expected of a biologically relevant species in vivo.
The mechanism for high fidelity of the protomer can be described based on a variety of biochemical data. The terminase protomer assembles into a ring tetramer in solution and the pre-assembled complex possesses site-specific (cos) maturation activity and it also efficiently packages duplex DNA, but in a non-specific manner. This reaction is independent of IHF and we presume that the ring tetramer can bind to any duplex end and then to a capsid to initiate DNA packaging. Terminase remains predominantly in the protomeric state at in vivo concentrations (100 nM) and the maturation and packaging activities of the protomer, while kinetically identical to the ring-tetramer, have a strict requirement for IHF. In this scenario, the protomer is devoid of maturation and packaging activities and assembly of the catalytically competent complexes occurs only at the cos site, mediated by IHF. Duplex nicking at cosN affords an intermediate in which terminase is tightly and specifically bound to the matured DL end and which only then binds to a procapsid to initiate packaging. This sequence of events bestows fidelity in the genome packaging reaction, as depicted in Scheme 1.
Regulation of the Packaging ATPase and a Model for Genome Packaging
The terminase protomer has a low basal ATPase activity while that of the isolated ring tetramer is robust (11, 12). We have proposed that this reflects assembly of the packaging ATPase catalytic site at the subunit interface of two protomers in the complex, in analogy to many hexameric ring helicases (11, 56, 57). We show here that non-specific DNA duplexes also stimulate ATPase activity, but at much lower and physiologically relevant terminase concentrations (100 nM). In contrast, duplexes that contain the cos-sequence do not stimulate ATPase activity and may in fact attenuate ATP hydrolysis in the presence of IHF. We interpret the ensemble of data presented here and in published work in the following model for genome packaging.
Non-specific DNA promotes the assembly of four protomers into the ring tetramer, similar to that we have observed in solution studies. We speculate that ring encircles the DNA and that the activated ATPase activity reflects a translocating complex that has engaged in a “one dimensional” search for a cos site sequence, a feature common to many site-specific DNA binding proteins (58, 59). IHF binds to a cos site in the concatemer and introduces a strong bend in the duplex; capture of the translocating complex by the bent duplex architecture engenders a stable, site-specifically bound maturation complex in which the packaging ATPase is down-regulated and in which cos-cleavage activity has been activated. Within this context, we have shown that terminase and IHF cooperatively bind to cos-DNA to afford a distinct and stable complex, while binding to non-specific duplexes yields multiple, diffuse bands in electrophoretic mobility shift studies (Sanyal and Catalano, in preparation) (15, 36). The activated maturation complex nicks the duplex, which is followed by procapsid binding. This affords the fully assembled motor complex which triggers strand separation and reactivates the packaging ATPase to fuel DNA translocation into the capsid shell§.
The model presented above is harmonious with published work and with the data presented here. It invokes coordinated interactions between the terminase subunits, cos-DNA, IHF, and the portal vertex of the capsid that regulate the assembly of a stable maturation complex and its transition to a dynamic packaging motor complex. A further complication is the observation that ATP binding to the assembly ATP binding site in TerS (i) regulates DNA binding interactions (13), (ii) down-regulates genome maturation activities (21), and (iii) activates the packaging ATPase site in TerL (29). Thus, complex allosteric regulation of the catalytic activities of terminase fine-tune each complex to its specific role – excision of a single genome from the concatemer and subsequent packaging of the duplex into the capsid shell.
The Protomer is the Biologically Relevant Species
Taken together, the biochemical features of the purified protomer closely match those expected of a biologically relevant species, as follows. First, the protomer is the prevalent species at in vivo concentrations; second, the kinetics of protomer assembly at cos and the subsequent DNA nicking reaction is much faster than previously reported; third, the procapsid binds to terminase only after it has matured the DL end to afford a motor with high packaging fidelity; and finally, ATP hydrolysis by the isolated protomer and in the maturation complex is weak but is strongly stimulated in the packaging motor complex (11). Thus, non-productive ATP hydrolysis is averted in the un-assembled protomer and in the maturation complex that must remain bound to cos, but the packaging ATPase is activated when required to power translocation. In sum, the isolated protomer is devoid of catalytic activity and the nuclease, strand-separation, ATPase, and DNA translocation activities are sequentially activated as appropriate to each nucleoprotein complex along the packaging pathway.
The Viral Genome Packaging Motors
Biological motors are essential to cellular vitality and serve a variety of roles. They are multimeric complexes that transduce the chemical energy of ATP hydrolysis to mechanical work. The terminase motors perform an equally important role in virus assembly and are among the most powerful biological motors characterized to date. Recent structural and single-molecule studies have yielded significant insight into the physical nature of the motors and have led to viable though conflicting models for the mechanism of DNA translocation by the complexes. Validation of these models requires solution based, biochemical interrogation of the catalytic properties of the enzymes. The present work thus complements the structural studies by providing a biochemical framework that describes the enzymology of the packaging motors.
It is clear that the functional terminase motors act as higher-order multimers of TerL and TerS subunits, but the nature of these complexes remains uncertain. Structural studies demonstrate that isolated TerS subunits assemble into ring structures composed of 8-12 subunits, depending on the virus studied (60-62). In contrast, the DNA binding domain of the lambda TerS subunit assembles into a stable dimer (63-65), which is consistent with the observed stoichiometry in the terminase protomer (TerL1•TerS2). Assembly of four lambda protomers affords a ring tetramer containing eight TerS subunits, which we presume are radially disposed in the packaging motor complex. Crystal structures have also been published for the isolated TerL subunit of phage T4 (66) and for the nuclease domains of phage P2 (67), phage SPP1 (68), and cytomegalovirus TerL subunits (69). These studies reveal that the TerL subunits are composed of two structural domains, an N-terminal translocation domain and a C-terminal maturation domain. Although there is no structural information available for the lambda TerL subunit, genetic and biochemical data indicate a similar structural and functional domain organization (14, 34).
CryoEM studies in the bacteriophage ϕ29 system suggest that the packaging motor is composed of five “ATPase” subunits (70). This virus represents a distinct class of dsDNA viruses that package monomeric genomes that uniquely utilize a “packaging RNA” (pRNA) as part of the functional motor complex and that do not utilize a TerS subunit, per se** (7, 8, 71). Adenoviruses utilize analogous genome replication and perhaps DNA packaging strategies (72). In contrast, lambda is representative of viruses such as herpesviruses and many bacteriophages that package genomes from a concatemeric DNA precursor (4, 8, 10, 14). In these cases, the terminase enzymes serve dual functions – excision of a single genome from the concatemer and concomitant packaging of the duplex into the capsid. These enzymes uniformly utilize a TerS subunit that is responsible for recognition of viral DNA and a TerL subunit that performs all of the maturation and packaging functions. Bacteriophage T4 terminase requires a small subunit for specific packaging of viral DNA in vivo, but it is dispensable for packaging non-specific duplexes in vitro (73). CryoEM structural studies of the T4 packaging motor suggest that the complex assembled from isolated TerL subunits is pentameric, similar to the phi29 “ATPase” complex described above.
Unfortunately, there is no high-resolution data for a fully assembled hetero-oligomeric terminase enzyme from any source. The isolation of a homogenous, well-behaved, and functional lambda terminase protomer composed of both TerS and TerL subunits in a well-defined stoichiometry has allowed detailed biochemical and biophysical characterization of the enzyme. The ensemble of biochemical data suggest that unlike the ϕ29 and T4 systems, the catalytically-competent maturation and motor complexes are composed of four terminase protomers assembled into a ring-like structure that encircles duplex DNA; this differs from the pentameric motors proposed in ϕ29 and T4. Notwithstanding, the essential features of the motors, including mechanochemical coupling of ATP hydrolysis to motor movement, cooperative interactions between the motor subunits during translocation, and the capacity to generate significant packaging forces, will certainly be recapitulated among all of the virus classes and the results presented here provide mechanistic insight into the enzymology of these fascinating biological motors.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of the Catalano laboratory for helpful discussions of this work. The authors further wish to thank Rishi Sanyal for providing the IHF and Shannon Kruse for providing procapsids used in these studies.
FUNDING: This work was supported by National Institute of Health grants GM088186 (CEC) and F32GM-905652 (BTA)
Abbreviations
- β-ME
β-mercaptoethanol
- cos
the cohesive end site of the bacteriophage lambda genome
- immature DNA
concatemeric lambda DNA
- mature DNA
genome length lambda DNA found within the viral capsid that contains the 12-base complementary single stranded ends
- TerL
the large terminase subunit, a.k.a. gpA
- TerS
the small terminase subunit, a.k.a. gpNu1
- terminase protomer
the homogenous holoenzyme composed of TerL and TerS subunits in a TerL1•TerS2 heterotrimer complex
- ring tetramer
the catalytically competent terminase complex composed of four protomers in a ring-like structure (TerL1•TerS2)4
- terminase mix
a pure, but structurally heterogeneous mixture of terminase protomers observed in purified protein preparations
Footnotes
SUPPLEMENTAL INFORMATION: The protocol for the synchronous DNA packaging assay, statistical analysis of the packaging ATPase data, and kinetic time course data for the maturation activities of the terminase protomer. This material is available free of charge online at http://pubs.acs.org.
Purified lambda terminase is composed of a homogenous 5.1 S species and a heterogeneous 13.3 S species that can be separated by gel filtration chromatography. The latter likely represents a tetramer of protomers with additional gpNu1 subunits non-specifically associated with the complex. The 13.3 S species dissociates to the protomer, which at elevated concentrations can be reassembled to a homogenous 14 S ring tetramer species in vitro.
Genome replication in the ϕ29-like viruses utilizes a protein-primed replication strategy and the product is a genome monomer that contains the initiator protein covalently attached at the 5′ end. This protein plays a role in DNA recognition by the packaging motor and may serve as a de facto TerS subunit.
We use the terms NTP and NDP to denote triphosphate and diphosphate nucleotides in a generic sense.
We note that the proposed translocating complex that engages in a one-dimensional search for cos is physically distinct from the packaging motor complex. In the latter case, the presumed ring tetramer is associated with the portal ring of a procapsid, which likely affects catalytic activity and mechanochemical properties of the motor.
REFERENCES
- 1.Knipe DM, Howley PM. Fields Virology. Fifth ed Lippincott-Williams, and Wilkins; New York, NY: 2007. [Google Scholar]
- 2.Calendar R, Abedon ST. The Bacteriophages. Oxford University Press; New York, N.Y.: 2006. [Google Scholar]
- 3.Ball A. Virus Replication Strategies. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott-Williams, and Wilkins; New York: 2007. pp. 119–138. [Google Scholar]
- 4.Catalano CE. Viral Genome Packaging Machines: An Overview. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, N.Y.: 2005. pp. 1–4. [Google Scholar]
- 5.Jardine PJ, Anderson DL. DNA Packaging in Double-Stranded DNA Phages. In: Calendar R, Abedon ST, editors. The Bacteriophages. 2nd ed Oxford University Press; New York, N.Y.: 2006. pp. 49–65. [Google Scholar]
- 6.Roizman B, Knipe DM, Whitley RJ. Herpes Simplex Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth ed Lippincott, Williams, and Wilkins; New York, NY: 2007. pp. 2501–2602. [Google Scholar]
- 7.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 8.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Micro. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 9.Furth M, Wickner S. Lambda DNA Replication. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. pp. 145–174. [Google Scholar]
- 10.Baines JD, Weller SK. Cleavage and Packaging of Herpes Simplex Virus 1 DNA. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, N.Y.: 2005. pp. 135–149. [Google Scholar]
- 11.Maluf NK, Yang Q, Catalano CE. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J Mol Biol. 2005;347:523–542. doi: 10.1016/j.jmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Catalano CE. A minimal kinetic model for a viral DNA packaging machine. Biochemistry. 2004;43:289–299. doi: 10.1021/bi035532h. [DOI] [PubMed] [Google Scholar]
- 14.Feiss M, Catalano CE. Bacteriophage Lambda Terminase and the Mechanism of Viral DNA Packaging. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, N.Y.: 2005. pp. 5–39. [Google Scholar]
- 15.Ortega ME, Catalano CE. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor. Biochemistry. 2006;45:5180–5189. doi: 10.1021/bi052284b. [DOI] [PubMed] [Google Scholar]
- 16.Medina E, Nakatani E, Kruse S, Catalano CE. Thermodynamic Characterization of Viral Procapsid Expansion into a Functional Capsid Shell. J. Mol. Biol. 2012;418:167–180. doi: 10.1016/j.jmb.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Yang Q, Maluf NK, Catalano CE. Packaging of a Unit-Length Viral Genome: The Role of Nucleotides and the gpD Decoration Protein in Stable Nucleocapsid Assembly in Bacteriophage Lambda. Journal of Molecular Biology. 2008;383:1037–1048. doi: 10.1016/j.jmb.2008.08.063. [DOI] [PubMed] [Google Scholar]
- 18.Imber R, Tsugita A, Wurtz M, Hohn T. Outer Surface Protein of Bacteriophage Lambda. Journal of Molecular Biology. 1980;139:277–295. doi: 10.1016/0022-2836(80)90131-x. [DOI] [PubMed] [Google Scholar]
- 19.Feiss M, Sippy J, Miller G. Processive action of terminase during sequential packaging of bacteriophage lambda chromosomes. J Mol Biol. 1985;186:759–771. doi: 10.1016/0022-2836(85)90395-x. [DOI] [PubMed] [Google Scholar]
- 20.Woods L, Terpening C, Catalano CE. Kinetic analysis of the endonuclease activity of phage lambda terminase: assembly of a catalytically competent nicking complex is rate-limiting. Biochemistry. 1997;36:5777–5785. doi: 10.1021/bi963044m. [DOI] [PubMed] [Google Scholar]
- 21.Chang JR, Andrews BT, Catalano CE. Energy Independent Helicase Activity of a Viral Genome Packaging. Biochemistry. 2012;51:391–400. doi: 10.1021/bi201604b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hang Q, Woods L, Feiss M, Catalano CE. Cloning, expression, and biochemical characterization of hexahistidine-tagged terminase proteins. J Biol Chem. 1999;274:15305–15314. doi: 10.1074/jbc.274.22.15305. [DOI] [PubMed] [Google Scholar]
- 23.Demeler B, van Holde KE. Sedimentation Velocity Analysis of Highly Heterogeneous Systems. Analytical Biochemistry. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 24.Demeler B, Saber H, Hansen JC. Identification and interpretation of complexity in sedimentation velocity boundaries. Biophysical Journal. 1997;72:397–407. doi: 10.1016/S0006-3495(97)78680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuck P. On the Analysis of Protein Self-Association by Sedimentation Velocity Analytical Ultracentrifugation. Analytical Biochemistry. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 26.Schuck P. Size-Distribution Analysis of Macromolecules by Sedimentation Velocity Ultracentrifugation and Lamm Equation Modeling. Biophysical Journal. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Catalano CE. Biochemical characterization of bacteriophage lambda genome packaging in vitro. Virology. 2003;305:276–287. doi: 10.1006/viro.2002.1602. [DOI] [PubMed] [Google Scholar]
- 28.Yang Q, Catalano CE, Maluf NK. Kinetic Analysis of the Genome Packaging Reaction in Bacteriophage Lambda. Biochemistry. 2009;48:10705–10715. doi: 10.1021/bi901016n. [DOI] [PubMed] [Google Scholar]
- 29.Woods L, Catalano CE. Kinetic characterization of the GTPase activity of phage lambda terminase: evidence for communication between the two "NTPase" catalytic sites of the enzyme. Biochemistry. 1999;38:14624–14630. doi: 10.1021/bi990866l. [DOI] [PubMed] [Google Scholar]
- 30.Tomka MA, Catalano CE. Kinetic characterization of the ATPase activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry. 1993;32:11992–11997. doi: 10.1021/bi00096a008. [DOI] [PubMed] [Google Scholar]
- 31.Hwang Y, Feiss M. Mutations affecting the high affinity ATPase center of gpA, the large subunit of bacteriophage lambda terminase, inactivate the endonuclease activity of terminase. J Mol Biol. 1996;261:524–535. doi: 10.1006/jmbi.1996.0480. [DOI] [PubMed] [Google Scholar]
- 32.Hwang Y, Catalano CE, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage Chi. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 33.Hwang Y, Feiss M. Mutations affecting lysine-35 of gpNu1, the small subunit of bacteriophage lambda terminase, alter the strength and specificity of holoterminase interactions with DNA. Virology. 1997;231:218–230. doi: 10.1006/viro.1997.8542. [DOI] [PubMed] [Google Scholar]
- 34.Ortega ME, Gaussier H, Catalano CE. The DNA maturation domain of gpA, the DNA packaging motor protein of bacteriophage lambda, contains an ATPase site associated with endonuclease activity. J Mol Biol. 2007;373:851–865. doi: 10.1016/j.jmb.2007.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosloff M, Selinger Z. Substrate Assisted Catalysis , Application to G Proteins. Trends in Biochemical Sciences. 2001;26:161–166. doi: 10.1016/s0968-0004(00)01748-5. [DOI] [PubMed] [Google Scholar]
- 36.Yang Q, Hanagan A, Catalano CE. Assembly of a nucleoprotein complex required for DNA packaging by bacteriophage lambda. Biochemistry. 1997;36:2744–2752. doi: 10.1021/bi9622682. [DOI] [PubMed] [Google Scholar]
- 37.Tomka MA, Catalano CE. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage lambda. J Biol Chem. 1993;268:3056–3065. [PubMed] [Google Scholar]
- 38.Murialdo H, Siminovitch L. The Morphogenesis of Bacteriophage Lambda. IV. Identification of Gene Products and Control of the Expression of the Morphogenetic Information. Virology. 1972;48:785–823. doi: 10.1016/0042-6822(72)90162-6. [DOI] [PubMed] [Google Scholar]
- 39.Gaussier H, Yang Q, Catalano CE. Building a virus from scratch: assembly of an infectious virus using purified components in a rigorously defined biochemical assay system. J Mol Biol. 2006;357:1154–1166. doi: 10.1016/j.jmb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Catalano CE. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catalano CE, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 42.Daniels D, Schroeder J, Szybalski W, Sanger F, Coulson A, Hong G, Hill D, Petersen G, Blattner F. Complete Annotated Lambda Sequence. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. [Google Scholar]
- 43.Davidson AR, Gold M. Mutations Abolishing the Endonuclease Activity of Bacteriophage Lambda Terminase Lie in Two Distinct Regions of the A gene, One of Which May Encode a "Leucine zipper" DNA-Binding Domain. Virology. 1992;189:21–30. doi: 10.1016/0042-6822(92)90677-h. [DOI] [PubMed] [Google Scholar]
- 44.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II Restriction Endonucleases: Structure and Mechanism. Cellular and Molecular Life Sciences. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 45.Lohman TM, Bjornson KP. Mechanisms of Helicase-Catalyzed DNA Unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 46.von Hippel PH, Delagoutte E. Macromolecular Complexes That Unwind Nucleic Acids. BioEssays. 2003;25:1168–1177. doi: 10.1002/bies.10369. [DOI] [PubMed] [Google Scholar]
- 47.Patel SS, Pandey M, Nandakumar D. Dynamic Coupling Between the Motors of DNA Replication: Hexameric Helicase, DNA Polymerase, and Primase. Current Opinion in Chemical Biology. 2011;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perucchetti R, Parris W, Becker A, Gold M. Late Stages in Bacteriophage Lambda Head Morphogenesis: in Vitro Studies on the Action of the Bacteriophage Lambda D-Gene and W-Gene Products. Virology. 1988;165:103–114. doi: 10.1016/0042-6822(88)90663-0. [DOI] [PubMed] [Google Scholar]
- 49.Chan S-H, Stoddard BL, Xu S.-y. Natural and Engineered Nicking Endonucleases— From Cleavage Mechanism to Engineering of Strand-Specificity. Nucleic Acids Res. 2011;39:1–18. doi: 10.1093/nar/gkq742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higgins RR, Becker A. Interaction of Terminase, the DNA Packaging Enzyme of Phage Lambda, with its cosDNA Substrate. Journal of Molecular Biology. 1995;252:31–46. doi: 10.1006/jmbi.1995.0473. [DOI] [PubMed] [Google Scholar]
- 51.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. Structure of the Replicative Helicase of the Oncoprotein SV40 Large Tumour Antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 52.Fletcher RJ, Shen J, Gomez-Llorente Y, San Martin C, Carazo JM, Chen XS. Double Hexamer Disruption and Biochemical Activities of Methanobacterium thermoautotrophicum MCM. Journal of Biological Chemistry. 2005;280:42405–42410. doi: 10.1074/jbc.M509773200. [DOI] [PubMed] [Google Scholar]
- 53.Oliveira L, Alonso JC, Tavares P. A Defined in Vitro System for DNA Packaging by the Bacteriophage SPP1: Insights into the Headful Packaging Mechanism. Journal of Molecular Biology. 2005;353:529–539. doi: 10.1016/j.jmb.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 54.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, Kottadiel VI, Vafabakhsh R, Dai L, Chemla YR, Ha T, Rao VB. A Promiscuous DNA Packaging Machine from Bacteriophage T4. PLoS Biol. 2011;9:e1000592. doi: 10.1371/journal.pbio.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niedenzu T, R√∂leke D, Bains G, Scherzinger E, Saenger W. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. Journal of Molecular Biology. 2001;306:479–487. doi: 10.1006/jmbi.2000.4398. [DOI] [PubMed] [Google Scholar]
- 57.Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal Structure of the Helicase Domain from the Replicative Helicase-Primase of Bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 58.von Hippel PH, Berg OG. Facilitated Target Location in Biological Systems. Journal of Biological Chemistry. 1989;264:675–678. [PubMed] [Google Scholar]
- 59.Givaty O, Levy Y. Protein Sliding along DNA: Dynamics and Structural Characterization. Journal of Molecular Biology. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 60.Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proceedings of the National Academy of Sciences. 2010;107:1971–1976. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Büttner CR, Chechik M, Ortiz-Lombard√≠a M, Smits C, Ebong I-O, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson AA. Structural basis for DNA recognition and loading into a viral packaging motor. Proceedings of the National Academy of Sciences. 2012;109:811–816. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proceedings of the National Academy of Sciences. 2012;109:817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 64.Bain DL, Berton N, Ortega M, Baran J, Yang Q, Catalano CE. Biophysical characterization of the DNA binding domain of gpNu1, a viral DNA packaging protein. J Biol Chem. 2001;276:20175–20181. doi: 10.1074/jbc.M100517200. [DOI] [PubMed] [Google Scholar]
- 65.Yang Q, de Beer T, Woods L, Meyer JD, Manning MC, Overduin M, Catalano CE. Cloning, expression, and characterization of a DNA binding domain of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999;38:465–477. doi: 10.1021/bi981271d. [DOI] [PubMed] [Google Scholar]
- 66.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 67.Roy A, Cingolani G. Structure of P22 Headful Packaging Nuclease. Journal of Biological Chemistry. 2012;287:28196–28205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA. Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase. EMBO Reports. 2009;10:592–598. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nadal M, Mas PJ, Blanco AG, Arnan C, Sol√† M, Hart DJ, Coll M. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proceedings of the National Academy of Sciences. 2010;107:16078–16083. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining Molecular and Domain Boundaries in the Bacteriophage Phi-29 DNA Packaging Motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson D, Grimes S. The phi29 DNA Packaging Motor: Seeking the Mechanism. In: Catalano CE, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, N.Y.: 2005. pp. 103–116. [Google Scholar]
- 72.Berk AJ. Adenoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth ed Lippincott, Williams and Wilkins; Philidelphia, PA: 2007. pp. 2355–2394. [Google Scholar]
- 73.Gao S, Rao VB. Specificity of Interactions among the DNA-packaging Machine Components of T4-related Bacteriophages. Journal of Biological Chemistry. 2011;286:3944–3956. doi: 10.1074/jbc.M110.196907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.