Abstract
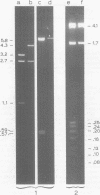
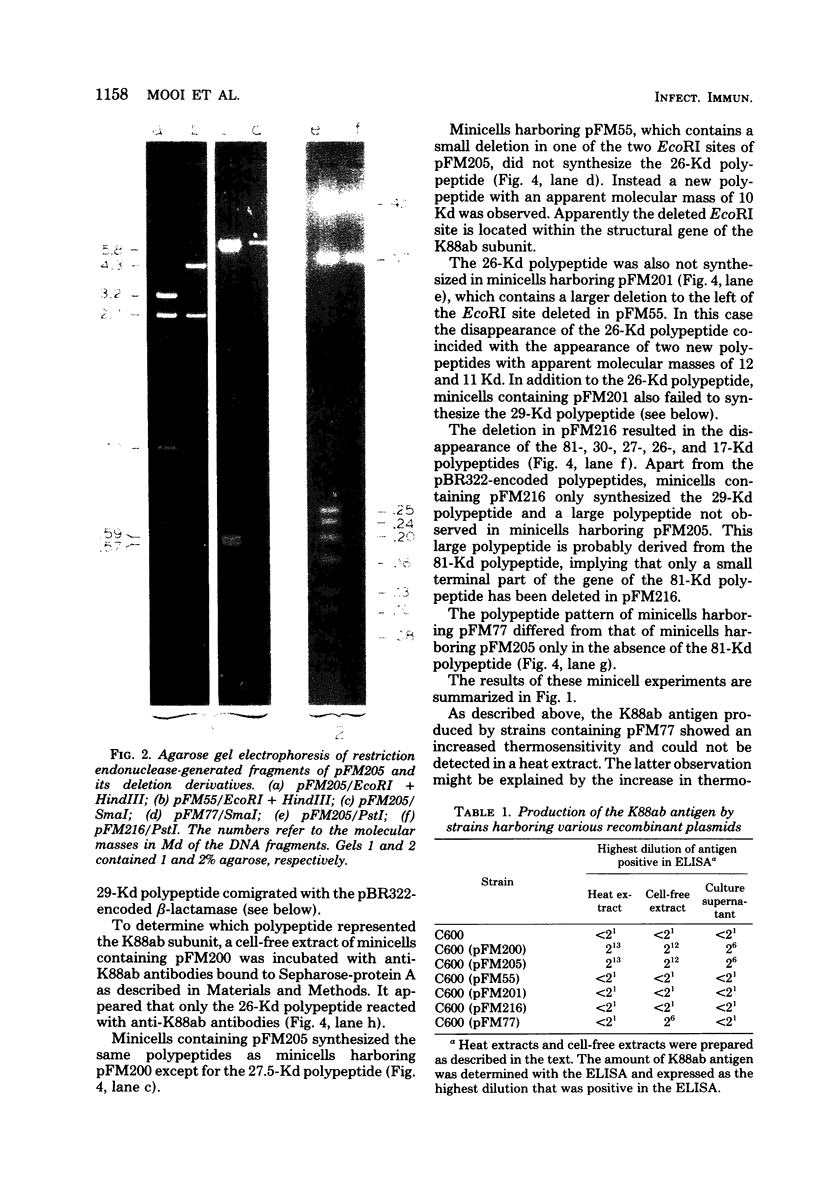
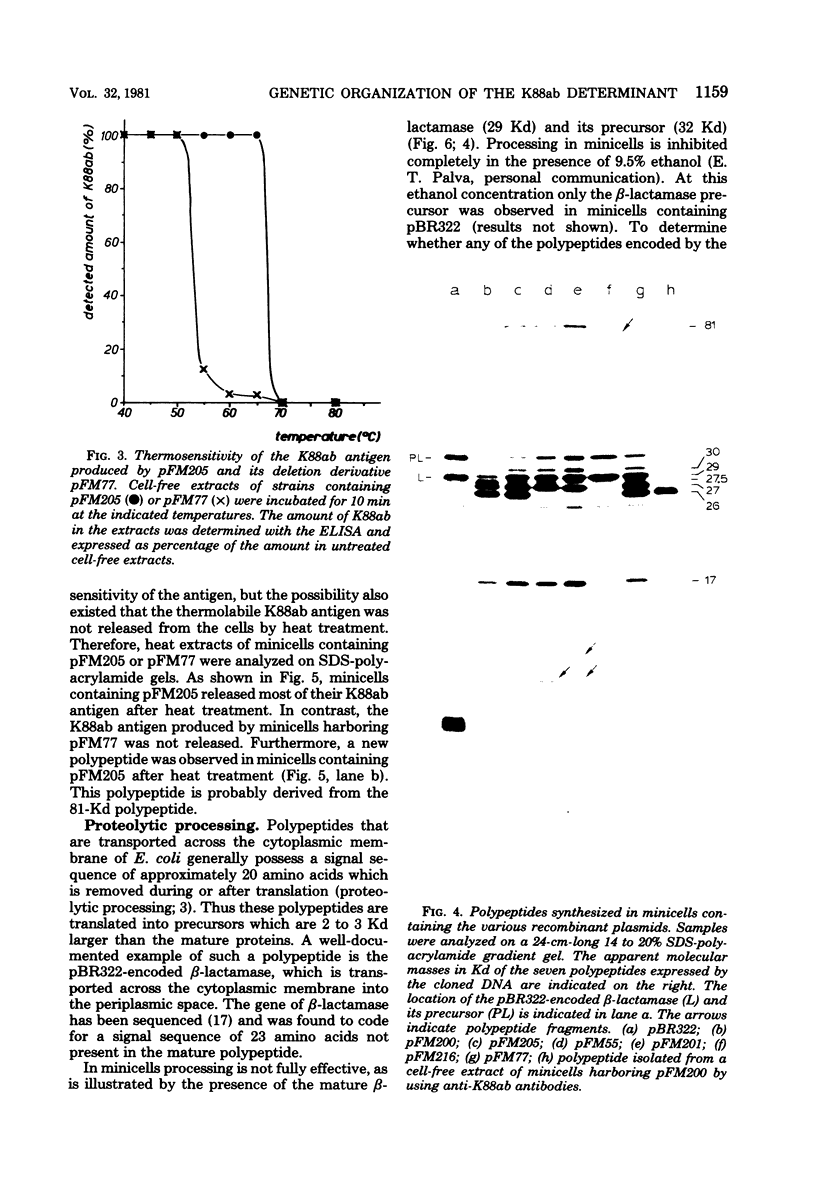
Escherichia coli K-12 minicells were used to study the expression of the genes located on plasmid pFM205, which contains the genetic determinant of the K88ab antigen. Plasmid pFM205 is composed of a 4,3-megadalton large deoxyribonucleic acid fragment derived from the wild-type K88ab plasmid pRI8801 (51 megadaltons) and the cloning vehicle pBR322. The K88ab deoxyribonucleic acid of pFM205 appeared to express six polypeptides with apparent molecular masses of 81, 30, 29, 27, 26, and 17 kilodaltons, respectively. These polypeptides account for approximately 85% of the coding capacity of the cloned deoxyribonucleic acid. The 26-kilodalton polypeptide was found to react with specific anti-K88ab antibodies and therefore represents the K88ab subunit. The K88ab subunit and at least two other polypeptides (81 and 17 kilodaltons) were translated into precursors which were about 2 kilodaltons larger than the mature proteins. Plasmid pFM205 was used to construct deletion mutants. By analyzing these mutants in minicells the genes of the six polypeptides could be located on the physical map of pFM205. It appeared that deletion of the gene of the 81-kilodalton polypeptide resulted in an altered conformation of the K88ab antigen.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertschinger H. U., Moon H. W., Whipp S. C. Association of Escherichia coli with the small intestinal epithelium. I. Comparison of enteropathogenic and nonenteropathogenic porcine strains in pigs. Infect Immun. 1972 Apr;5(4):595–605. doi: 10.1128/iai.5.4.595-605.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Davis B. D., Tai P. C. The mechanism of protein secretion across membranes. Nature. 1980 Jan 31;283(5746):433–438. doi: 10.1038/283433a0. [DOI] [PubMed] [Google Scholar]
- Dougan G., Saul M., Twigg A., Gill R., Sherratt D. Polypeptides expressed in Escherichia coli K-12 minicells by transposition elements Tn1 and Tn3. J Bacteriol. 1979 Apr;138(1):48–54. doi: 10.1128/jb.138.1.48-54.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Guinée P. A., Jansen W. H. Behavior of Escherichia coli K antigens K88ab, K88ac, and K88ad in immunoelectrophoresis, double diffusion, and hemagglutination. Infect Immun. 1979 Mar;23(3):700–705. doi: 10.1128/iai.23.3.700-705.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halegoua S., Inouye M. Translocation and assembly of outer membrance proteins of Escherichia coli. Selective accumulation of precursors and novel assembly intermediates caused by phenethyl alcohol. J Mol Biol. 1979 May 5;130(1):39–61. doi: 10.1016/0022-2836(79)90551-5. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mooi F. R., de Graaf F. K., van Embden J. D. Cloning, mapping and expression of the genetic determinant that encodes for the K88ab antigen. Nucleic Acids Res. 1979 Mar;6(3):849–865. doi: 10.1093/nar/6.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- ORSKOV I., ORSKOV F., SOJKA W. J., WITTIG W. K ANTIGENS K88AB(L) AND K88AC(L) IN E. COLI. A NEW O ANTIGEN: 0147 AND A NEW K ANTIGEN: K89(B). Acta Pathol Microbiol Scand. 1964;62:439–447. doi: 10.1111/apm.1964.62.3.439. [DOI] [PubMed] [Google Scholar]
- Orskov I., Orskov F. Episome-carried surface antigen K88 of Escherichia coli. I. Transmission of the determinant of the K88 antigen and influence on the transfer of chromosomal markers. J Bacteriol. 1966 Jan;91(1):69–75. doi: 10.1128/jb.91.1.69-75.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. L., West R. W., Heyneker H. L., Bolivar F., Boyer H. W. Characterizing wild-type and mutant promoters of the tetracycline resistance gene in pBR313. Nucleic Acids Res. 1979 Jul 25;6(10):3267–3287. doi: 10.1093/nar/6.10.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]