Abstract
To establish the zebrafish as a model for investigating the methylation pathway of drug metabolism, we embarked on the molecular cloning of the zebrafish catechol O-methyltransferase (COMT). By searching the GenBank database, a zebrafish nucleotide sequence encoding a putative COMT was identified. Based on the sequence information, we designed and synthesized oligonucleotides corresponding to its 5’- and 3’-coding regions of this zebrafish COMT. Using the first-strand cDNA reverse-transcribed from the total RNA isolated from a 3-month-old adult female zebrafish as the template, the cDNA encoding the zebrafish COMT was PCR-amplified. The recombinant zebrafish COMT protein was subsequently expressed in and purified from BL21 (DE3) Escherichia coli cells transformed with the pGEX-2TK expression vector harboring the zebrafish COMT cDNA. Upon enzymatic characterization, purified COMT displayed methylating activity toward dopamine, dopa, and catecholestrogens, as well as three representative catechol drugs, methyldopa, dobutamine, and isoproterenol. A reverse transcription-polymerase chain reaction (RT-PCR) analysis revealed developmental stage-dependent expression of the zebrafish COMT during embryonic development and throughout the larval stage onto maturity. These results provide a foundation for investigating the involvement of COMT-mediated methylation in protection against the adverse effects of catechol drugs and other xenobiotic catechols during the developmental process.
Keywords: Catechol O-methyltransferase, developmental expression, methylation, molecular cloning, zebrafish
Introduction
It is generally known that the metabolism of drugs and other xenobiotics may proceed through two phases, with the Phase I reactions involving the generation of functional groups that may subsequently be used in the Phase II conjugations [1]. Prominent among the Phase II conjugation reactions are methylation, sulfonation, acetylation, glucuronidation, and glutathione conjugation [2]. In addition to their involvement in drug metabolism, some of the Phase II conjugation reactions such as methylation, sulfonation, and glucuronidation have been reported to also play a role in the metabolism of key endogenous compounds such as catecholamines and steroid/thyroid hormones [3–7].
Enzymatic methylation of catecholamines was first discovered by Axelrod et al. [8, 9]. The responsible enzyme was identified as the catechol O-methyltransferase (COMT) which catalyzes the methylation reactions using S-adenosyl-L-methionine as the methyl group donor [2, 10, 11]. The enzyme has since been shown to be capable of methylating not only the endogenous catecholamines and catecholestrogens, but also many catechol drugs such as levodopa, carbidopa, benserazide, apomorphine, dobutamine, isoprenaline (isoproterenol), rimiterol, inamrinone, and isoetharine [10–12]. In humans, there exist two isoforms of COMT, membrane-bound and soluble, that are encoded by mRNAs derived from the same gene through differential transcription/translation [13, 14]. Both the membrane-bound and soluble COMTs have been detected in various human tissues, albeit at different ratios [15]. The major physiological function of COMT-mediated methylation is generally thought to be for the deactivation of biologically-active or chemically-reactive endogenous as well as xenobiotic catechols [6, 10, 11, 16]. For catechol drugs, COMT therefore plays a dual role in undesirably lowering the efficacy of catechol drugs, as in the case of levodopa used in treating Parkinson’s disease, and in furnishing a protection mechanism against their potential adverse effects.
Zebrafish has in recent years emerged as a popular animal model for a wide range of studies [17–19]. Its advantages, compared with mouse, rat, or other vertebrate animal models, include the small size, availability of a relatively large number of eggs, rapid external development of virtually transparent embryos, and short generation time. These unique characteristics make the zebrafish an excellent model for a systematic investigation of the developmental stage-dependent and cell type/tissue/organ-specific expression, as well as physiological involvement of the COMT particularly with regard to the homeostasis of catecholamines and catecholestrogens and the detoxification of xenobiotic catechols including catechol drugs. A prerequisite for using the zebrafish in these studies, however, is the identification and functional characterization of the zebrafish COMT.
We report in this communication the identification of a COMT from zebrafish. The enzymatic activities toward a variety of endogenous and xenobiotic catecholic compounds were examined. To gain insight into its involvement in vivo, the developmental stage-dependent expression of the zebrafish COMT was investigated.
Materials and Methods
Materials
Dopamine, epinephrine, l-3,4-dihydroxyphenylalanine (l-Dopa), methyl-Dopa, S-adenosyl-L-methionine (AdoMet), sodium dodecyl sulfate (SDS), sodium acetate, 2-morpholinoethanesulfonic acid (MES), 3-(N-morpholino)propanesulfonic acid (MOPS), N-2-hydroxylpiperazine-N’-2-ethanesulfonic acid (HEPES), 3-[N-tris-(hydroxymethyl)methylamino]-propanesulfonic acid (TAPS), 2-(cyclohexylamino)ethanesulfonic acid (CHES), 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), Trizma base, dithiothreitol (DTT), carbidopa, isoproterenol, apomorphine, dobutamine, and isopropyl β-d-thiogalactopyranoside (IPTG) were products of Sigma Chemical Company. 2-Hydroxy-E1, 4-hydroxy-E1, 2-hydroxy-E2, and 4-hydroxy-E2 were products of Steraloids, Inc. TRI Reagent was from Molecular Research Center, Inc. Total RNA from a 3-month-old female zebrafish was prepared using the TRI Reagent based on the procedure described previously [20]. Taq DNA polymerase was a product of Promega Corporation. Takara Ex Taq DNA polymerase was purchased from Fisher Scientific. T4 DNA ligase and Bam HI restriction endonuclease were from New England Biolabs. Oligonucleotide primers were synthesized by MWG Biotech. pSTBlue-1 AccepTor Vector Kit and BL21 (DE3) competent cells were from Novagen. Protein molecular weight standards were from Fermentas Life Sciences. pGEX-2T glutathione S-transferase (GST) gene fusion vector, GEX-5’ and GEX-3’ sequencing primers, and glutathione-Sepharose 4B were products of GE Healthcare. DyNAmo HS SYBR Green qPCR Kit was from New England Biolabs. Cellulose thin-layer chromatography (TLC) plates were products of EM Science. [14C]-labeled AdoMet was from American Radiolabeled Chemicals, and Ecolume scintillation cocktail was from MP Biomedicals. All other reagents were of the highest grades commercially available.
Cloning, bacterial expression, and purification of recombinant zebrafish COMT
By searching the GenBank database, a zebrafish cDNA (GenBank Accession No. CR457452.12) encoding a putative COMT was identified. To generate the cDNA for subcloning into the pSTBlue-1 vector, sense and antisense oligonucleotide primers designed based on 5’- and 3’- regions of the coding sequence were synthesized (Table 1). Using these primer sets, PCR was carried out under the action of EX Taq DNA polymerase, with the first-strand cDNA reverse-transcribed from the total RNA isolated from a 3-month-old adult female zebrafish as the template. Amplification conditions were 2 min at 94°C and 25 cycles of 94°C for 30 s, 60°C for 35 s, and 72°C for 45 s, followed by a 5-min incubation at 72°C. The final reaction mixture was applied onto a 1% agarose gel, separated by electrophoresis, and visualized by ethidium bromide staining. The PCR product band detected was excised from the gel, and the DNA therein was isolated by spin filtration. Purified PCR product was cloned into the pSTBlue-1 vector and verified for authenticity by nucleotide sequencing [21]. To amplify a truncated cDNA encoding a “soluble-form” of the zebrafish COMT, another set of sense and antisense primers (Table 1) was used in a PCR reaction with pSTBlue-1 harboring the full-length zebrafish COMT cDNA (see above) as the template. Amplification conditions were the same as described above. At the end of the PCR reaction, the PCR product was purified, subjected to Bam HI restriction, and subcloned into Bam HI-restricted pGEX-2TK vector. To express the recombinant zebrafish COMT, competent Escherichia coli BL21 (DE3) cells transformed with pGEX-2TK harboring the COMT cDNA were grown in 1 L LB medium supplemented with 60 µg/ml ampicillin. After the cell density reached 0.6 OD600nm, IPTG (0.1 mM final concentration) was added to induce the production of recombinant zebrafish COMT. After an overnight induction at room temperature, the cells were collected by centrifugation and homogenized in 25 ml ice-cold lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA) using an Aminco French Press. Twenty µl of 10 mg/ml aprotinin (a protease inhibitor) was added to the crude homogenate. The crude homogenate was subjected to centrifugation at 10,000 × g for 15 min at 4°C. The supernatant collected was fractionated using 2.5 ml of glutathione-Sepharose, and the bound GST-fusion protein was eluted by an elution buffer (50 mM Tris-HCl, pH 8.0, plus 10 mM reduced glutathione) at 4°C or treated with 3 ml of a thrombin digestion buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2) containing 5 unit/ml bovine thrombin at room temperature. Following a 15-min incubation with constant agitation, the preparation was subjected to centrifugation. The recombinant zebrafish COMT was analyzed for purity by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to enzymatic characterization.
Table 1.
Oligonucleotide primers used for the cDNA cloning of zebrafish COMT and for the quantitative real-time RT-PCR analysis of the developmental stage-dependent expression of the zebrafish COMT
| Target sequence |
Sense and antisense oligonucleotide primers used | |
|---|---|---|
| I. For cDNA cloning*: | ||
| COMT | Sense: | 5’ -CGCGGATCCATGCTGTGGGTTGTGTTGGCAGTGGTGGTG-3’ |
| (Full-length) | Antisense: | 5’ -CGCGGATCCTCATCCTAAGAAGACCGATTTCTCCAGGCC-3’ |
| COMT | Sense: | 5’ -CGCGGATCCCACGATGTGGTCCATCAGCGGCTCCTGAAC-3’ |
| (Truncated) | Antisense: | 5’ -CGCGGATCCTCATCCTAAGAAGACCGATTTCTCCAGGCC-3’ |
| II. For real-time RT-PCR analysis**: | ||
| COMT | Sense: | 5’ -TCACGACCACAGCGCATCT-3’ |
| Antisense: | 5’ -CCCACATTCATGGCCCATT-3’ | |
| β-Actin | Sense: | 5’ -CGAGCTGTCTTCCCATCCA-3’ |
| Antisense: | 5’ -TCACCAACGTAGCTGTCTTTCTG-3’ | |
Recognition sites of BamHI restriction endonuclease in the oligonucleotides are underlined. Initiation and termination codons for translation are in bold type.
The sense and antisense oligonucleotide primer sets listed were verified by BLAST Search to be specific for the zebrafish COMT or β-actin nucleotide sequence.
Enzymatic assay
The methylating activity of purified recombinant zebrafish COMT was assayed using radioactive [14C]-labeled AdoMet as the methyl group donor. The standard assay mixture, with a final volume of 20 µl, contained 50 mM TrisHCl buffer at pH 7.5, 0.1 mM [14C]-labeled S-adenosyl-L-methionine, 5 mM DTT, 1.5 mM MgCl2, 1 mM substrate. Controls with DMSO or water, in place of substrate, were also prepared. The reaction was started by the addition of the enzyme, allowed to proceed for 60 min at 28°C, and terminated by the addition of 10 µl of 1 N HCl. The precipitates formed were cleared by centrifugation, and the supernatant was subjected to the analysis of [14C]methylated product using a previously developed TLC procedure [22], with n-butanol/isopropanol/88% formic acid/water (3:1:1:1; by volume) as the solvent system. To examine the pH-dependence of the methylation of 2-OH-E1 or dobutamine, different buffers (50 mM Mes at pH 5.5 or 6.5; Hepes at pH 7.5; Taps at pH 8.5; Ches at pH 9.5; Caps at pH 10.5 or 11.5), were used in the reactions with 1 mM of each substrate.
Analysis of the development stage-dependent expression of the zebrafish COMT
Quantitative real-time PCR was employed to investigate the developmental stage-dependent expression of the zebrafish COMT. For use as templates in real-time PCR, first-strand cDNA was reverse-transcribed from total RNAs isolated from zebrafish embryos and larvae at different developmental stages, and adult (3-months-old male or female) fish. Oligonucleotide primers (Table 1) for quantitative real-time PCR were designed using Primer Express software (Applied Biosystems, Foster City, CA) based on the following parameters: primer size between 18 and 22 base pairs; primer Tm range between 58°C and 62°C; and GC content between 50% and 60%. The nucleotide sequences for respective target PCR products were blasted to confirm their specificity. PCR amplification was performed using the Eppendorf mastercycler® ep realplex system. Reactions were carried out in triplicate. The reaction mixtures (25 µl final volume) contained 12.5 µl of SYBR® 2X Master mix, 500 nM each of sense and antisense oligonucleotide primers, and 1 µl (1.25 ng) of the DNA template. Reaction conditions were as follows: 15 min at 95°C for initial denaturation followed by 94°C for denaturation, 25 sec at 56°C for annealing and 30 sec at 72°C for extension. The expression values obtained were normalized against those from the control zebrafish β-actin.
Miscellaneous methods
SDS-PAGE was performed on 12% polyacrylamide gels using the method of Laemmli [23]. Protein determination was based on the method of Bradford [24] with bovine serum albumin as the standard.
Statistical analysis
Student’s t-test was used for the analysis of the specific activities of the zebrafish COMT toward catecholic compounds (cf. Table 2). Analysis of variance (ANOVA) was used for the analysis of pH-dependence data (cf. Figure 4). ANOVA with subsequent group comparisons using the Dunnett’s test at the 0.001 level of significance (99.9% confidence interval) was used for the analysis of real-time PCR data (cf. Figure 5).
Table 2.
Specific activities of the zebrafish COMT toward catecholic compounds
| Substrate | Specific Activity (nmol/min/mg)a,b |
|---|---|
| L-DOPA | 5.44 ± 0.51# |
| Methyl-DOPA | 0.20 ± 0.05# |
| Carbidopa | 2.19 ± 0.22# |
| Dopamine | 37.43 ± 1.07* |
| Epinephrine | 18.94 ± 0.96* |
| Isoproterenol | 41.04 ± 1.24* |
| Apomorphine | 0.529 ± 0.18# |
| 2-OH-E1 | 157.33 ± 6.67* |
| 4-OH-E1 | 46.31 ± 3.46# |
| 2-OH-E2 | 144.57 ± 4.29* |
| 4-OH-E2 | 85.80 ± 1.12* |
| Dobutamine | 128.54 ± 5.66* |
Data represent means ± S.D. derived from four experiments.
Statistical significance versus the activity of control is indicated by * with p<0.0001 or # with p<0.005. The sample size (n) analyzed was 8.
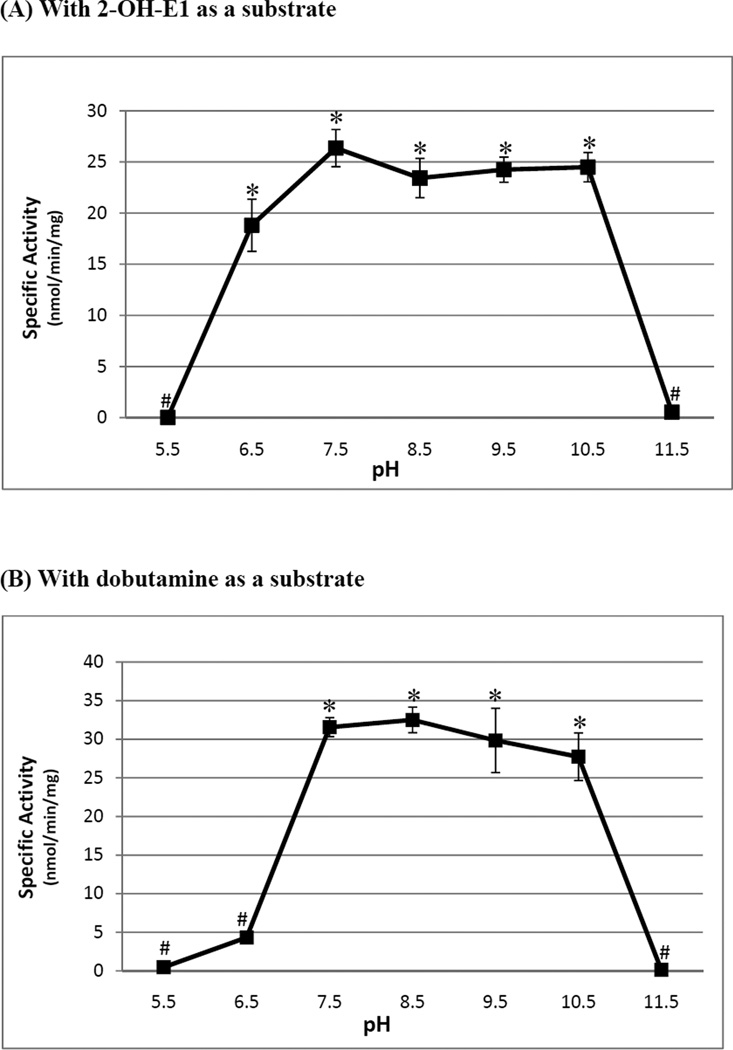
Figure 4.
pH-dependency of the methylating activity of the zebrafish COMT with (A) 2-hydroxy-E1 and (B) dobutamine as substrate. The enzymatic assays were carried out under standard assay conditions as described in Materials and Methods, using different buffer systems as indicated. The data represent calculated mean ± SD derived from three experiments. Statistical significance versus the activity of control is indicated by * with p<0.001 or # with p>0.001 for the 2-hydroxy-E1- or dobutamine-sulfating activity of the zebrafish COMT. The sample size (n) analyzed was 8.
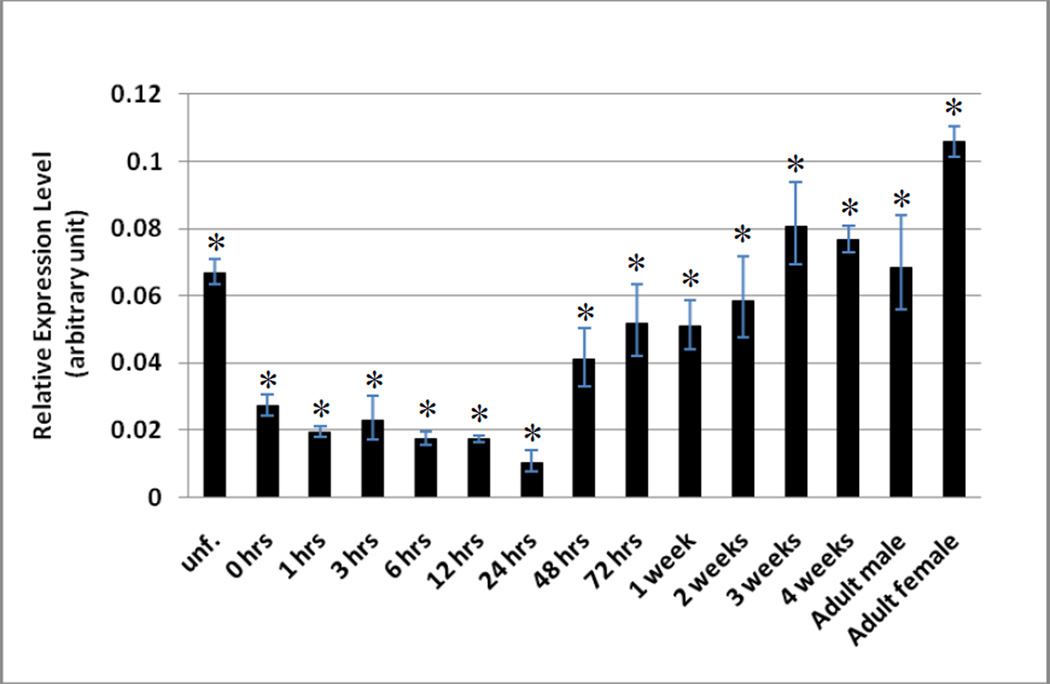
Figure 5.
Developmental stage-dependent expression of the zebrafish COMT. The expression of the COMT mRNA at different stages during embryogenesis and larval development onto maturity was analyzed using quantitative real-time PCR. Samples analyzed were unfertilized zebrafish eggs, zebrafish embryos during the zygote period (0-hour post-fertilization (pf)), cleavage period (1-hour pf), blastula period (3-hour pf), gastrula period (6-hour pf), neurula/segmentation period (12-hour pf), pharyngula period (24-hour pf), and hatching period (48- and 72-hour pf), 1, 2, 3, 4-week-old zebrafish larvae, and 3-month-old male or female zebrafish. The zebrafish COMT mRNA levels were quantified in arbitrary units, normalized against β-actin signal, as described in the Materials and Methods. The data represent calculated mean values derived from three experiments. Statistical significance versus the result for the control is indicated by * with p<0.001. The sample size (n) analyzed was 8.
Results and Discussion
To develop a zebrafish model for systematically investigating the physiological involvement of the Phase II detoxifying/drug-metabolizing enzymes including the COMT, a prerequisite is the identification of the different Phase II enzymes that are present in the zebafish. We have earlier embarked on the molecular cloning and characterization of the zebrafish cytosolic sulfotransferases (SULTs) that led to the identification of a number of enzymes that belong to different SULT gene families [25–34]. In the current study, we report the identification, characterization and ontogeny of a zebrafish COMT.
Molecular cloning of the zebrafish COMT
By searching the GenBank database, a zebrafish nucleotide sequence (GenBank Accession No. CR457452) encoding a putative COMT was identified. The cDNA, amplified by RT-PCR and cloned into the pGEX-2T vector, was subjected to nucleotide sequencing for authenticity [21]. The nucleotide sequence obtained was submitted to the GenBank database under the Accession No. HM997189. Figure 1 shows the nucleotide and deduced amino acid sequences of the newly identified zebrafish COMT. The open reading frame encompasses 768 nucleotides and codes for a 256-amino acid polypeptide, with a predicted molecular weight of 29,410. Sequence analysis based on a BLAST pairwise search revealed that the deduced amino acid sequence of the zebrafish COMT displays 55%, 57%, and 58% identity to human, rat, and mouse COMT, respectively. Figure 2 shows the alignment of the zebrafish COMT amino acid sequence with those of the membrane-bound form of human, rat, and mouse COMTs [35–37]. Similar to the other three COMTs, the zebrafish COMT also contains a transmembrane sequence (as underlined), indicating its being likely a membrane protein. More over, a number of conserved amino acid residues previously shown to be important for AdoMet-binding (E140, D191, K194 for the human COMT; as indicated by solid round dots) or catechol-binding (W88, P224, E249, and Y250 for the human COMT; as indicated by solid triangles) [38–40] are also found for the zebrafish COMT. These results therefore showed further the considerable conservation with regard to the critical amino acid residues between the zebrafish and mammalian COMTs.
Figure 1.
Nucleotide and deduced amino acid sequences of the zebrafish COMT cDNA. Nucleotides are numbered in the 5’ to 3’ direction.
Figure 2.
Alignment of the amino acid sequences of the zebrafish, human, rat, and mouse COMTs. Residues conserved among the four enzymes are bracketed. Amino acid residues previously shown to be important for AdoMet-binding are indicated by solid round dots (●); and those important for catechol-binding are indicated by solid triangles (▲). The transmembrane regions of the four enzymes are underlined.
Expression, purification, and characterization of recombinant zebrafish COMT
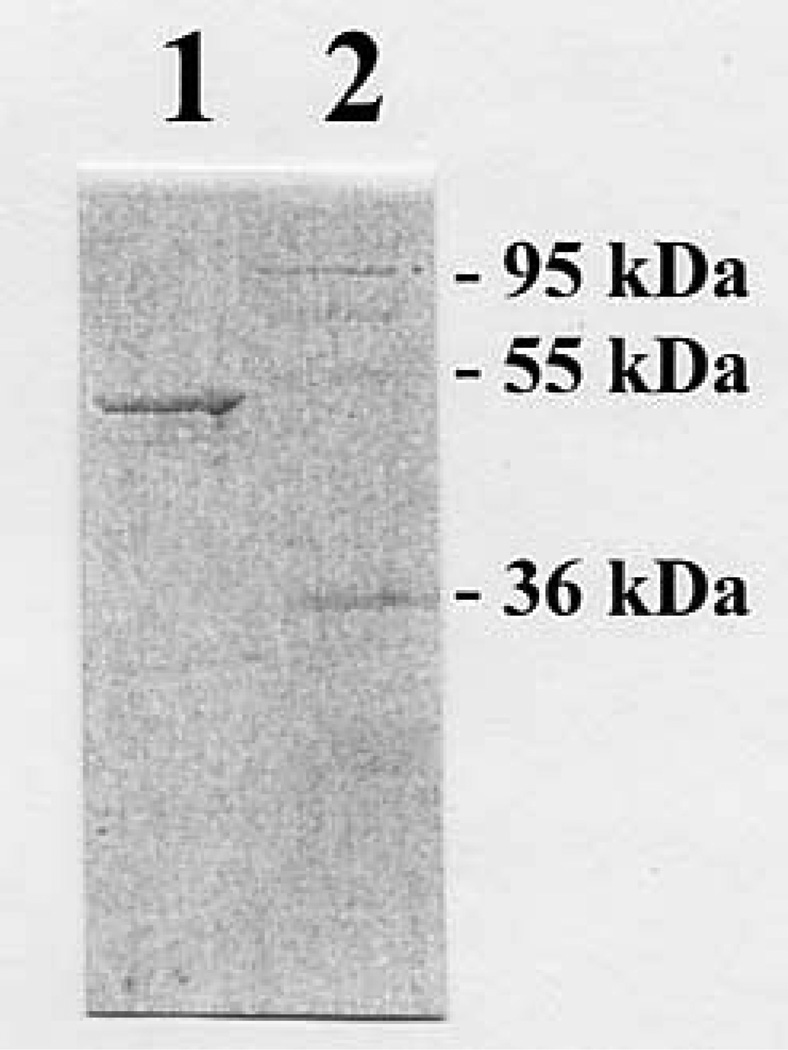
To avoid potential problems frequently encountered in the expression and purification of membrane proteins, we opted to express the zebrafish COMT in a soluble form. Sense primer corresponding to an N-terminal sequence immediately following the transmembrane domain (cf. Figure 2) and antisense primer corresponding to the C-terminal end of the open reading frame were used to PCR-amplify a truncated COMT cDNA, which was cloned into the pGEX-2T prokaryotic expression vector. pGEX-2T harboring zebrafish COMT cDNA was then transformed into E. coli BL21 (DE3) cells for the expression of recombinant enzyme. As shown in Figure 3, the GST-fusion protein form of the recombinant zebrafish COMT, purified from the E. coli extract, migrated at ~48 kDa position upon SDS-PAGE. It should be mentioned that, while the free form of the zebrafish COMT generated upon thrombin digestion was also prepared, the enzyme exhibited lower and unstable methylating activity in comparison with the GST-COMT fusion protein (data not shown). The GST-COMT fusion protein, therefore, was used for the enzymatic characterization. (The specific activities determined in the following studies were corrected for the molecular mass (23.9 kDa) of the GST moiety, in the fusion protein form of the enzymes.) A pilot experiment first revealed that the zebrafish COMT displayed strong methylating activity toward 2-hydroxy-E1. A subsequent pH-dependence experiment showed that the zebrafish COMT displayed a broad pH optimum (spanning pH 6.5 through 10.5) with 2-hydroxy-E1 as the substrate (Figure 4A). With dobutamine as the substrate, the enzyme exhibited a pH optimum spanning pH 7.5 through 10.5 (Figure 4B). A variety of endogenous and xenobiotic catecholic compounds were subsequently tested as substrates for the enzyme. The activity data obtained are compiled in Table 2. Among the substrates tested, the zebrafish COMT displayed strong methylating activities toward the catecholestrogens (2-OH-E1, 4-OH-E1, 2-OH-E2, and 4-OH-E2) and dobutamine. Lower, but significant activities were also detected toward other endogenous (L-Dopa, dopamine, epinephrine, and epinephrine) and xenobiotic (methyl-Dopa, carbidopa, isoproterenol, and apomorphine) catecholic compounds tested as substrates.
Figure 3.
SDS gel electrophoretic pattern of the purified recombinant zebrafish COMT. SDS-PAGE was performed on a 12% gel, followed by Coomassie blue staining. Samples analyzed in lanes 1 and 2 were GST-COMT fusion protein and protein molecular weight markers.
Developmental stage-dependent expression of the zebrafish COMT
To gain insight into the timing of the physiological involvement of the zebrafish COMT, the expression of the mRNA encoding COMT was examined from embryogenesis to maturity using RT-PCR. As shown in Figure 5A, a significant level of the coding mRNA of COMT, was detected in unfertilized eggs (lane 1), indicating clearly its maternal origin. Upon fertilization, however, there was a considerable decrease in the level of the COMT mRNA, which remained low throughout the early embryonic development until the pharyngula period (24-hour pf; lane 7). Upon hatching (48- and 72-hour pf; lanes 8 and 9), the COMT mRNA showed a dramatic increase, which remained high into the late (4-week) larval stage (lane 13). In adult zebrafish, there appeared to be a decrease in the level of COMT mRNA. Interestingly, the level of COMT mRNA was considerably lower in male fish (lane 14) than in female fish (lane 15). In contrast to the developmental stage-dependent expression of the zebrafish COMT, β-actin, a housekeeping protein was found to be consistently expressed throughout the entire developmental process (Figure 5B). While it will be necessary to examine the level of COMT protein using, for example, Western blot analysis, these results provided the initial clues to the developmental stage-dependent expression of the zebrafish COMT. That the low level of COMT mRNA during the early embryonic development may imply that there may correspondingly be low level of the COMT enzyme in place to mediate the methylation of catecholic compounds, leading to their inactivation and/or facilitated removal from the body.
In conclusion, we have cloned, expressed, and purified a novel zebrafish COMT enzyme, and examined its enzymatic properties, as well as developmental stage-dependent expression. This study represents part of an overall effort to establish the zebrafish as a model for investigating the Phase II metabolism of xenobiotics. More work is warranted in order to achieve this goal.
Acknowledgments
This work was supported in part by a National Institutes of Health grant GM085756 and a startup fund from College of Pharmacy, The University of Toledo.
Abbreviations
- COMT
catechol O-methyltransferase
- AdoMet
S-adenosyl-L-methionine
- RT-PCR
reverse transcription-polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gonzalez FJ, Tukey RH. Drug metabolism. In: Brunton LL, Lazo JS, Parker KL, editors. Goodman & Gilman’s The Pharmacological Basis of Therapeutics. New York: McGraw-Hill; 2006. pp. 71–91. [Google Scholar]
- 2.Mulder GJ. Conjugation reactions in drug metabolism. London: Taylor and Francis, Ltd.; 1990. [Google Scholar]
- 3.Falany C, Roth JA. Properties of human cytosolic sulfotransferases involved in drug metabolism. In: Jeffery EH, editor. Human Drug Metabolism: From Molecular Biology to Man. Boca Raton, FL: CRC Press; 1993. pp. 101–115. [Google Scholar]
- 4.Weinshilboum R, Otterness D. Sulfotransferase enzymes in conjugation-deconjugation reactions. In: Kaufmann FC, editor. Drug Metabolism and Toxicity. Berlin: Springer-Verlag; 1994. pp. 45–78. [Google Scholar]
- 5.Coughtrie MW. Sulfation through the looking glass--recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- 6.Zhu BT. Catechol-O-Methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: importance in pathophysiology and pathogenesis. Curr. Drug Metab. 2002;3:321–349. doi: 10.2174/1389200023337586. [DOI] [PubMed] [Google Scholar]
- 7.King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr. Drug Metab. 2000;1:143–161. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- 8.Axelrod J, Senoh S, Witkop B. O-Methylation of catechol amines in vivo. J. Biol. Chem. 1958;233:697–701. [PubMed] [Google Scholar]
- 9.Axelrod J, Tomchick R. Enzymatic O-methylation of epinephrine and other catechols. J. Biol. Chem. 1958;233:702–705. [PubMed] [Google Scholar]
- 10.Axelrod J. Methylation reactions in the formation and metabolism of catecholamines 15 and other biogenic amines. Pharmacol. Rev. 1966;18:95–113. [PubMed] [Google Scholar]
- 11.Lautala P, Ulmanen I, Taskinen J. Molecular mechanisms controlling the rate and specificity of catechol O-methylation by human soluble catechol O-methyltransferase. Mol. Pharmacol. 2001;59:393–402. doi: 10.1124/mol.59.2.393. [DOI] [PubMed] [Google Scholar]
- 12.Taskinen J, Ethell BT, Pihlavisto P, Hood AM, Burchell B, Coughtrie MW. Conjugation of catechols by recombinant human sulfotransferases, UDP-glucuronosyltransferases, and soluble catechol O-methyltransferase: structure-conjugation relationships and predictive models. Drug Metab. Dispos. 2003;31:1187–1197. doi: 10.1124/dmd.31.9.1187. [DOI] [PubMed] [Google Scholar]
- 13.Lundström K, Salminen M, Jalanko A, Savolainen R, Ulmanen I. Cloning and characterization of human placental catechol-O-methyltransferase cDNA. DNA Cell Biol. 1991;10:181–189. doi: 10.1089/dna.1991.10.181. [DOI] [PubMed] [Google Scholar]
- 14.Tenhunen J, Salminen M, Lundström K, Kiviluoto T, Savolainen R, Ulmanen I. Genomic organization of the human catechol O-methyltransferase gene and its expression from two distinct promoters. Eur. J. Biochem. 1994;223:1049–1059. doi: 10.1111/j.1432-1033.1994.tb19083.x. [DOI] [PubMed] [Google Scholar]
- 15.Lundström K, Tenhunen J, Tilgmann C, Karhunen T, Panula P, Ulmanen I. Cloning, expression and structure of catechol-O-methyltransferase. Biochim. Biophys. Acta. 1995;1251:1–10. doi: 10.1016/0167-4838(95)00071-2. [DOI] [PubMed] [Google Scholar]
- 16.Weinshilboum RM. Pharmacogenomics: catechol O-methyltransferase to thiopurine S-methyltransferase. Cell. Mol. Neurobiol. 2006;26:539–561. doi: 10.1007/s10571-006-9095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beis D, Stainier DY. In vivo cell biology: following the zebrafish trend. Trends Cell Biol. 2006;16:105–112. doi: 10.1016/j.tcb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Aleström P, Holter JL, Nourizadeh-Lillabadi R. Zebrafish in functional genomics and aquatic biomedicine. Trends Biotechnol. 2006;24:15–21. doi: 10.1016/j.tibtech.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Ingham PW. The power of the zebrafish for disease analysis. Hum. Mol. Genet. 2009;18:R107–R112. doi: 10.1093/hmg/ddp091. [DOI] [PubMed] [Google Scholar]
- 20.Sugahara T, Liu C-C, Carter G, Pai TG, Liu M-C. cDNA cloning, expression, and functional characterization of a zebrafish SULT1 cytosolic sulfotransferase. Arch. Biochem. Biophys. 2003;414:67–73. doi: 10.1016/s0003-9861(03)00172-3. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu MC, Lipmann F. Decrease of tyrosine-O-sulfate-containing proteins found in rat fibroblasts infected with Rous sarcoma virus or Fujinami sarcoma virus. Proc. Natl. Acad. Sci. USA. 1984;81:3695–3698. doi: 10.1073/pnas.81.12.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda S, Liu C-C, Takahashi S, Suiko M, Chen L, Snow R, Liu M-C. Identification of a novel estrogen-sulfating cytosolic SULT from zebrafish: molecular cloning, expression, characterization, and ontogeny study. Biochem. Biophys. Res. Commun. 2005;330:219–225. doi: 10.1016/j.bbrc.2005.02.152. [DOI] [PubMed] [Google Scholar]
- 26.Liu M-Y, Yang YS, Sugahara T, Yasuda S, Liu M-C. Identification of a novel zebrafish SULT1 cytosolic sulfotransferase: cloning, expression, characterization, and developmental expression study. Arch. Biochem. Biophy. 2005;437:10–19. doi: 10.1016/j.abb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 27.Yasuda S, Kumar AP, Liu M-Y, Sakakibara Y, Suiko M, Chen L, Liu M-C. Identification of a novel thyroid hormone-sulfating cytosolic sulfotransferase, SULT1 ST5, from zebrafish. FEBS J. 2005;272:3828–3837. doi: 10.1111/j.1742-4658.2005.04791.x. [DOI] [PubMed] [Google Scholar]
- 28.Yasuda S, Liu M-Y, Yang YS, Snow R, Takahashi S, Liu M-C. Identification of novel hydroxysteroid-sulfating cytosolic SULTs, SULT2 ST2 and SULT2 ST3, from zebrafish: cloning, expression, characterization, and developmental expression. Arch. Biochem. Biophys. 2006;455:1–9. doi: 10.1016/j.abb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Liu TA, Bhuiyan S, Snow R, Yasuda S, Yasuda T, Yang YS, Williams FE, Liu M-Y, Suiko M, Carter G, Liu M-C. Identification and characterization of two novel cytosolic sulfotransferases, SULT1 ST7 and SULT1 ST8, from zebrafish. Aquat. Toxicol. 2008;89:94–102. doi: 10.1016/j.aquatox.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Yasuda T, Yasuda S, Williams FE, Liu M-Y, Sakakibara Y, Bhuiyan S, Snow R, Carter G, Liu M-C. Characterization and ontogenic study of novel steroid-sulfating SULT3 sulfotransferases from zebrafish. Mol. Cell. Endocrinol. 2008;294:29–36. doi: 10.1016/j.mce.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Sugahara T, Liu C-C, Pai TG, Liu M-C. Molecular cloning, expression, and functional characterization of a novel zebrafish cytosolic sulfotransferase. Biochem. Biophys. Res. Commun. 2003;300:725–730. doi: 10.1016/s0006-291x(02)02915-7. [DOI] [PubMed] [Google Scholar]
- 32.Sugahara T, Liu C-C, Carter G, Pai TG, Liu M-C. cDNA cloning, expression, and functional characterization of a zebrafish SULT1 cytosolic sulfotransferase. Arch. Biochem. Biophys. 2003;414:67–73. doi: 10.1016/s0003-9861(03)00172-3. [DOI] [PubMed] [Google Scholar]
- 33.Sugahara T, Liu C-C, Pai TG, Collodi P, Suiko M, Sakakibara Y, Nishiyama K, Liu M-C. Sulfation of hydroxychlorobiphenyls. Molecular cloning, expression, and functional characterization of zebrafish SULT1 sulfotransferases. EurJBiochem. 2003;270:2404–2411. doi: 10.1046/j.1432-1033.2003.03608.x. [DOI] [PubMed] [Google Scholar]
- 34.Sugahara T, Yang Y-S, Liu C-C, Pai TG, Liu M-C. Sulphonation of dehydroepiandrosterone and neurosteroids: molecular cloning, expression, and functional characterization of a novel zebrafish SULT2 cytosolic sulfotransferase. Biochem. J. 2003;375:785–791. doi: 10.1042/BJ20031050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GenBank Accession # NP_000745
- 36.GenBank Accession # NP_036663
- 37.GenBank Accession # NP_001104532
- 38.Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 39.Rutherford K, Le Trong I, Stenkamp RE, Parson WW. Crystal structures of human 108V and 108M catechol O-methyltransferase. J. Mol. Biol. 2008;380:120–130. doi: 10.1016/j.jmb.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 40.Tsuji E, Okazaki K, Isaji M, Takeda K. Crystal structures of the apo and holo form of rat catechol-O-methyltransferase. J. Struct. Biol. 2009;165:133–139. doi: 10.1016/j.jsb.2008.11.012. [DOI] [PubMed] [Google Scholar]