Abstract
Ribosome synthesis involves dynamic association of ribosome biogenesis factors with evolving pre-ribosomal particles. Rio2 is an atypical protein kinase required for pre-40S subunit maturation. We report the crystal structure of eukaryotic Rio2 with bound ATP/Mg2+. Unexpectedly, the structure reveals a phosphoaspartate intermediate with ADP/Mg2+ in the active site, typically found in Na+-, K+- and Ca2+-ATPases. Consistent with this finding, ctRio2 exhibits a robust ATPase activity in vitro. In vivo, Rio2 docks on the ribosome with its active site occluded, and its flexible loop positioned to interact with the pre-40S subunit. Moreover, Rio2 catalytic activity is required for its dissociation from the ribosome, a necessary step in pre-40S maturation. We propose that phosphoryl transfer from ATP to Asp257 in Rio2’s active site and subsequent hydrolysis of the aspartylphosphate could be a trigger to power late cytoplasmic 40S subunit biogenesis.
The atypical serine protein kinase, Rio2, is essential for the maturation of the 40S subunit of the eukaryotic ribosome 1–3. Eukaryotic ribosome biogenesis is characterized by transcription of a single ribosomal RNA (rRNA) transcript by RNA Pol I that via endo- and exo-nucleolytic processing yields 3 out of the 4 rRNA molecules that constitute the mature ribosome. In eukaryotes, ribosome maturation involves more than 200 non-ribosomal trans-acting factors that are necessary for RNA modification and folding and ribosomal protein assembly, in addition to RNA cleavage 4,5. This process, which begins during transcription of the rDNA loci, is a highly coordinated stepwise process that begins in the nucleolus and ends in the cytoplasm4,5. Rio2 is essential in yeast and present in all archaea and eukaryotes, and its depletion results in a late pre-40S biogenesis defect 1–3. Furthermore, in higher eukaryotes its kinase activity was implicated in factor recycling6. However, none of these recycled factors have been clearly established as Rio2 substrates. Moreover, according to the recently published cross-linking and cryo-EM studies of a late pre-40S intermediate7,8, raises the possibility that Rio2 autophosphorylation or subsequent dephosphorylation could be a driving force for factor recycling. Therefore, the molecular mechanism of the role of Rio2 and its catalytic activity in small subunit biogenesis progression still needs to be established.
Rio2 is a serine protein kinase capable of in vitro autophosphorylation, albeit with low activity1,9. Previously determined structures of Rio2 from Archaeoglobus fulgidus (afRio2) demonstrated that it contains a smaller trimmed version of a canonical eukaryotic protein kinase (ePK) domain (called the RIO domain) and an N-terminal winged helix (wHTH) domain10. The ePK domains are bilobed with the mostly β-sheet N-terminal lobe containing the phosphate-binding loop (P-loop), and the helical C-terminal lobe containing the catalytic and metal binding loops. The ATP molecule is bound with magnesium in the cleft between the two domains. The archaeal Rio2 (aRio2) lacked some key features of canonical ePKs, including the activation loop and the last two helices of the C-terminal lobe, which provide much of the surface for peptide substrate binding in ePKs. However, comparison of the sequences of aRio2 and the eukaryotic Rio2 (eRio2) reveals eukaryote-specific conserved extension sequences2 (see Supplementary Fig. 1a).
Recent cryo-electron microscopy (cryo-EM) maps of the yeast pre-40S resulted in the localization of Rio2 in the pre-ribosome. However, a reliable positioning of the molecule on the pre-40S complex was not provided by this study, in part because the only available Rio2 structure was the much smaller afRio2. Insight into the mechanism of Rio2 kinase-driven function in 40S ribosome biogenesis therefore requires an atomic resolution structure of eukaryotic Rio2 and structure-based mechanistic probes to link catalytic activity to function during 40S assembly. In order to address this gap, we have chosen to study Rio2 from Chaetomium thermophilum (ct), a eukaryotic thermophile, whose proteins are thermostable11. ctRio2 is highly homologous to yeast and higher eukaryotic Rio2 orthologs, which all have eukaryotic-specific features including the (conserved) C-terminal extension absent in afRio2 structure (Supplementary Fig. 1a).
We report the X-ray crystal structure of ctRio2, both free and bound to ATP-Mg2+, to 2.5 and 2.2 Å respectively. The structures revealed the presence of an additional alpha helix in the conserved eukaryotic C-terminal extension sequence and an unusual phophoaspartate intermediate, typically observed in P-type ATPases 12,13. In agreement, we show that ctRio2, in contrast to a bona fide protein kinase, predominantly acts as an ATPase in vitro. In addition, the ctRio2 structure was docked into cryo-EM maps of the yeast pre-40S particle. The best fit shows contact between the pre-40S subunit and all three domains of the Rio2 kinase, and occlusion of the active site entrance such that it is inaccessible to a potential external substrate when bound. Furthermore, functional analysis of mutants of the yeast Rio2 demonstrates that catalytic activity is required for its own disassociation but not association with the pre-ribosome. Overall our study suggests a model of an ATPase-dependent role of Rio2 on the evolving pre-40S subunit.
RESULTS
ctRio2 structure reveals a phosphoaspartate intermediate
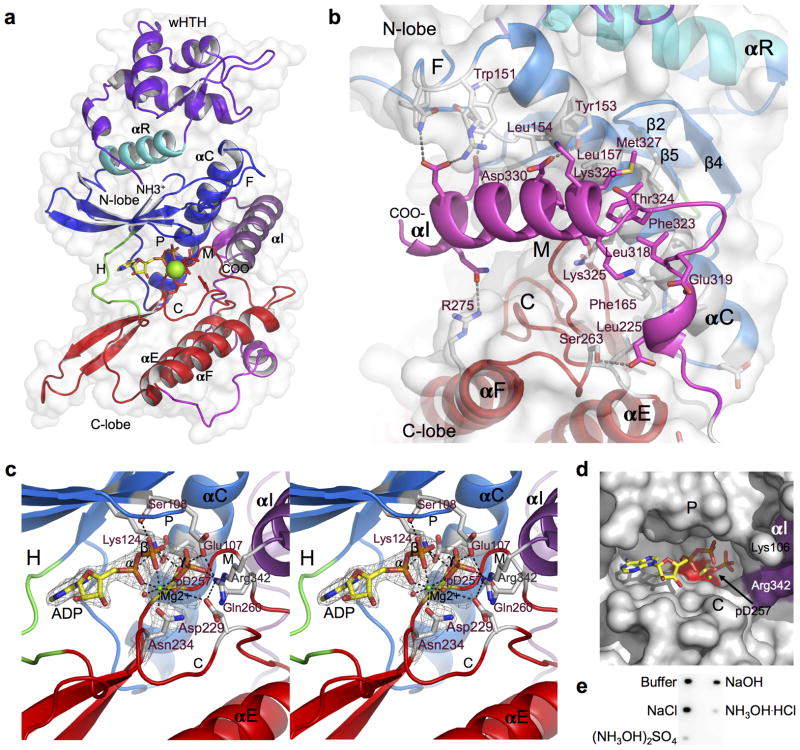
Rio2 from Chaetomium thermophilum (ct) was chosen as a target for structural and functional investigation in this study as it complements the otherwise non-viable yeast rio2 null mutant (Supplementary Fig. 1b), which allowed us to integrate structural investigations with functional studies in yeast. ctRio2 readily crystallized with one molecule per asymmetric unit, and its structure with bound ATP-Mg2+ was determined to 2.2 Å resolution (for data and refinement statistics see Table 1). In addition, the structure of the free ctRio2 protein was solved to 2.5 Å resolution. Rio2 consists of an N-terminal winged-helix-turn-helix domain (wHTH), a well-known nucleic acid binding motif, followed by a RIO domain (Fig. 1a). Like canonical ePKs, the Rio2 RIO domain houses an N- and C-lobe linked by a short hinge region, with nucleotide and Mg2+ bound in the cleft between these two domains. The N-lobe consists of a single anti-parallel β-sheet containing 5 β-strands (β1–5), and a long α-helix, known as αC14. The positioning of αC is critical to the activity of the protein kinase15. A highly conserved Rio2-specific flexible region with no secondary structure known as the “flexible loop” is located between β3 and αC of the N-lobe. The C-terminal lobe consists of two alpha helices, αE and αF and a long β-hairpin (β6–7). The catalytic loop with the conserved catalytic residues Gln234 and Asp229 (scRio2 Gln234, Asp229 respectively) resides between the end of αE and β6, while the metal binding loop containing the conserved Asp257 (scRio2 Asp253) connects the end of β7 and the start of αF. Gln234 and Asp257, invariant in protein kinases, are responsible for binding to Mg2+, while Asp229, also invariant, is the proposed catalytic base for deprotonation of the substrate serine hydroxyl group. In addition, the eukaryote-specific C-terminal extension in ctRio2 consists of an extended loop with two small helices followed by a long α-helix (named αI), which is wedged between the N- and C-lobes close to the active site (Fig. 1a). The interaction between αI and the kinase domain is extensive and consists mainly of hydrophobic contact with conserved residues from αC helix (Fig. 1b). The end ofαI approaches the active site of the enzyme near the tri-phosphate moiety. This helix, though in a drastically different position, appears related in sequence to αI of aRio2 10. In aRio2, αI packs underneath αE and αF, but in ctRio2, a roughly 20 amino acid insertion allows its repositioning (see Supplementary Fig. 2a).
Table 1.
Data collection and refinement statistics (molecular replacement)
| ctRio2/ATP/Mg2+ | ctRio2/APO | |
|---|---|---|
| Data collection | ||
| Space group | H3 | H3 |
| Cell dimensions | ||
| a = b, c (Å) | 119.62, 71.78 | 117.40, 71.29 |
| α=β, γ (°) | 90.00, 120.00 | 90.00, 120.00 |
| Resolution (Å) | 40.0-2.20 (2.28-2.20)* | 40.0-2.48(2.59-2.48) |
| Rsym | 0.067 (0.539)1 | 0.074 (0.516)1 |
| I / σI | 13.1 (2.19)1 | 12.8 (1.83)1 |
| Completeness (%) | 100 (100)1 | 97.9 (99.1)1 |
| Redundancy | 3.2 (3.2)1 | 2.9 (2.9)1 |
| Refinement | ||
| Resolution (Å) | 34.5-2.20 (2.26-2.20) | 30.0-2.48 (2.60-2.48) |
| No. reflections | 19451 (1942) | 12,674 (1284) |
| Rwork / Rfree | 16.8/22.6 (20.7/24.1)1 | 18.3/24.4 (26.5/33.8)1 |
| No. atoms | 2835 | 2671 |
| Protein | 2695 | 2630 |
| Ligand/ion | 36 | 0 |
| Water | 104 | 41 |
| B-factors (average; Å2) | 53.6 | 56.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.008 |
| Bond angles (°) | 1.18 | 1.16 |
One crystal was used for each data set. Values in parentheses are for highest-resolution shell.
Figure 1. Crystal structure of the ctRio2 kinase.
(a) Overall structure of the Chaetomium thermophilum ctRio2 kinase. The cartoon representation is colored according to Rio2 sub-domains. The phosphate-binding, flexible, hinge, catalytic and metal-binding loops are labeled as P, F, H, C and M, respectively. The RIO domain specific N-terminal helix is labeled αR, and the eukaryote-specific C-terminal extension helix is labeled αI. Mg2+ is shown as a green sphere, and ATP and catalytic residues are shown in stick representation. (b) Detailed view of αI (magenta) showing extensive interaction between the N- and C-lobes of the RIO domain. Interacting residues (stick) labeled in purple are highly conserved among eukaryotes. (c) Detailed view of the active site of ctRio2 with ADP-Mg2+ and the phosphoaspartate intermediate (p-D257). 2Fo-Fc electron density contoured at 2.0 σ is shown in grey mesh. Black dashed lines connect atoms within hydrogen or coordinate bond distance of each other. (d) Surface view of active site containing ADP (stick), phosphoaspartate (stick) and Mg2+ (green sphere). The phosphate group is buried in a pocket near αI (purple surface). e) Blot of 32P-labeled ctRio2 after treatment with assay buffer, or 0.5 M NaCl, NaOH or hydroxylamine salts ((NH3OH)2SO4, or NH3OH·HCl).
Comparison of the ATP-bound and free ctRio2 show only very small shifts of the wHTH and N-lobe (overall RMSD of 0.36 Å for 302 Cα), but it suggests some flexibility in positioning of the wHTH domain relative to the RIO domain (Supplementary Fig. 2b). In addition, changes in the positioning of the catalytic and metal-binding loops are observed. The majority of the conformational changes can be attributed to interactions between the nucleotide and these loops. Most of the interactions with nucleotide seen in afRio2 are conserved in ctRio2, but instead of two metal ions (as observed in afRio2), only one is bound to ctRio2 (Supplementary Fig. 2c). Unexpectedly, in the ctRio2::ATP complex the nucleotide is hydrolyzed, with clear density for ADP-Mg2+ and a phosphoaspartate residue resulting from transfer of the γ-phosphate to Asp257 (scRio2 Asp253), the canonical kinase invariant metal-binding aspartate14 (Fig. 1c, d, Supplementary Fig. 2d, e). This phosporylated aspartate is not seen in the APO ctRio2 structure. A phosphoaspartate intermediate in serine kinases is rare, but known to occur in “P-type” ATPases, bacterial chemotaxis responsive phospho-acceptor proteins and small molecule phosphotransferases12,16,17. The α, β-, and transferred phosphates are in contact with invariant Lys124, Ser108, and Glu107 and Gln260, respectively (Lys123, Ser107, Glu106 and Gln256 in scRio2; Fig. 1c). The α- and β-phosphates are further bound by Mg2+, which also coordinates the now phosphoaspartate at Asp257 and the catalytic loop invariant Asn234 (Asp253 and Asn234 in scRio2). In ctRio2, the positioning of the transferred phosphate relative to the catalytic base Asp229 (Asp229 in yeast) is similar to that seen in PKA18, with a phosphorus atom to Asp229 carboxyl oxygen distance of 3.94 Å. This suggests that the phosphate is positioned for transfer, but the presence of a phosphoaspartate intermediate suggests a different mechanism than that thought to function in canonical ser/thr protein kinases 19.
Since Rio2 kinase is known to autophosphorylate1,9, our crystallographic observation of a phosphoaspartate may be also relevant to the behavior of the molecule in solution. Hence, we tested whether autophosphorylated Rio2 obtained after incubation with 32P-γ-ATP could also carry 32P-γ-phosphate on Asp257. Acyl-phosphates, but not phosphate attached to ser/thr, are known to be sensitive to hydroxylamine treatment or high pH12. To determine whether the Rio2 phosphoaspartate intermediate is present in solution, 32P-labeled ctRio2 was incubated with 0.5 M hydroxylamine or NaOH and assayed for loss of the radiolabel. As shown in Figure 1e, treatment with hydroxylamine caused a loss of 32P-labeling, compared to treatment with 0.5 M NaCl, indicating the presence of an acyl-phosphate chemical linkage within Rio2 in aqueous solution.
ctRio2 can act as an ATPase in vitro
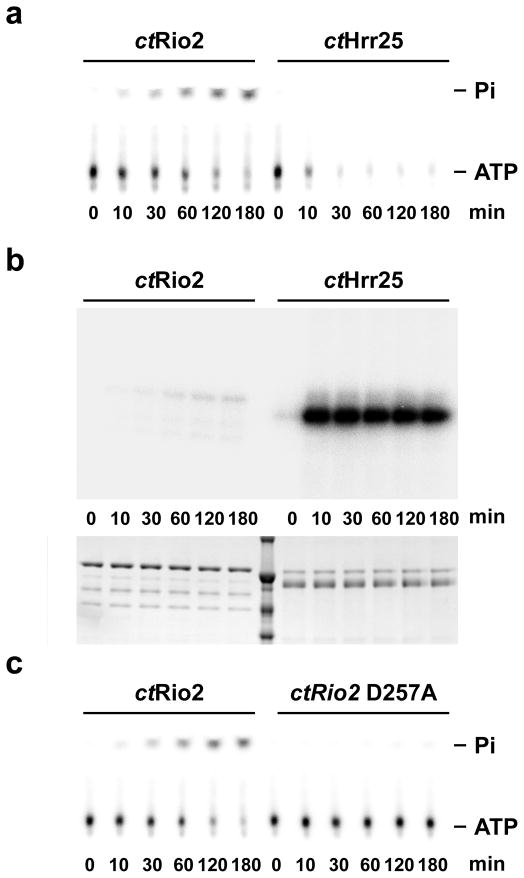
The presence of an acyl-phosphate in Rio2 suggests either a different mechanism than previously thought for functioning as a canonical ser/thr protein kinase or that the phosphate can be also transferred to a water molecule, as occurs in the case of P-loop ATPases. This latter mechanism would result in Rio2-catalyzed hydrolysis of ATP to generate ADP and free γ-phosphate. We therefore assayed purified ctRio2 in single turnover measurements using 32P-γ-phosphate labeled ATP, and compared the amounts of released radioactive γ-phosphate by thin-layer chromatography versus autophosphorylation analyzed by SDS-PAGE and autoradiography. As shown in Supplementary Figure 3a, wild-type ctRio2 readily hydrolyzes ATP in a protein concentration-dependent manner, with ATP disappearance and concomitant stoichiometric 32P-γ-phosphate appearance (Fig. 2a). In a control experiment, we used a well-characterized protein kinase, ctHrr25, involved in 40S subunit biogenesis20, which shows ATP consumption without 32P-γ-phosphate release, but strong autophosphorylation of the Hrr25 protein (see Fig. 2a, b). In the applied in vitro single turnover assay, we estimate that Rio2 autophosphorylation contributes to less than 0.1% of the total ATP hydrolysis (data not shown). Mutation of ctRio2 at the known catalytic residues Asp257 and Asp229 to A results in loss of ATP hydrolysis independently of loss of nucleotide binding, whereas, in agreement with its role in nucleotide binding, K124A exhibits a decreased affinity to nucleotide and consequently decrease in catalytic activity (see Fig. 2c and Supplementary Fig. 3b,c). The turnover rates of ATP hydrolysis measured for the wild-type and mutant enzymes under saturating concentrations of ATP indicate that wild-type Rio2 exhibits measurable ATPase activity with a turnover rate of 0.91 ± 0.05 min−1 and mutation of the catalytic aspartate residues D229A and D257A reduces the turnover rate to 0.011 ± 0.002 and 0.035 ± 0.017 min−1 respectively. The turnover rate for the K124A mutation is 0.19 ± 0.01 min−1, indicating that catalysis, as well as binding, is affected. All together our structural and in vitro studies support the model that Rio2 can function as an ATPase.
Figure 2. ctRio2 has ATPase activity in vitro.
The amount of released free phosphate (Pi) (a and c) or phosphorylated protein (b) was analyzed in single-turnover experiments using 1μM of the indicated purified recombinant protein. (a) and (c) ATP and Pi were separated by thin-layer chromatography. (b) the amount of phosphorylated protein was analyzed by SDS-PAGE. Dried gels and TLC plates were exposed overnight on a phosphorimager screen (see experimental procedures for a full description). Note that 100 times more of the reactions were loaded in panel b in comparison to panel a and c.
ctRio2 active site is occluded when bound to the pre-40S
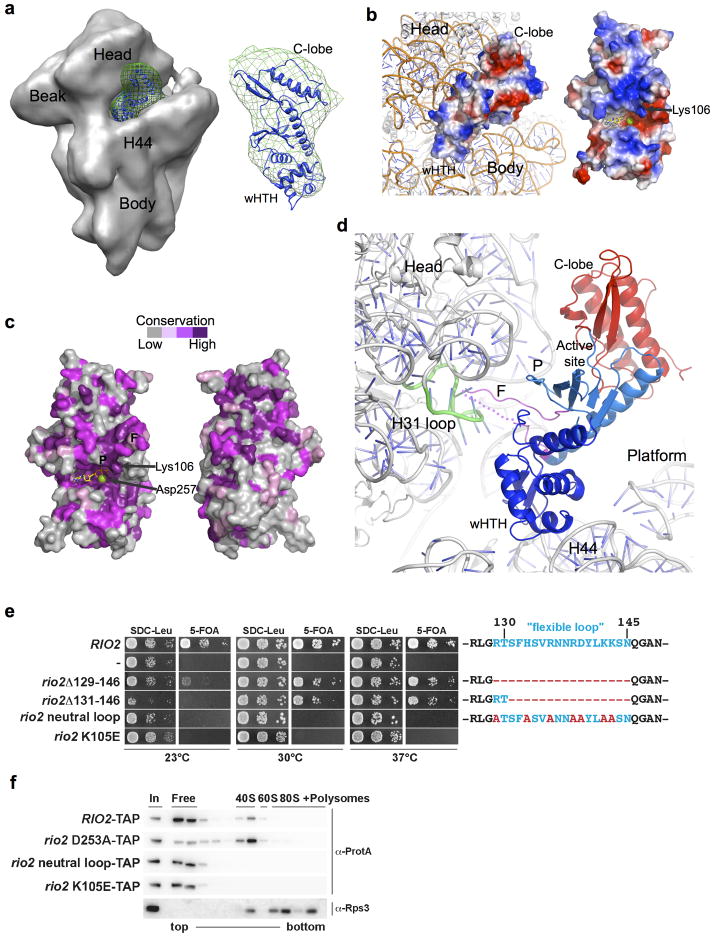
To understand how Rio2 functions in the context of the nascent 40S subunit, we docked ctRio2 into recently reported cryo-EM density of the yeast pre-40S particle8. In this report, the afRio2 structure was docked into difference density generated by subtracting the cryo-EM maps of Rio2-depleted from Rio2-containing pre-40S particles. Cryo-EM Rio2 difference density was generated using EMD maps EMD-1923, EMD-1925, EMD-1927 following previously described protocols21 and the ctRio2 model was fit using the Molecular Dynamics Flexible Fit protocol within NAMD22,23. In our attempts to fit eukaryotic ctRio2, which is larger than afRio2 and more similar to the scRio2, into the same difference density, the best fit was obtained with the active site positioned towards the rRNA, which would occlude access by outside substrate (Fig. 3a; cross correlation coefficient (CC) of 0.93 for ctRio2 compared to 0.90 for afRio2 (see Supplementary Fig. 4). This positioning differs from the reported orientation of afRio2 for which the active site is away from the rRNA and accessible, but is strongly supported by positive charge distribution and conserved residues on the rRNA contact surface of ctRio2 (Fig. 3b,c). The C-terminal extension was not included in the Rio2 model docked in the pre-ribosome, as it protruded extensively from the difference density in all orientations tested, suggesting it is likely in altered position when the molecule is docked. The ctRio2 interface with the rRNA involves both the wHTH and RIO domains, with the wHTH lodged in a groove between head and body, near the top of rRNA helix 44, and the RIO domain interacting primarily with the head of the pre-40S subunit (Fig. 3b,d). Moreover, the Rio2 flexible loop also faces the pre-40S surface and is positioned to interact in the groove between head and body (Fig. 3d). Due to crystal contacts, this ~20 amino acid long flexible loop (residue 126-148) in contrast to afRio210 is well-ordered in the ctRio2 structure (except for residues 137-140) and protrudes from the compact kinase fold (see Fig. 1a). In this way, the flexible loop may penetrate deeply into a cleft of the pre-40S head, in close proximity to helix 31 in the mature 18S rRNA (Fig. 3d). Interestingly, a recent study revealed that Rio2 is preferentially cross-linked to the terminal loop of helix 31 of the 18S rRNA, a region accommodating initiator eIF1 in the pre-initiation complex or A-site tRNA7.
Figure 3. Positioning of Rio2 kinase into the cryo-EM density map of the yeast pre-40S subunit.
(a) ctRio2 (blue cartoon) positioned in difference electron density (green mesh) generated from comparison of maps for Rio2-TAP containing (EMD-1925; shown) and Rio2-depleted pre-40S particles (EMD-1927) reported by Strunk and coworkers8. (b) Docking of ctRio2 on the yeast 40S particle (PDB:3O2Z)30 suggests extensive contact of all Rio2 domains with rRNA. A view of the rRNA-bound surface is shown for comparison of charge distribution, indicating a concentration of positive charge (blue). (c) Strong conservation of residues (violet) on the rRNA-contacting surface of ctRio2, in contrast to the opposite surface. (d) Close-up of Rio2’s flexible loop in the cleft between the 40S head and body near helix 31. Dashed line indicates flexible loop regions lacking electron density. (e) Yeast growth is inhibited by mutations of Rio2 residues involved pre-40S binding. RIO2 shuffle strain transformed with LEU2-carrying plasmids harboring wild-type RIO2, no RIO2 (−) and the indicated rio2 flexible loop mutations or rio2 K105E, were spotted on SDC-Leu and SDC plus 5-FOA plates (to shuffle out wild-type pURA3-RIO2). (f) Rio2’s flexible loop and Lys105 are required for efficient binding to the pre-40S subunit. Sucrose gradient (5–50%) centrifugation analysis of whole cell lysates prepared from RIO2 wild-type strain expressing plasmid-derived RIO2-TAP constructs. Fractions were analyzed by SDS-PAGE and Western blotting using the indicated antibodies. The soluble pool (free) and the position of 40S, 60S, and 80S ribosomes are indicated.
To confirm the validity of this docking model, mutations were made in residues of the RIO kinase domain predicted to interact with the pre-40S in the docked model, and the effect on viability and pre-40S subunit binding were determined (Fig. 3e,f). The flexible loop is predicted by the model to have extensive contact with the pre-40S particle, and hence may contribute to binding. Deletion of the flexible loop caused a slow growth phenotype, particularly at lower temperatures (Fig. 3e). However, point mutations in the flexible loop generated a stronger defect, exemplified by the Rio2 neutral loop mutant that does not support cell growth at any temperature (Fig. 3e). In a similar way mutation of the invariant lysine scRio2 K105E failed to complement the loss of function of Rio2. The Rio2 neutral loop and rio2 K105E were C-terminally TAP-tagged and co-expressed in wild-type cells. Both TAP-tagged alleles, rio2 neutral loop mutant and rio2 K105E, were expressed at similar levels when compared to wild-type Rio2-TAP (see Fig. 3f and data not shown). However, the mutants failed to be recruited to the pre-40S subunit, whereas another mutant rio2 D253A (involved in the catalytic reaction) became efficiently trapped to the pre-40S particle (Fig. 3f). These equivalent mutations were also generated in ctRio2 (K106E and neutral loop), and the purified recombinant proteins were shown to bind and hydrolyze ATP (see Supplementary Fig 3b and c). Importantly these mutants were normally expressed both in yeast and E.coli, and were catalytically active in vitro, suggesting that Rio2 binding is independent of its catalytic activity and that as predicted by our pre-40S::ctRio2 docking model, these residues are involved in contact with the pre-ribosome. Finally, this data suggests that the primary structure of the flexible loop is crucial for efficient Rio2 kinase recruitment to the pre-40S particle. Accordingly, the change of the positively charged flexible loop to neutral may interfere with its ability to contact important elements within the 18S rRNA. In contrast, a Rio2 protein lacking the loop still can bind to the pre-40S particle, albeit less efficiently, via other interacting domains (e.g. RIO domain, wHTH).
Given that cryo-EM docking suggests that all Rio2 domains directly contact the surface of the nascent 40S subunit, we tested the requirement of other Rio2 domains for their in vivo function. Deletion of the wHTH domain (rio2Δ2-86) or the entire C-terminal extension (rio2Δ316-425) yielded non-functional proteins that do not support cell growth Supplementary Fig. 5a). However, shorter C-terminal truncations were functional and complemented the rio2Δ strain, but yielded synthetic lethal phenotypes when combined with mutations in LTV120 (Supplementary Fig. 5b). These data indicate that Rio2’s C-terminal extension is required in the absence of LTV1, suggesting functional overlap with this non-essential ribosome processing factor. However the exact molecular role of the Rio2 C-terminal region in 40S biogenesis needs to be further established.
Rio2’s catalytic activity stimulates its own pre-40S release
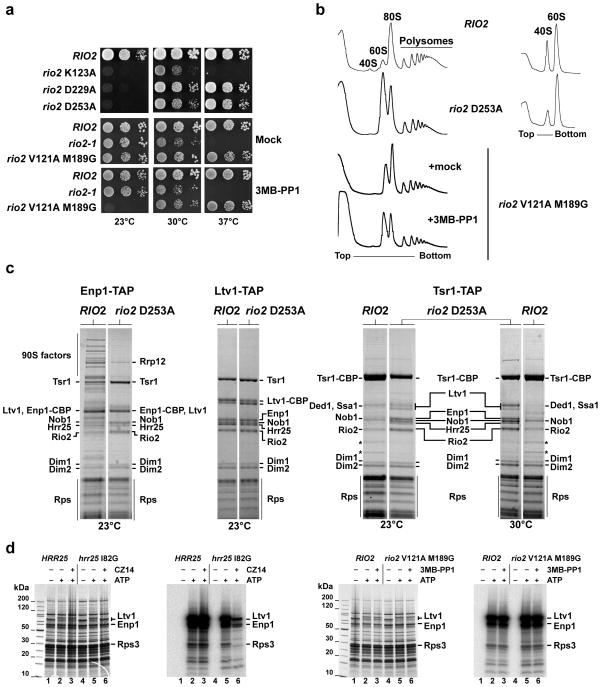
To determine whether Rio2’s dynamic association with the nascent 40S subunit is controlled by its catalytic activity, we analyzed several scRio2 kinase mutants, rio2 D229A, rio2 D253A and rio2 K123A, which contained mutations in catalytic residues predicted to affect catalysis, metal binding and triphosphate interaction, respectively (see Fig. 1 and 2 and Supplementary Fig. 1 and 3). All these mutants supported cell growth, but yielded a cold-sensitive phenotype (Fig. 4a). However, the rio2 K123A was also impaired in growth at 30°C and 37°C, suggesting that the likely decrease in ATP affinity associated with the loss of this residue results in a more severe defect (Fig. 4a). Based on structural data, we generated another scRio2 catalytic mutant, which is responsive for a bulky ATP analog24. In this case, two space-creating gatekeeper mutations, V121A and M189G, were introduced into the ATP-binding pocket of scRio2 (Val122 Met189 in ctRio2; Supplementary Fig. 5), which made Rio2 in vitro and in vivo susceptible to the ATP analog 3MB-PP1. Similar to the other Rio2 catalytic mutants, rio2 V121A M189G cells showed a cold-sensitive growth inhibition upon addition of 3MB-PP1 (Fig. 4a). Importantly, the rio2 catalytic mutants, as well as the chemical mutant poisoned by 3MB-PP1, exhibited inhibition of ribosomal 40S subunit production (Fig. 4b).
Figure 4. Rio2’s catalytic activity are required for 40S subunit biogenesis.
(a) Rio2 kinase mutants are cold-sensitive. Serial dilutions of yeast strains expressing wild-type RIO2 or the indicated kinase mutations were spotted on YPD plates and incubated at the indicated temperatures. For the series analyzing rio2 V121A M189G, it was spotted onto YPD plates containing 0.5% DMSO (Mock) or (25μM 3MB-PP1; solved in DMSO). (b) Ribosomal subunit/polysome profile analysis of the rio2 D253A kinase-dead and the 3MB-PP1 sensitive rio2 V121A M189G mutant. Whole cell lysates from cells incubated for 4 hr at 23°C were separated on a 7–40% sucrose gradient. The A260nm profiles of the derived sucrose gradient fractions are depicted. (c) Maturation of late pre-40S particles is inhibited in the rio2 D253A kinase-dead mutant. Tandem affinity-purification of pre-40S particles was performed with the indicated bait from yeast expressing either wild-type RIO2 or rio2 D253A and shifted to the indicated temperature for 4 hr. The final EGTA eluates were analyzed by SDS-PAGE and Coomassie staining. Labeled protein bands were identified by mass spectrometry.*, Rpl3 and Rpl4. (d) Comparison of Hrr25-dependent and Rio2-dependent phosphorylation of biogenesis factors associated with isolated pre-40S particles. Ltv1-TAP was affinity-purified from HRR25/RIO2 wild-type strain, the CZ14-sensitive hrr25 I82G mutant and the 3MB-PP1-sensitive rio2 V121A M189G mutant. Subsequently, these isolated pre-40S particles carrying either wild-type or mutant kinases were subjected to an in vitro phosphorylation assay20 (see also Supplementary Note). The γ–P32 labeled pre-40S particles were analyzed by SDS-PAGE/Coomassie staining, followed by autoradiography of dried gels.
These findings prompted us to analyze how these mutations affect the dynamic formation of 40S subunits. We affinity-purified different pre-40S particles from the rio2 D253A mutant and compared the pattern of co-enriched factors to that of wild-type cells (Fig. 4c). Pre-40S particles isolated via Ltv1-TAP from rio2 D253A or wild-type cells were indistinguishable. Enp1-TAP particles contain early 90S and late pre-40S factors when isolated from wild-type cells3, but exhibited a predominant ‘late’ composition with loss of early 90S factors when derived from rio2 D253A cells (Fig. 4c). In the case of Tsr1-TAP, purification from wild-type cells resulted in enrichment of a relatively late particle carrying Rio2, Tsr1, Nob1, Dim1 and Dim2 (Fig. 4c). In comparison, isolation from rio2 D253A cells demonstrated that additional factors including Ltv1, Enp1 and Hrr25 become trapped with affinity-purified Tsr1-TAP (Fig. 4c). Since Ltv1, Enp1 and Hrr25 are involved in Rps3 assembly and beak formation and Rio2 binds at the opposite site of the head8 (Fig. 2), it is conceivable that these events are more coordinated than previously thought and that Rio2’s catalytic activity might ‘crosstalk’ to these nearby pre-40S biogenesis factors by triggering their dissociation.
We took further advantage of the analogue-sensitive Rio2 mutant to perform in vitro phosphorylation assays using isolated pre-40S particles with bound biogenesis factors as potential substrates (isolated via Ltv1-TAP20). As positive control, we utilized another analog-sensitive kinase mutant, hrr25 I82G, which is responsive to the analog inhibitor CZ14 (Supplementary Fig. 5) Hrr25 kinase associated with pre-40S particles phosphorylates the pre-40S factors Ltv1 and Enp1, and ribosomal Rps3 in vivo20, and addition of CZ14 to isolated pre-40S particles carrying hrr25 I82G blocked ATP-induced phosphorylation of Ltv1, Enp1 and Rps3 (Fig. 4d). However, no change in the in vitro phosphorylation pattern compared to wild-type was observed when pre-40S particles harboring rio2 V121A M189G were incubated with 3MB-PP1 and ATP (Fig. 4d). These data support the model that Rio2’s active site is masked and inaccessible to substrate when bound to the pre-40S subunit.
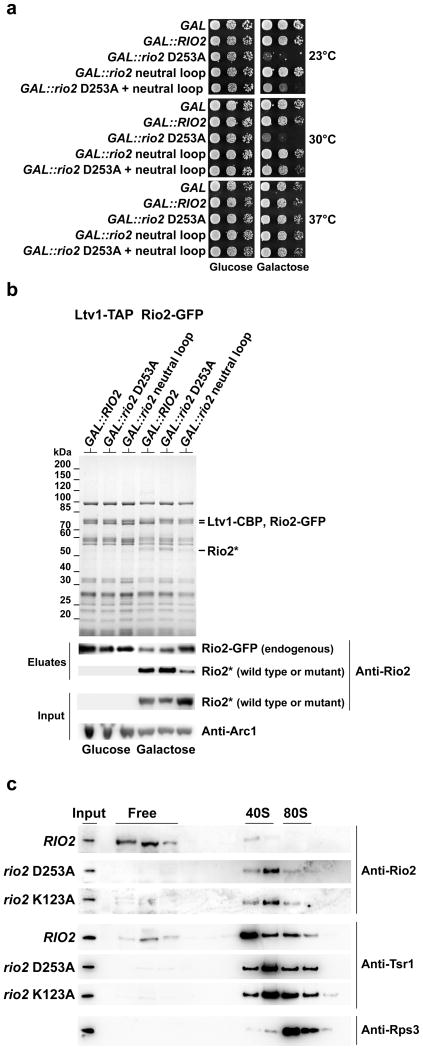
Since Rio2-dependent phosphorylation of pre-40S particle proteins was not observed, another mechanism that employs Rio2 catalytic function is indicated. Notably, GAL promoter-dependent overexpression of the rio2 catalytic mutant (rio2 D253A), but not the rio2 neutral loop mutant, caused a dominant-lethal growth defect in RIO2 wild-type cells (Fig. 5a). This phenotype agrees well with another finding that overproduced GAL::rio2 D253A but not overproduced GAL::rio2 neutral loop can efficiently replace endogenous Rio2 in pre-40S particles (Fig. 5b). However, when both mutations were intragenically combined, the dominant effect was partially suppressed (Fig. 5a). These data imply that the negative phenotype observed with the catalytic mutant requires an intact binding interface, which indicates that Rio2 catalytic mutants must be efficiently bound to exert their effect. Therefore, catalytic activity may be required for dissociation from the pre-40S. This would agree with the cold-sensitive phenotype of the catalytic mutants, which would be explained by slower dissociation rates with decreasing temperature. In order to directly assay for this, we followed trapping of Rio2 catalytic mutants in pre-40S particles in the absence of endogenous Rio2. Whereas wild-type Rio2 exists both as free protein and bound to pre-40S particles (Fig. 5c), the catalytic mutants rio2 D253A and rio2 K123A were no longer detected as free but are trapped on pre-40S particles (Fig. 5c). Furthermore, the free pool of another pre-40S factor, Tsr1, was diminished in these rio2 mutants (Fig. 5c). This indicates that Rio2’s catalytic activity is required for its own release from the late pre-40S particle, and thus influences the progression and recycling of other late pre-40S processing factors.
Figure 5. Rio2’s kinase activity is required for its own recycling from the pre-40S subunit.
(a) Overexpression of the rio2 D253A kinase dead mutant induces a dominant-negative growth defect. Serial dilutions of a wild-type yeast strain carrying the empty GAL vector (GAL), or the same plasmid with galactose-inducible GAL::RIO2, GAL::rio2 D253A, GAL::rio2 neutral loop and GAL::rio2 D253A+neutral loop (intragenically combined), respectively, were spotted on SDC-Ura (glucose, repressed) and SGC-Ura (galactose, induced) plates and incubated at the indicated temperatures for 3 (30 and 37°C) or 5 (23°C) days. (b) Recruitment of overproduced wild-type and mutant Rio2 proteins to pre-40S particles. Tandem affinity-purifications of pre-40S particle using Ltv1-TAP as bait were performed from rio2Δ strain complemented by Rio2-GFP and carrying the indicated RIO2 allele under the control of the GAL promoter. Expression of the indicated allele was repressed (glucose) or induced (galactose) for 6 hr at 23°C. The EGTA eluates were analyzed by SDS-PAGE and Coomassie staining or Western blotting using the indicated antibodies. Endogenous Rio2-GFP and GAL-induced untagged wild-type and mutant Rio2 proteins were detected with anti-Rio2 antibodies. (c) Rio2 kinase dead mutant proteins become trapped on pre-40S subunits. Whole cell extract from the indicated sample shifted for 4hr at 23°C were prepared and separated on a 5–50% sucrose gradient. Gradient fractions were analyzed by SDS-PAGE and Western blotting using the indicated antibodies.
DISCUSSION
The combination of detailed structural, biochemical and in vivo studies of eukaryotic Rio2 described in this report provide several important clues to understand its function in ribosome biogenesis.
First of all, we observed an unusual phosphoaspartate intermediate in the crystal structure, and we showed that phosphate transferred to Rio2 in solution is sensitive to incubation with hydroxylamine, a characteristic of acyl-phosphates. This finding supports the presence of a stabilized phospho-intermediate, which, if taken in analogy to P-type ATPases, requires an additional step to result in phosphate release 25. To our knowledge, this type of intermediate has only been shown for one other serine protein kinase thus far, the alpha kinase domain from myosin heavy chain kinase A, in which the phosphate is also transferred to the metal binding aspartate26. The presence of this intermediate indicated the ability of the enzyme to catalyze ATP hydrolysis, which could be subsequently verified by experimental analysis. In fact, in single turnover assays in which the concentrations of ATP is at least 10 times less the concentration of enzyme, ~99% of the total phosphate released was found to be free phosphate, unlike Hrr25, for which <1% of the total phosphate was found to be free. Thus, we suggest that Rio2, at least in its isolated form, acts as an ATPase rather than a kinase.
In good agreement with an ATPase dependent role of Rio2 in ribosome biogenesis, our model for the Rio2/pre-40S complex positions all three domains of Rio2 in contact with the pre-40S subunit, suggests that extensive interactions are made by the subfamily-specific flexible loop, and shows an occlusion of the entrance to the active site. The model is supported by the observed loss of interaction with the pre-40S upon charge mutations of the RIO domain, as well as conservation of surface residues and electrostatic charge distribution. One implication of this model is that Rio2 is positioned to sense changes in repositioning of the head of the pre-40S relative to the body domain, which is known to occur by comparison of the pre-40S particle and the structure of the mature yeast ribosome8. Another implication of the model is that Rio2 does not function as a protein kinase when bound to the pre-40S subunit, an idea further supported by our observation that phosphorylation patterns on the pre-40S do not change when Rio2 is inactive. Moreover, we have identified by mass-spectrometry phospho-serine residues after incubation of purified recombinant scRio2 with ATP. However, the systematic mutations of these autophosphorylation sites to alanine or aspartic acid alone or in combination do not show any phenotype in vivo (S.F-C and E.H. unpublished results).
Despite the inaccessibility of the active site, functional data reported here strongly indicates that catalytic activity of Rio2 is necessary for dissociation of Rio2 from the pre-40S particle. Catalytic mutants generally remain trapped on the pre-40S subunit and display a cold-sensitive phenotype, which can be explained by decreased dissociation at cold temperatures. Mutations that result in decreased binding to the ribosome result in loss of complementation. These data support a model, in which ATP hydrolysis facilitates dissociation from the pre-40S.
Surprisingly, Rio2 ATP hydrolysis dependent release is only essential for cell growth at lower temperature, however our results suggest that in the absence of Rio2 catalytic activity at higher temperature, pre-40S progression and Rio2 released remain kinetically delayed. Moreover, Rio2 catalytic activity becomes essential for cell growth at any temperature in combination with viable mutations of several late pre-40S factors (S.F.-C. and E. H. unpublished results). However, it is possible that at higher temperature spontaneous dissociation of Rio2 from the pre-40S occurs frequently enough to allow for viability, or that an alternative “safety” mechanism is predominantly active at higher temperature that allows, or stimulates, Rio2’s release. Nevertheless, in Rio2 catalytic mutants, the basal ATPase activity is not completely abolished. Therefore, this remaining enzymatic activity will be influenced on one hand by the temperature, and will be regulated on the other hand suggesting that a trigger could stimulate Rio2’s basal enzymatic activity of several fold, thus enabling Rio2 activity in vivo to a level sufficient to support growth at higher temperature. In future work, it will be interesting to determine the in vivo molecular mechanism regulating Rio2 ATPase activity.
Finally, the structure of an eukaryotic Rio2 reveal the presence of a C-terminal extension helix that binds between the N- and C-terminal lobes of the kinase domain, a peculiar new feature. Auto-inhibitory helices positioned similarly in the kinase domain or as an extension of the C-terminal domain have been identified in ePKs such as Src Kinase, Ribosomal S6 Kinase (RSK2) and c-Abl tyrosine kinase27–29. The positioning of this helix near the active site results in partial shielding of the γ-phosphate binding pocket. Although our data shows that the helix is not essential for growth, the synthetic lethality observed with Δltv1 suggests a role in coordination with other ribosome processing factors. However the exact molecular function of this helix remain to be clarified. It is also important to note that this helix was not included in the docking of the Rio2 protein on the pre-ribosome, since initial attempts resulted in the positioning of the helix outside the cryo-EM density. It would therefore be plausible that the helix is repositioned upon docking, perhaps to contact nearby regions of the pre-40S or that the observed positioning occur in another pre-40S intermediate.
In conclusion, our data indicate that Rio2 is not a classical substrate kinase but rather functions as an ATPase which with a key structural role to bind and protect a ‘neuralgic’ region on the pre-40S particle from premature recruitment of the translation machinery and possibly sense or promote conformational changes in the pre-40S. However, Rio2 is strategically positioned between the head and body of nascent 40S subunit to sense structural changes occurring during pre-40S factor modification/release and late pre-rRNA processing. We propose that the kinase domain of the evolutionary ancient family of RIO kinases has evolved not to transfer phosphate onto other substrates but to exploit conformation-dependent ATP-hydrolysis as a molecular switch to drive through 40S biogenesis.
Online Methods
Yeast strains and yeast genetic methods
The Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table 1. All strains, unless otherwise specified, are derivatives of BY4741 (Euroscarf). Gene disruption and C-terminal tagging were performed as previously described31–33. Preparation of media, yeast transformation, and genetic manipulations were done according to standard procedures.
Plasmid constructs
All recombinant DNA techniques were performed according to standard procedures using Escherichia coli DH5α for cloning and plasmid propagation. Site-directed mutagenesis was performed by overlap extension PCR. All cloned DNA fragments generated by PCR amplification were verified by sequencing. Plasmids used in this study are listed in Supplementary Table 2. ctRio2 was amplified from Chaetomium thermophilum cDNA11 and cloned into appropriate yeast or E. coli expression vectors.
Expression and purification of full-length ctRio2
Full-length ctRio2 was expressed using pT7-ctRio2 vector in E. coli Rosetta™-DE3 (pLysS) cells. Transformed cells were used to inoculate an overnight 100 ml culture containing both 100 μg/ml ampicillin and 34 μg/ml chloramphenicol at 37°C. The overnight culture was diluted 1/100 into LB with antibiotics. The cultures were incubated at 37°C until they reached OD600nm of 0.5, and induced with 1 mM IPTG for overnight expression at 18°C. The cells were collected by centrifugation at 5,000 rpm for 30 min and stored frozen at −80°C. Frozen pellets were resuspended in 10 ml of buffer A (50 mM Tris, pH 8.0, 200 mM NaCl, 10% glycerol, 2.5 mM MgCl2, 0.05% β-mercaptoethanol (BME) per gram of cell pellet. After resuspension, 5 mg of DNAse, 1 mg of RNase A, and 20 μl of RNase T1 (100,000 U/ml) per 5 g of cell pellet were added to the resuspended cells. One Complete™, EDTA free, protease inhibitor cocktail tablet (Roche) per 50 ml, 0.3 X Bugbuster™ and 0.1 mg of lysozyme/ml was added to lyse the cells with stirring at 4°C for 30 min. The lysate was centrifuged at 16,000 rpm for 1 hr and the supernatant containing the soluble protein was filtered through a 0.22 μm filter, then passed over a 5 ml His-Trap™ column (GE Healthcare) pre-equilibrated with Buffer A. Buffer B (50 mM Tris, pH 8.0, 200 mM NaCl, 10% glycerol, 2.5 mM MgCl2, 1 M imidazole, 0.05% BME) was used to elute the protein. 3% Buffer B was used to elute ctRio2 from the column in 5 ml fractions were collected. The combined fractions were dialyzed against Buffer A overnight at 4°C in the presence of TEV protease at a concentration of 0.1 mg/ml. The cleaved protein was subjected to a second pass over the His-Trap™ column equilibrated in Buffer A. The cleaved protein was collected in the column flow-through. The collected fractions were then passed over a 5 ml Hi-Trap Q column pre-equilibrated with the loading buffer, Buffer C, (50 mM Tris pH 8.0, 200 mM NaCl, 2.5 mM MgCl2, 10% glycerol, 0.05% BME). The protein was eluted using a gradient from Buffer C to Buffer D (50 mM Tris pH 8.0, 1 M NaCl, 2.5 mM MgCl2, 10% glycerol, 0.05% β-mercaptoethanol) over 100 ml. Fractions containing pure ctRio2 were combined and dialyzed against 1 liter of Buffer E (10 mM Tris pH 8.0, 150 mM NaCl, 2.5 mM MgCl2, and 0.5% BME). The ctRio2 was concentrated to 10 mg/ml. Selenomethionine substituted protein was grown using M9 SeMET High-Yield™ Growth Media Kit (Shanghai Medicilon Inc.) following the instructions provided and purified using the procedure outlined above.
Single turnover reactions
Single turnover experiments were essentially performed as previously described34. In brief, the indicated amounts of proteins (see Supplementary Note for detailed purification procedure) were incubated with a final concentration of 50 nM ATP containing 750 nCi of γ32P-labeled ATP (Hartman Analytic 6000 Ci/mmol). For thin-layer chromatography (TLC) experiments the reactions were stopped at the indicated time points, by rapidly mixing 5% of the reaction volume with 18 volumes of perchloric acid (1 M) and directly neutralized by addition of 6 volumes of potassium acetate (8 M) and stored in liquid nitrogen. Reactions were centrifuge at 14,000rpm for 10 min at room temperature. 1.6% of every time points was loaded on Polygram Cel 300 PEI (Macherey-Nagel) TLC plates and developed with 350 mM KH2PO4 buffer for 45–60 min and dried.
For SDS-PAGE analysis, 10% of the reaction was stopped by addition of 1 volume of urea containing SDS sample buffer (10% SDS, 200 mM Tris-HCl pH 6.8, 1 mM EDTA, 8 M Urea, 1% BME) and store in liquid nitrogen before loading the samples on a Nu-PAGE 4–12% gradient gel (Invitrogen). TLC and SDS gel were exposed overnight on a Phosphorimager screen (BAS-MS 2040 Fujifilm), and read with a FLA-7000 (Fujifilm). Quantifications were perfomed with ImageJ and described in details in Supplementary Note.
Steady state ATP hydrolysis assays
Proteins were purified as described above. A coupled-enzyme assay system was used to monitor ADP release under saturating concentrations of ATP. Reactions (1 mL) were performed at 37°C in a 1 mL disposable cuvette (Fisher) containing 25 mM HEPES pH 7.5, 5 mM KCl, 5 mM MgCl2, 0.2% BME, 0.2 mM phosphoenol pyruvate (Sigma), 0.4 mM NADH (Sigma), 500 μM ATP (Sigma), pyruvate kinase/lactate-dehydrogenase (4 μl at 900–1400 U/mL; Sigma) and 0.46 mM Rio2 enzyme. The rate conversion of NADH to NAD+ was determined by monitoring decrease in absorbance at 350 nm in a temperature-controlled, UV/Vis spectrophotometer. The rate of ADP turnover was calculated by the following:
where ε350nm, NADH is the extinction coefficient for NADH at 350 nM (5770 M−1 cm−1) and [Rio2] is the concentration of Rio2 enzyme.
Hydroxylamine sensitivity assay
Wild-type ctRio2 (5 μg) was autophosphorylated by incubation for 10 mins at 37°C in 50 μl reaction buffer containing 50 mM HEPES pH 7.5, 50 mM NaCl, 2 mM MgCl2, 0.2% BME, and 2 μCi γ-32P-labeled ATP (Perkin-Elmer, 10 μCi/μL). The proteins were precipitated using cold trichloroacetic acid (TCA) added to the reactions to a total concentration of 10%. The samples were centrifuges and the supernatants were discarded. 100 μl of either reaction buffer without ATP, 0.5 M NaCl, 0.5 M (NH3OH)2SO4, or NH3OH·HCl or 0.5 M NaOH, was used to resuspend each pellet. The reactions were incubated at room temperature for 30 minutes. The reactions were blotted to nitrocellulose membrane using a vacuum manifold and washed 5 times in phosphate-buffered saline. The blot was used to expose a phosphorimager screen (Molecular Dynamics), which was then scanned after 4 hours using a Storm imager (Molecular Dynamics).
Crystallisation of ctRio2
Purified recombinant proteins were crystallized by the sitting-drop vapour-diffusion method. The purified Rio2 protein samples were screened for crystallization conditions using commercially available crystal screens. A total of 384 conditions were tested in three different protein-to-well solution ratios (in 0.6 μl drops). Screens were incubated at 20°C. ctRio2 was screened with and without 2 mM ATP and 2.5 mM MgCl2. Diffraction quality crystals were obtained initially in 0.2 M magnesium chloride hexahydrate, 0.1 M Bis-Tris, pH 5.5, 25% (w/v) PEG 3350 for both. In optimized conditions, (0.1 M Tris pH 8.5 and 16% PEG 3350), crystals grew large enough for X-ray diffraction studies after 4–5 days at 20°C.
Data Collection and Structure Determination
ATP-complexed and APO ctRio2 crystals were flash frozen in mother liquor containing 25% ethylene glycol. Diffraction data were collected at 100K with a MAR300 CCD detector at the NE-CAT beamline 22-ID located at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL, USA). X-ray data collection and refinement statistics are shown in Supplementary Table 1. The data were integrated and merged using HKL200035. The structure of ATP ctRio2 was solved by molecular replacement using the known structure of afRio2 (PDB ID 1ZAO) and the AutoMR program from the PHASER crystallography software suite36. The structure of APO ctRio2 was solved by using the structure determined in the presence of ATP as a model. Rfree was monitored by using 8% of the reflections as a test set. Table 1 contains the data collection and refinement statistics of the data. The figures that depict the structures of ctRio2 were created using PYMOL37.
Cryo-EM Density Fitting
Cryo-electron microscopy data obtained for the Rio2-TAP and Rio2-depleted yeast pre-40S particles8 (EMD-1927 and EMD-1925) were downloaded from the Electron Microscopy Databank. Difference maps were generated by subtraction after alignment and scaling, and the density previously identified as belonging to Rio2 was used to fit the ctRio2 model. ctRio2 was rigid body fit in three orientations using Chimera and compared to determine the best fit. The fit for the three orientations is shown in Supplementary Fig. 3. The molecular dynamics flexible fit (MDFF) protocol in NAMD v2.822,23 (with VMD v1.9)38 was used to estimate the optimal repositioning of the domains of ctRio2 to allow a better fit into the difference density. ctRio2 was modeled with explicit solvent and ions were added to neutralize the system and maintain and ionic strength of 0.1 M within VMD prior to the fit. Domain restraints were used to define the wHTH, N-lobe and C-lobes, and the extension helix αI was omitted from the MDFF protocol due to poor fit of this region into the difference electron density and for clarity. The resulting domain fit after simulations calculated over 0.4 ns (until root mean square deviation (RMSD) convergence), is shown in Supplementary Fig. 3.
Supplementary Material
Acknowledgments
We thank K. Shokat (Howard Hughes Medical Institute and Department of Cellular and Molecular Pharmacology, University of California, San Francisco, CA 94158, USA) for providing the ATP analog compounds CZ14 and 3MB-PP1, K. Karbstein (Scripps Research Institute, Jupiter, FL 33458, USA) for providing anti-Tsr1 antibody, S. Amlacher for providing Chaetomium thermophilum cDNA, E. Thomson and S. Griesel for providing ctHrr25 expression vector (Biochemistry Center, University of Heidelberg, 69120 Heidelberg, Germany) J. Lechner and his team for mass spectrometry, G. Lorimer (Department of Chemistry and Biochemistry, University of Maryland, College Park, MD 20742) for assistance and advice in steady state kinetic measurements, and M. Gnädig for her excellent technical assistance. Data collection was conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, supported by grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8 P41 GM103403-10) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. This work was funded by the postdoctoral fellowship from the Medical Faculty of the University of Heidelberg to S.F.-C., the German Research Council (DFG Hu363/10-4) to E.H. and National Institutes of Health National Cancer Institute grant (K22CA123152) to N.L.-L.
Footnotes
Accession codes Atomic coordinates and structure factors for the crystal structures have been deposited in the Protein Data Bank under accession numbers 4GYG (APO-Rio2) and 4GYI (pRio2-ADP-Mg2+).
Author Contributions S.F.-C., T.S., and E.H. conceived the experiments. S.F.-C., T.S. and A.M.W. constructed all plasmids; yeast strains and carried out all yeast genetic experiments. S.F.-C. performed all the sucrose gradient analysis, tandem-affinity purifications of yeast proteins, single-turnover and nucleotide binding analysis. T.S. performed in vitro phosphorylation experiments on purified pre-40S. S.F.-C. and A.M.W. performed the first biochemical characterization of ctRio2. V.S. and E.C. optimized and performed protein purification and identified and refined crystallization conditions. V.S. and N.L.-L determined the crystal structures. M.D. performed hydroxylamine phosphate release assay and steady-state rate determination, using purified protein provided by H.L. N.L.-L performed ctRio2::40S docking analysis. E.H. and N.L.-L. supervised the work; S.F.-C., N.L.-L., and E.H. wrote the manuscript. All authors commented on the manuscript.
The authors declare no competing financial interests.
References
- 1.Geerlings TH, Faber AW, Bister MD, Vos JC, Raue HA. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae. J Biol Chem. 2003;278:22537–45. doi: 10.1074/jbc.M300759200. [DOI] [PubMed] [Google Scholar]
- 2.Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:2083–95. doi: 10.1128/MCB.23.6.2083-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–80. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henras AK, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–59. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 6.Zemp I, et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol. 2009;185:1167–80. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010;29:2026–36. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strunk BS, et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–53. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaRonde-LeBlanc N, Guszczynski T, Copeland T, Wlodawer A. Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes. FEBS J. 2005;272:2800–10. doi: 10.1111/j.1742-4658.2005.04702.x. [DOI] [PubMed] [Google Scholar]
- 10.LaRonde-LeBlanc N, Wlodawer A. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure. 2004;12:1585–94. doi: 10.1016/j.str.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Amlacher S, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–89. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 12.Post RL, Kume S. Evidence for an aspartyl phosphate residue at the active site of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem. 1973;248:6993–7000. [PubMed] [Google Scholar]
- 13.Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282–95. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 14.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989;264:21770–8. [PubMed] [Google Scholar]
- 17.Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;273:14107–12. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 18.Zheng J, et al. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–61. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 19.Parang K, Cole PA. Designing bisubstrate analog inhibitors for protein kinases. Pharmacol Ther. 2002;93:145–57. doi: 10.1016/s0163-7258(02)00184-5. [DOI] [PubMed] [Google Scholar]
- 20.Schafer T, et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–5. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 21.Aaronson RP, Blobel G. On the attachment of the nuclear pore complex. J Cell Biol. 1974;62:746–754. doi: 10.1083/jcb.62.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–83. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–72. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 25.Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–66. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 26.Ye Q, Crawley SW, Yang Y, Cote GP, Jia Z. Crystal structure of the alpha-kinase domain of Dictyostelium myosin heavy chain kinase A. Sci Signal. 2010;3:ra17. doi: 10.1126/scisignal.2000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–38. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 28.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–71. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 29.Malakhova M, et al. Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2. Nat Struct Mol Biol. 2008;15:112–3. doi: 10.1038/nsmb1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 31.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–62. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 32.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–29. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 34.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry. 2001;40:15224–33. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology, Macromolecular Crystallography, part A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delano W. The Pymol molecular graphics system. Delano Scientific; 2002. [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.