Abstract
We report an algorithm designed for the calibration of low resolution peptide mass spectra. Our algorithm is implemented in a program called FineTune which corrects systematic mass measurement error in one minute, with no input required besides the mass spectra themselves. The mass measurement accuracy for a set of spectra collected on an LTQ-Velos improved 20-fold from −0.1776 ± 0.0010 m/z to 0.0078 ± 0.0006 m/z after calibration (avg +/− 95% confidence interval). The precision in mass measurement was improved due to the correction of non-linear variation in mass measurement accuracy across the m/z range.
Keywords: Mass measurement accuracy, shotgun proteomics, linear ion trap
Introduction
The field of proteomics has experienced significant growth in the past decade due to advancements in mass spectrometer instrumentation and computational tools for data interpretation. Instrument scan speed, dynamic range, sensitivity, resolution, and mass measurement accuracy (MMA) continue to improve, allowing for more comprehensive analysis of complex protein digests.
Systematic mass measurement error (SMME) is typically corrected by routine instrument calibration. External calibration is performed by analyzing a standard with molecules of known elemental composition and calibrating the instrument to match the measurements of the calibrant ions to their known mass-to-charge (m/z). Ion species covering a wide range of m/z are often used for calibration because systematic mass measurement error can vary with m/z. Over time, the mass calibration drifts, requiring periodic recalibration. Even the most sophisticated instruments will have some systematic and/or random mass measurement error specifically if they are not properly calibrated.
Mass measurement accuracy can be further improved by analyzing calibrants spiked into each sample (i.e., internal calibration). Internal calibration is usually coupled with high-resolution mass analyzers (e.g., TOF1–2 , FTICR3–4, Orbitrap5) where instrument factors such as space charge effects, electric fields, peak intensity, and mass analyzer temperature vary during the course of an analysis. These factors ultimately cause mass measurements to deviate. On trapping instruments, the implementation of automatic gain control helps alleviate the mass deviations due to space charge by controlling for total ion population6. As a result, internal calibration is not necessarily needed in these instruments to achieve high MMA and can be detrimental to performance due to the loss in sensitivity and dynamic range associated with introducing a calibrant ion.
Computational calibration techniques aim to match the robustness of internal calibration without requiring the addition of specific calibrants. To calibrate precursor data, ion species known to be present in a sample can be used as internal calibrants in lieu of spiked in calibrants. Peptides expected to be present in a data set a priori2, 7–8, peptides confidently identified by database searching9–13, or commonly observed contaminants can be used in this manner14. These techniques improve mass measurement accuracy but will likely struggle with low resolution data due to the difficulty of unambiguously mapping theoretical ion species to low resolution precursor features. Charge state pairs can be used to detect frequency shifts in precursor spectra without a priori knowledge of sample content15–17. Unfortunately, the reliance of this technique on resolving charge-state precludes its application to low-resolution data where resolving power is too low to resolve isotopic peaks for charge state determination. Monoisotopic, singly-charged peak masses in peptide mass fingerprinting data collected on MALDI-TOF instruments can be calibrated without a priori knowledge of the sample content18–20.
Fewer computational techniques exist for the calibration of MS/MS spectra. Fragmentation spectra can be calibrated de novo by detection of type-1 peak edges21 or by analysis of trypsin auto-lysis products22. Confident peptide-spectrum matches from a database search can be used to calibrate MS/MS data as well23, although such methods can be computationally expensive and require protein annotations from a genome sequence.
Herein, we demonstrate that low resolution ion trap instruments can be subject to systematic mass measurement error even after external calibration. We implement a novel method for de novo calibration of peptide MS/MS data collected on low resolution instruments capable of improving mass measurement accuracy and mass measurement precision quickly (< 1 minute) without the need for protein annotations or even knowledge of the organism(s) being analyzed. The algorithm is implemented in a freely-available, open-source software package named “FineTune”. We demonstrate improved mass measurement accuracy after calibration with FineTune for MS/MS spectra acquired on both an LTQ and LTQ-Velos mass spectrometer by analyzing mass measurement error in confident peptide-spectrum matches pre and post-calibration. The robustness of FineTune is demonstrated by successfully calibrating data sets with only a small percentage of the total spectra used as input for the calibration. The impact of calibrating MS/MS spectra on database search results is tested using the Mascot, SEQUEST, and X!Tandem search algorithms.
Materials & Methods
Collection of data for testing the de novo calibration algorithm
A S. cerevisiae sample digest was analyzed by LC-MS/MS on an LTQ-Orbitrap-Velos (Thermo Fisher Scientific) hybrid mass spectrometer. Peptides were separated by reversed-phase high performance liquid chromatography (RP-HPLC) across a 100 minute linear acetonitrile gradient on a 40 cm column with 75 μM inner-diameter. Mass spectra were collected using a top-13 data dependent acquisition scheme with precursor scans acquired in the orbitrap (60,000 Resolving Power @ 400 m/z) in profile mode. Peptide fragmentation by resonance collision induced dissociation (CID) and subsequent mass analysis was executed in the dual pressure linear-ion trap in centroid mode. Dynamic exclusion was enabled with a 50 entry exclusion list and 180 second exclusion time.
A S. cerevisiae sample digest was analyzed in a similar manner on an LTQ-FTICR (Thermo Fisher Scientific) hybrid mass spectrometer. A 125 minute linear acetonitrile gradient was used. Mass spectra were collected using a top-5 data dependent acquisition scheme with precursor scans acquired on the FT-ICR (50,000 resolution @ 400 m/z) in profile mode. Fragmentation spectra were acquired by resonance CID in the linear ion trap in centroid mode. Dynamic exclusion was enabled with a 50 entry list and 30 second exclusion time.
Generation of a theoretical fragment ion map for MS/MS spectra
Previous studies demonstrate that polypeptide masses occupy “allowable regions” of the mass range of width ~0.2 m/z spaced apart by about 1.00045475 m/z24. We made a similar observation by analyzing fragment ion masses in spectra from our C. elegans Bibliospec library25.
Every MS/MS spectrum and matching peptide sequence was extracted from the C. elegans Bibliospec library (Version 5.1) of confident peptide spectrum matches. For each of these spectra, a theoretical MS/MS spectrum was generated using monoisotopic amino acid masses for singly-charged b and y ions. Intensities of theoretical fragment ions were matched to the intensity of the nearest peak in the experimental spectrum within +/− 0.5 m/z. The spectra were then binned with a bin width of 0.009995454567 m/z and summed to generate a theoretical fragment ion map (Figure 1). This bin width was selected to be amenable to applying the discrete fast Fourier transform (FFT) to model the location of the peaks in the theoretical ion map using a sinusoid function (see De novo calibration of MS/MS data using the theoretical fragment ion map). Using a bin width of 0.009995454567 m/z ensures that one of the bins of the discrete FFT will correspond to the component of the data with a period of 1.00045475 m/z which is the average distance between peaks in the theoretical fragment ion map24, 26–30.
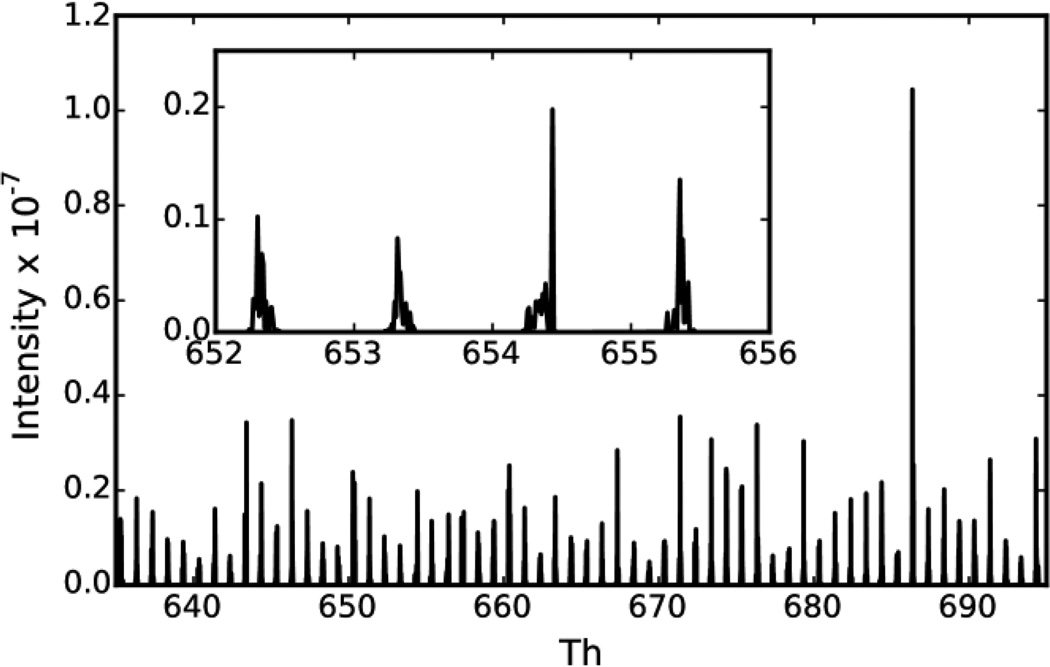
Figure 1. Theoretical fragment ion map.
The theoretical fragment ion map is plotted for m/z 635–695. The map was generated by summing over 100,000 fragmentation spectra. The peaks repeating every ~1.0005 m/z represent regions where a singly charged b or y-ion is theoretically likely to occur in a peptide fragmentation spectrum. The inset is a zoomed view of a subset of the fragment ion map. Notably, there are repeating regions in m/z-space where these peaks do not occur due to most peptides having a similar mass excess.
De novo calibration of MS/MS data using the theoretical fragment ion map
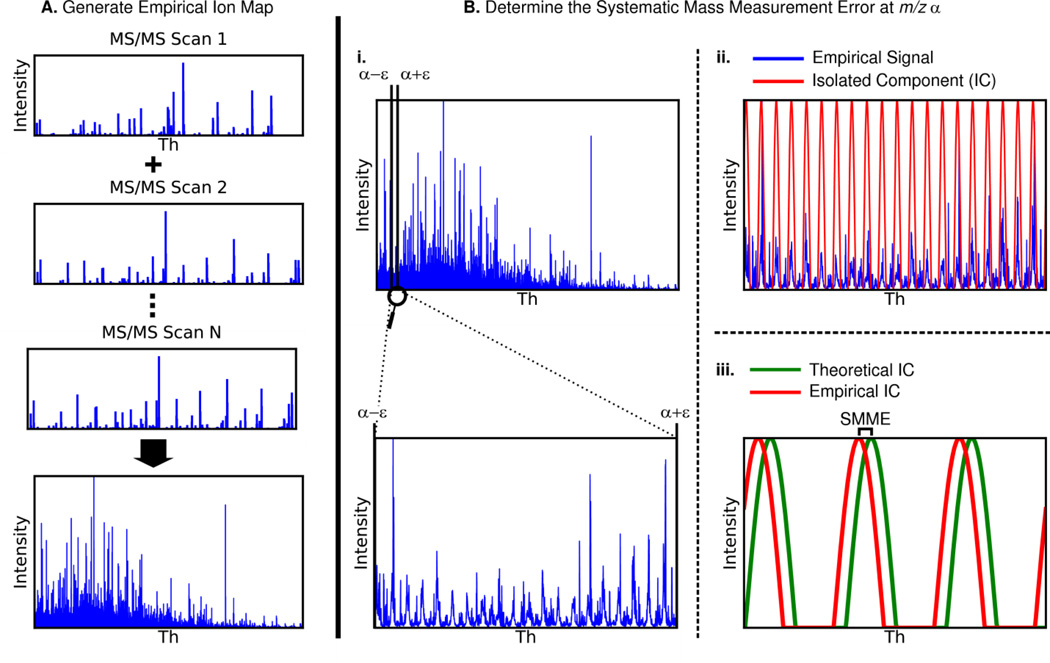
The technique used to calibrate MS/MS spectra is summarized in Figure 2. MS/MS spectra are binned with a bin width of 0.009995454567 m/z and summed to generate the observed fragment ion map. Misalignment between the observed and theoretical fragment ion maps is due to systematic mass measurement error (SMME). Therefore, the systematic mass measurement error can be determined by calculating the m/z shift required to align the observed map to the theoretical one. Because SMME can vary with respect to m/z, the SMME is calculated at an interval of every 20 m/z.
Figure 2. Steps for de novo calibration of MS/MS spectra.
A) All MS/MS spectra in the input file are binned and added to each other to generate an empirical fragment ion map for the file. This step only happens once for each file being calibrated. B) Systematic mass measurement error (SMME) is detected in 20 m/z intervals along the m/z range of the empirical fragment ion map.
- i) Analyze a slice of the empirical ion map with boundaries α +/− ε such that there is adequate signal for detection of SMME. The full empirical ion map (top) and a zoomed in slice of the empirical ion map (bottom) are shown.
- ii) Apply a discrete fast Fourier transform (DFFT) to this slice to isolate the frequency component of the signal with period 1.00045475 m/z (red). This isolated sinusoid component fits the observed peak clusters (blue).
- iii) Use the same technique as in steps i and ii to isolate the same frequency component from the theoretical ion map. The difference in phases of the empirical and theoretical frequency components is the systematic mass measurement error at m/z α.
Once SMME has been detected at each 20 m/z interval, use linear interpolation to determine the SMME at any m/z between these intervals.
To calculate the SMME at m/z α, a subset of both fragment ion maps are analyzed between α-ε and α+ε. The value ε is the minimum value for which the total ion current in the experimental fragment ion map between α-ε and α+ε exceeds 2.5 × 109 ions/sec and ε ≥ 20 m/z. If the values α−ε or α+ε are outside of the range of observed m/z values, the center of the window is shifted. Adjusting the window width in this manner compensates for variability in signal intensity with respect to m/z.
To reduce the impact of noise and multiply-charged peaks on the algorithm, a 218 point discrete fast Fourier transform is used to determine the phase of the signal component with period 1.00045475 m/z for both the theoretical (ϴ t) and experimental (ϴe) ion map subsets. This is essentially a computationally efficient method of fitting a sinusoid to the observed signal. The SMME is the difference in the phase of these two frequency components (ϴ e - ϴ t). After SMME has been calculated for every 20 m/z interval, the points are interpolated linearly to allow for the reporting of SMME at any m/z.
FineTune was coded in C++ and compiled with GNU gcc-4.3.3 on a 64-bit system running Linux kernel v. 2.6.29.6. FineTune uses the Boost Build system to allow for cross-platform compilation (tested on Linux and Windows). FineTune can read and write mzML, mzXML, MGF, MS2, CMS2, and BMS2 formats (http://proteowizard.sourceforge.net/formats.shtml). Additionally, the Windows version can read Agilent, Bruker, Thermo, Waters, and AB-Sciex vendor formats. Source code and binaries are available at the MacCoss lab website (http://proteome.gs.washington.edu/software/finetune).
Calculation of mass measurement error by database searching
The systematic mass measurement error detected by FineTune is compared to that detected by analyzing confident peptide spectrum matches (PSMs) identified by SEQUEST31 using a target-decoy strategy, and post-processed by Percolator (v 1.14)32. If precursor scans were collected on a high-resolution instrument, Bullseye33 is used to determine more accurate precursor masses prior to database searching. A threshold for peptide-level FDR of ≤0.01 is applied to the set of PSMs. For each PSM, the mass measurement error for each theoretical singly charged b- and y-ion with a matching peak within +/− 0.5 m/z is determined. Mass measurement accuracy is reported as the mean of the mass measurement error and its 95% confidence interval. When comparing mass measurement error before and after de novo calibration, the same spectra analyzed prior to calibration are analyzed post-calibration.
Results
A theoretical fragment ion map for peptide fragmentation spectra
Mass excess is the nominal mass (i.e., mass number) of an atom subtracted from the exact mass. The twenty amino acids coded by the standard genetic code have a very similar mass excess which causes the masses of polypeptides to group together into regions of the mass range spaced roughly 1.0005 m/z apart28 (Figure 1). The regions of the mass range between these mass clusters have been termed “forbidden zones”29 because it is theoretically impossible for the mass of a polypeptide built from these twenty amino acids to fall in these zones.
To visualize the distribution of “allowable” fragment ion masses, MS/MS spectra in the C. elegans BiblioSpec spectrum library were summed (Figure 1). Only annotated MS/MS peaks were included in the summation, and each peak’s m/z was corrected to match its theoretical m/z. Supplementary Figure 1A shows a “zoomed out” view of the summed spectrum. Fragment ions between 200 and 800 m/z contribute roughly the same intensity to the summed spectrum throughout this range despite the greater number of fragment ions at the lower end of this range (Supplementary Figure 1B). Above 800 m/z, the summed intensity of fragment ions gradually decreases with increasing m/z.
De novo calibration corrects systematic mass measurement error
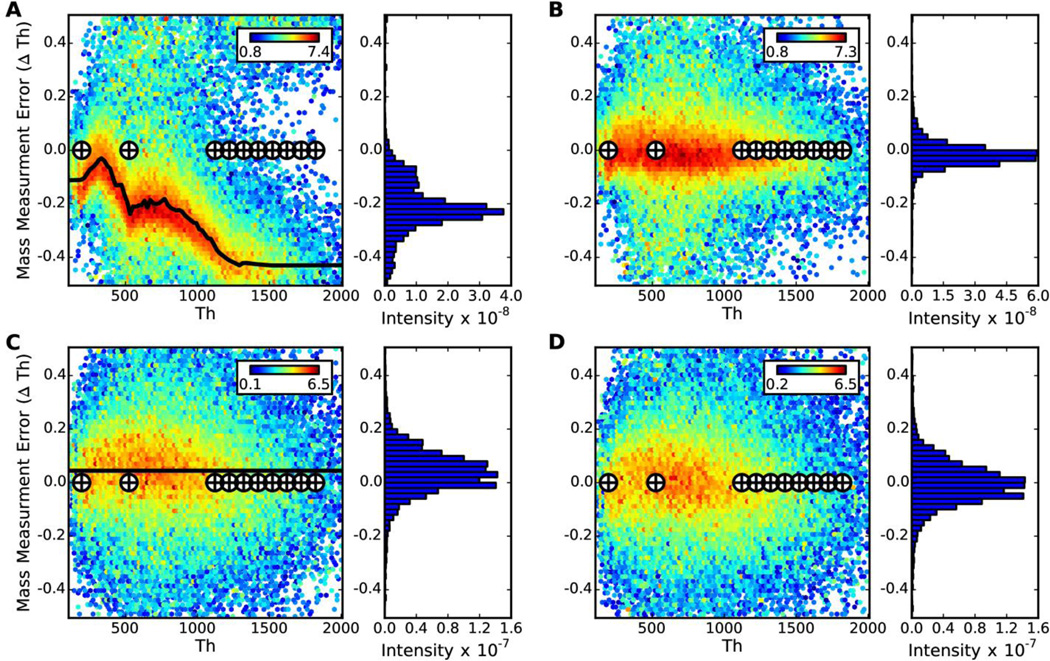
FineTune was tested on 44,944 low resolution MS/MS spectra acquired by a shotgun LC-MS/MS analysis of S. cerevisiae lysate on an LTQ-Orbitrap-Velos (Thermo Fisher Scientific, San Jose, CA) mass spectrometer. The recalibration determined de novo matches very closely to that determined by analyzing 6,628 confident (q≤0.01) peptide spectrum matches from a database search (Figure 3A–B). The mean mass measurement error prior to de novo calibration is −0.1776 ± 0.0010 m/z (95% confidence interval) due to calibration drift. After calibration with FineTune, the mean mass measurement error is 0.0078 ± 0.0006 m/z, thus the precision is improved by about half in addition to the improvement in mass accuracy (Figure 3A–B). The mass is uncharacteristically poor in this dataset to illustrate the capabilities of FineTune. This poor mass calibration is caused by detector and thus automatic gain control miscalibration resulting in significant space charge effects (Figure 3A). In a more common scenario, the mass error does not vary so drastically with m/z (Supplementary Figure 2A) and thus the improvement in the mass error spread will be more subtle. Additionally, the mass error between the caffeine and MRFA ions as well as between the MRFA and the first ultramark ion on the LTQ-Velos (Figure 3A, Supplementary Figure 2A) is a result of the absence of m/z calibrants in that region and a non-linear response between the RF ejection frequency and m/z. The mass calibration has since been improved by the instrument manufacturer using a new calibration procedure and will be available in future releases of the Velos and Velos Pro Tune software (personal communication Jae Schwartz).
Figure 3. Robust de novo detection of systematic mass measurement error.
In A) and C) the mass measurement error detected by FineTune is compared to that detected by analyzing peptide-spectrum matches from a database search on the LTQ-Orbitrap-Velos (6,628 PSMs) and LTQ-FTICR (4,961 PSMs) data respectively. The heatmap in the background is a twodimensional histogram of the mass error determined by comparing experimental fragment ion masses to theoretical masses from confident peptide-spectrum matches. The log10(total intensity) for peaks falling in any bin is mapped to color as indicated in the colorbar. The black curve on top of the heatmap is the systematic mass measurement error detected by FineTune. The cross hairs indicate the theoretical m/z of the calibrant ions in the manufacturer-supplied calibration mix. Next to each two-dimensional histogram is a one-dimensional histogram of mass measurement error detected from the confident peptide spectrum matches. B) and D) show the mass measurement error after using FineTune to calibrate the LTQ-Orbitrap-Velos and LTQ-FTICR data respectively. The data for these plots was generated using the same set of peptide-spectrum-matches as for A) and C) but with calibrated fragment ion masses.
FineTune was also tested on 21,433 MS/MS spectra acquired on the linear ion trap of an LTQ-FTICR (Thermo Fisher) mass spectrometer, improving mean mass measurement error from 0.0444 ± 0.0011 m/z to 0.0055 ± 0.0011 m/z (Figure 3C–D). In contrast to the LTQ-Orbitrap-Velos data shown above, these data have very little systematic mass measurement error (SMME); the mass error distribution is centered close to zero and there is almost no variation in SMME with m/z (Figure 3C). FineTune corrects for the slight m/z-, independent shift in SMME and importantly does not detect any false trends in SMME from noise in the data indicating that FineTune is applicable to data with extreme or subtle SMME.
De novo calibration is robust to a reduction in signal
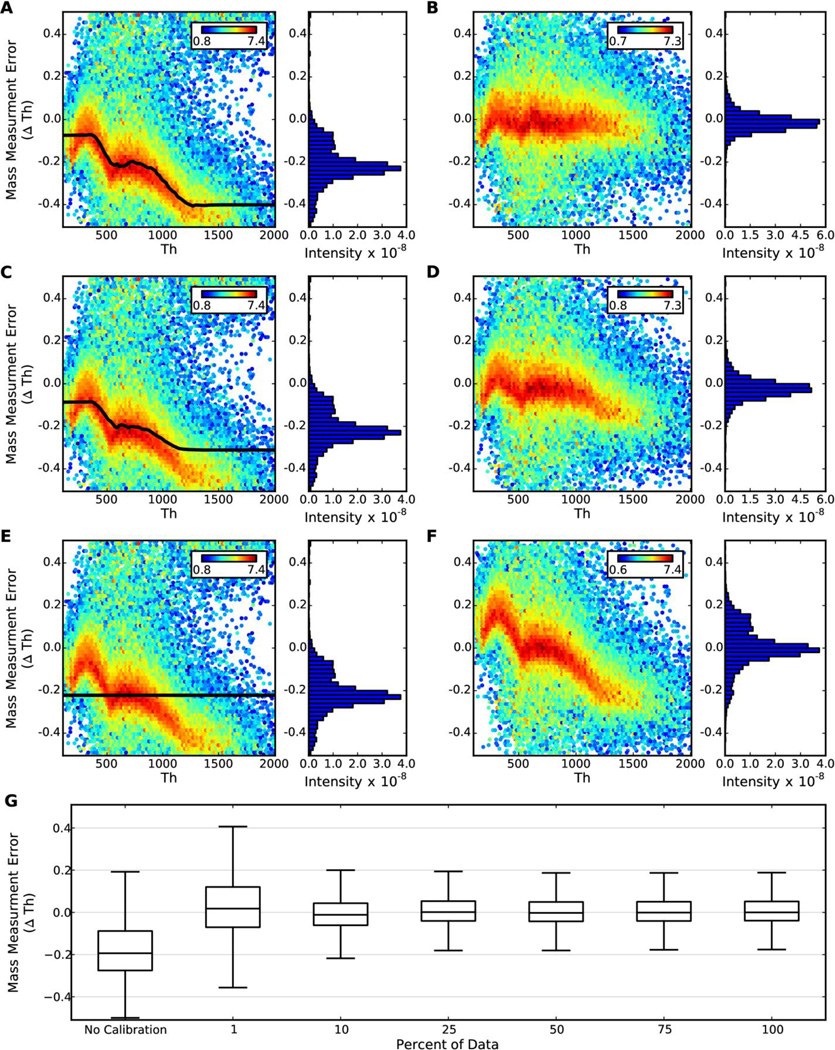
To test the response of FineTune to a reduction in signal (i.e. few peptide MS/MS spectra), FineTune was applied to the LTQ-Orbitrap-Velos data with varying numbers of MS/MS spectra removed. For each MS/MS spectrum in the data, a random number between 1 and 100 was generated, and if the number was greater than or equal to a threshold, the spectrum was removed from the data set. Therefore, if a threshold of 60 is applied, the resulting data should contain about 60% of the original spectra.
Five “thinned” data sets were generated containing ~75%, 50%, 25%, 10%, and 1% of the 44,944 original MS/MS spectra from the LTQ-Orbitrap-Velos data (Figure 4). Even after removing 90% of the spectra, FineTune is extremely robust in high-signal regions of the data. In low-signal regions (e.g. m/z > 1200) SMME is still improved albeit not completely corrected (Figure 4C–D). Once 99% of the spectra are removed, FineTune detects the average mass measurement error, but not the m/z-dependent variation in systematic mass measurement error (Figure 4E–F). The mass accuracy is still improved, but the mass precision remains the same. The mean mass measurement error is −0.1776 ± 0.0010 for the uncalibrated data. The mean mass measurement error is improved in all of the aforementioned cases to 0.0162 +/− 0.0009, −0.0097 +/− 0.0006, 0.0078 +/− 0.0006, 0.0045 +/− 0.0006, 0.0064 +/− 0.0006, and 0.0078 +/− 0.0006 m/z when removing 99%, 90%, 75%, 50%, 25% and 0% of the spectra respectively (Figure 4G). Importantly, we have yet to find a case where FineTune negatively impacts the data.
Figure 4. De novo calibration is robust to a reduction in signal.
Mass measurement error heatmaps were generated using all 6,628 significant peptide spectrum matches in the LTQ-Orbitrap-Velos data as described in Figure 3 and Materials & Methods. A), C), and E) depict the systematic mass measurement error detected by FineTune using 25%, 10%, and 1% of the (44,944) MS/MS spectra from the original file, respectively; B), D), and F) depict the mass measurement error after correcting for the systematic mass measurement error detected in figures A), C), and E) respectively. G) shows the median mass measurement error from the 6,628 PSMs with no calibration and after calibration by FineTune with varying percentages of the original spectra as input. The whiskers are 1.5 times the inner quartile.
De novo calibration and database search results
The calibrated and uncalibrated (control) LTQ-Orbitrap-Velos data was searched using X!Tandem (Cyclone 2010.12.01.1). All data was first processed by Bullseye and then searched against S. cerevisiae sequence (target) and reversed sequence (decoy) databases with a precursor mass tolerance of 10 ppm. The fragment ion tolerance for the searches was varied between ±0.01Da and ±0.5 Da. The expectation values from target and decoy search results were used to determine the number of unique peptides at a false discovery rate (FDR) threshold of 0.01.
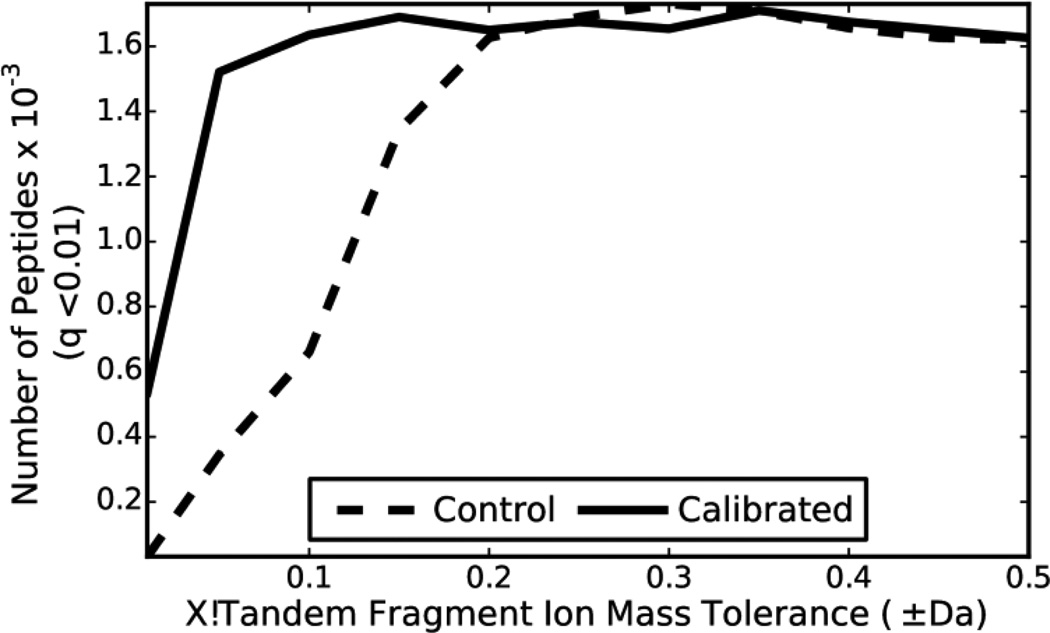
At fragment ion tolerances less than ±0.2 Da, the calibrated data returns many more peptide identifications than the data that has not been calibrated (Figure 5). For example, at a fragment ion tolerance of ±0.1 Da, the calibrated data returns 2.5 times more peptide identifications. However, at fragment ion tolerances greater than ±0.2 Da, calibrating the data does not increase the number of peptide identifications. Calibrated data had a similar, but far more subtle impact on Mascot search results. (Supplementary Figure 3). The impact of calibration on SEQUEST results is less straightforward due to how spectra are binned and is discussed in the Supplementary Results section. Using calibrated data and the optimal fragment ion tolerance/binning parameters SEQUEST, Mascot and X!Tandem identify 3,755; 2,622; and 1,710 unique peptides respectively at a q-value ≤ 0.01.
Figure 5. De novo calibration improves X!Tandem search results.
The LTQ-Orbitrap-Velos data was searched with X!Tandem using a target-decoy strategy to determine q values for each unique peptide from reported expect scores. The number of unique peptides identified by X!Tandem for the control and calibrated data with q ≤ 0.01 was compared using various fragment ion tolerances in X!Tandem.
Discussion
FineTune enables the de novo, unsupervised calibration of MS/MS spectra acquired on low resolution instrumentation. The algorithm corrects non-linear systematic mass measurement error and does not negatively-impact a data set even when it is trained on only 1% of the spectra in the data set. This robustness, speed, low memory (~10 Mb) and low processor requirements (the bottleneck is typically file I/O) make FineTune suitable for integration into an unsupervised data processing pipeline run on every data set acquired. Calibration can then be tracked over time which aids in the scheduling of instrument calibrations, especially for newer instruments which tend to drift out of calibration quickly. Additionally, the algorithm can be applied retroactively to old data that a researcher may suspect was collected on an improperly calibrated instrument.
Calibration of fragment ion masses had little impact on peptide identifications from three popular database searching algorithms. FineTune improved search results for X!Tandem at small fragment ion mass tolerance values. One might expect the search with fragment ion tolerance ±0.2 Da to perform better than larger fragment ion tolerances because ±0.2 Da is roughly the width of the fragment ion mass error distribution for the calibrated data (Figure 3B). Following this line of reasoning, when the fragment ion tolerance is made greater than ±0.2 Da the only effect would be to increase the number of incorrect peaks under consideration when calculating the match score for any particular peptide spectrum match. However, it appears that the scoring function used by X!Tandem (Figure 5) and Mascot (Supplementary Figure 3) are both unaffected by this phenomenon. Calibration does not greatly improve Mascot results at any fragment ion tolerance.
These algorithms are classified as database search algorithms due to their reliance on a database of candidate protein sequences to test each spectrum against. De novo search algorithms are designed to interpret spectra without a database of candidate sequences. Because FineTune does not rely on sequence information, it is a natural complement to de novo search algorithms often used when protein sequence information is unreliable, incomplete, or non-existent. De novo search algorithms can benefit from improved fragment ion mass accuracy34–35. Thus, in future work we will assess the impact of de novo calibration on de novo spectrum interpretation.
FineTune corrects systematic mass measurement error reliably by only making corrections when enough signal is present in the data to justify these corrections. FineTune improved the mass measurement accuracy of every data set tested to <0.01 m/z. This reliability makes FineTune suitable for use as an unsupervised preprocessor applied to any collection of MS/MS spectra prior to database searching. While improved mass measurement accuracy sometimes improves database search results (up to 40% increase in peptide identifications), the data presented herein indicates that database search algorithms could be optimized to take better advantage of more accurate fragment ion mass measurements. In future experiments, we look to expand the algorithm to be able to correct mass measurement error trends that vary with retention time as well as test the impact of calibration on de novo spectrum interpretation algorithms.
Supplementary Material
Acknowledgements
This work is supported in part by National Institutes of Health Grants P41 GM103533, F31 AG037265, R01 DK069386, and the University of Washington's Proteomics Resource (UWPR95794).
The authors acknowledge Jesse D. Canterbury for helpful discussion regarding the calibration algorithm and LTQ-Velos Pro calibration routine, Jae Schwartz and Philip M. Remes for helpful discussion regarding the m/z error distribution observed on the LTQ-Velos, and Vagisha Sharma and Barbara Frewen for help with extracting MS/MS spectra from Bibliospec libraries.
References
- 1.Beavis RC, Chait BT. High-accuracy molecular mass determination of proteins using matrix-assisted laser desorption mass spectrometry. Analytical chemistry. 1990;62(17):1836–1840. doi: 10.1021/ac00216a020. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter EF, Ferguson PL, Tang K, Smith RD. Proteome analyses using accurate mass and elution time peptide tags with capillary LC time-of-flight mass spectrometry. Journal of the American Society for Mass Spectrometry. 2003;14(9):980–991. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 3.Henry KD, Quinn JP, McLafferty FW. High-resolution electrospray mass spectra of large molecules. Journal of the American Chemical Society. 1991;113(14):5447–5449. [Google Scholar]
- 4.Hannis JC, Muddiman DC. A dual electrospray ionization source combined with hexapole accumulation to achieve high mass accuracy of biopolymers in Fourier transform ion cyclotron resonance mass spectrometry. Journal of the American Society for Mass Spectrometry. 2000;11(10):876–883. doi: 10.1016/S1044-0305(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 5.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Molecular & cellular proteomics : MCP. 2005;4(12):2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Syka JE, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. Journal of proteome research. 2004;3(3):621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 7.Strittmatter EF, Rodriguez N, Smith RD. High mass measurement accuracy determination for proteomics using multivariate regression fitting: application to electrospray ionization time-of-flight mass spectrometry. Analytical chemistry. 2003;75(3):460–468. doi: 10.1021/ac026057g. [DOI] [PubMed] [Google Scholar]
- 8.Tolmachev AV, Monroe ME, Jaitly N, Petyuk VA, Adkins JN, Smith RD. Mass measurement accuracy in analyses of highly complex mixtures based upon multidimensional recalibration. Analytical chemistry. 2006;78(24):8374–8385. doi: 10.1021/ac0606251. [DOI] [PubMed] [Google Scholar]
- 9.Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, Eling WM, Hall N, Waters AP, Stunnenberg HG, Mann M. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419(6906):537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 10.Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, Cox J, Foster LJ, Heck AJ, Blagoev B, Andersen JS, Mann M. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. Journal of proteome research. 2010;9(1):393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 11.Petyuk VA, Mayampurath AM, Monroe ME, Polpitiya AD, Purvine SO, Anderson GA, Camp DG, 2nd, Smith RD. DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Molecular & cellular proteomics : MCP. 2010;9(3):486–496. doi: 10.1074/mcp.M900217-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petyuk VA, Jaitly N, Moore RJ, Ding J, Metz TO, Tang K, Monroe ME, Tolmachev AV, Adkins JN, Belov ME, Dabney AR, Qian WJ, Camp DG, 2nd, Smith RD. Elimination of systematic mass measurement errors in liquid chromatography-mass spectrometry based proteomics using regression models and a priori partial knowledge of the sample content. Analytical chemistry. 2008;80(3):693–706. doi: 10.1021/ac701863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kil YJ, Becker C, Sandoval W, Goldberg D, Bern M. Preview: a program for surveying shotgun proteomics tandem mass spectrometry data. Analytical chemistry. 2011;83(13):5259–5267. doi: 10.1021/ac200609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Molecular & cellular proteomics : MCP. 2006;5(7):1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Bruce JE, Anderson GA, Brands MD, Pasa-Tolic L, Smith RD. Obtaining more accurate Fourier transform ion cyclotron resonance mass measurements without internal standards using multiply charged ions. Journal of the American Society for Mass Spectrometry. 2000;11(5):416–421. doi: 10.1016/S1044-0305(00)00096-9. [DOI] [PubMed] [Google Scholar]
- 16.Kaiser NK, Anderson GA, Bruce JE. Improved mass accuracy for tandem mass spectrometry. Journal of the American Society for Mass Spectrometry. 2005;16(4):463–470. doi: 10.1016/j.jasms.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 18.Gras R, Muller M, Gasteiger E, Gay S, Binz PA, Bienvenut W, Hoogland C, Sanchez JC, Bairoch A, Hochstrasser DF, Appel RD. Improving protein identification from peptide mass fingerprinting through a parameterized multi-level scoring algorithm and an optimized peak detection. Electrophoresis. 1999;20(18):3535–3550. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3535::AID-ELPS3535>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 19.Wool A, Smilansky Z. Precalibration of matrix-assisted laser desorption/ionizationtime of flight spectra for peptide mass fingerprinting. Proteomics. 2002;2(10):1365–1373. doi: 10.1002/1615-9861(200210)2:10<1365::AID-PROT1365>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Wolski WE, Farrow M, Emde AK, Lehrach H, Lalowski M, Reinert K. Analytical model of peptide mass cluster centres with applications. Proteome science. 2006;4:18. doi: 10.1186/1477-5956-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan B, Pan C, Olman VN, Hettich RL, Xu Y. A graph-theoretic approach for the separation of b and y ions in tandem mass spectra. Bioinformatics. 2005;21(5):563–574. doi: 10.1093/bioinformatics/bti044. [DOI] [PubMed] [Google Scholar]
- 22.Gentzel M, Kocher T, Ponnusamy S, Wilm M. Preprocessing of tandem mass spectrometric data to support automatic protein identification. Proteomics. 2003;3(8):1597–1610. doi: 10.1002/pmic.200300486. [DOI] [PubMed] [Google Scholar]
- 23.Matthiesen R, Bunkenborg J, Stensballe A, Jensen ON, Welinder KG, Bauw G. Database-independent, database-dependent, and extended interpretation of peptide mass spectra in VEMS V2.0. Proteomics. 2004;4(9):2583–2593. doi: 10.1002/pmic.200300792. [DOI] [PubMed] [Google Scholar]
- 24.Nefedov AV, Mitra I, Brasier AR, Sadygov RG. Examining troughs in the mass distribution of all theoretically possible tryptic peptides. Journal of proteome research. 2011;10(9):4150–4157. doi: 10.1021/pr2003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Analytical chemistry. 2006;78(16):5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- 26.Gay S, Binz PA, Hochstrasser DF, Appel RD. Modeling peptide mass fingerprinting data using the atomic composition of peptides. Electrophoresis. 1999;20(18):3527–3534. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3527::AID-ELPS3527>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 27.Yates JR, Iii, Eng JK, Clauser KR, Burlingame AL. Search of sequence databases with uninterpreted high-energy collision-induced dissociation spectra of peptides. Journal of the American Society for Mass Spectrometry. 1996;7(11):1089–1098. doi: 10.1016/S1044-0305(96)00079-7. [DOI] [PubMed] [Google Scholar]
- 28.Mann M. Useful Tables of Possible and Probable Peptide Masses. Atlanta, GA: Annual Conference on Mass Spectrometry and Allied Topics, Atlanta, GA, American Society of Mass Spectrometry; 1995. [Google Scholar]
- 29.Frahm JL, Howard BE, Heber S, Muddiman DC. Accessible proteomics space and its implications for peak capacity for zero-, one- and two-dimensional separations coupled with FT-ICR and TOF mass spectrometry. Journal of mass spectrometry : JMS. 2006;41(3):281–288. doi: 10.1002/jms.1024. [DOI] [PubMed] [Google Scholar]
- 30.Demirev PA, Zubarev RA. Probing combinatorial library diversity by mass spectrometry. Analytical chemistry. 1997;69(15):2893–2900. doi: 10.1021/ac970049w. [DOI] [PubMed] [Google Scholar]
- 31.Eng JK, McCormack AL, Yates JR., Iii An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh EJ, Hoopmann MR, MacLean B, MacCoss MJ. Comparison of database search strategies for high precursor mass accuracy MS/MS data. Journal of proteome research. 2010;9(2):1138–1143. doi: 10.1021/pr900816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grossmann J, Roos FF, Cieliebak M, Liptak Z, Mathis LK, Muller M, Gruissem W, Baginsky S. AUDENS: a tool for automated peptide de novo sequencing. Journal of proteome research. 2005;4(5):1768–1774. doi: 10.1021/pr050070a. [DOI] [PubMed] [Google Scholar]
- 35.Horn DM, Zubarev RA, McLafferty FW. Automated de novo sequencing of proteins by tandem high-resolution mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(19):10313–10337. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.