Abstract
Salicylic acid (SA) is widely recognized as a key player in plant immunity. While several proteins have been previously identified as the direct targets of SA, SA-mediated plant defense signaling mechanisms remain unclear. The Nature paper from Xinnian Dong's group demonstrates that the NPR1 paralogues NPR3 and NPR4 directly bind SA, and this binding modulates their interaction with NPR1 and thereby degradation of this key positive regulator of SA-mediated defense, shedding important new insight into the mechanism(s) of SA-mediated, NPR1-dependent plant defense signal transduction.
Salicylic Acid (SA) and its derivatives (e.g., aspirin) have long been recognized for their medicinal properties as non-steroidal anti-inflammatory drugs and as pain and fever relievers. An increasing number of studies show that SA also can delay and/or prevent the development of several cancers, cardiovascular diseases, and strokes1,2. While several SA protein targets have been identified in mammalian cells1, their molecular and physiological modes of action remain unclear. Thus, efforts to dissect SA's mechanisms of action continue to rely on identifying additional protein targets. Indeed, SA was recently shown to bind and activate AMP-activated protein kinase, helping to explain some of its disease-preventing effects3.
SA is naturally produced in plants, and it plays diverse roles in growth, development, and responses to abiotic stresses4. Additionally, SA is widely recognized as a key player in multiple layers of plant disease resistance, including basal resistance, effector-triggered immunity (ETI, also termed resistance gene-mediated resistance) and systemic acquired resistance (SAR)5. To decipher SA-mediated plant defense signaling mechanisms, several SA-binding proteins (SABPs) have been identified, including a catalase, cytosolic ascorbate peroxidase, chloroplastic carbonic anhydrase, and methyl salicylate esterase. Extensive study of the latter protein revealed its essential role in SAR5. However, despite identification of the aforementioned SABPs, SA's signaling mechanisms remain unclear. Considering SA's many roles in plants, these SABPs may constitute only a small portion of SA's targets; moreover, the SA receptor remained to be found.
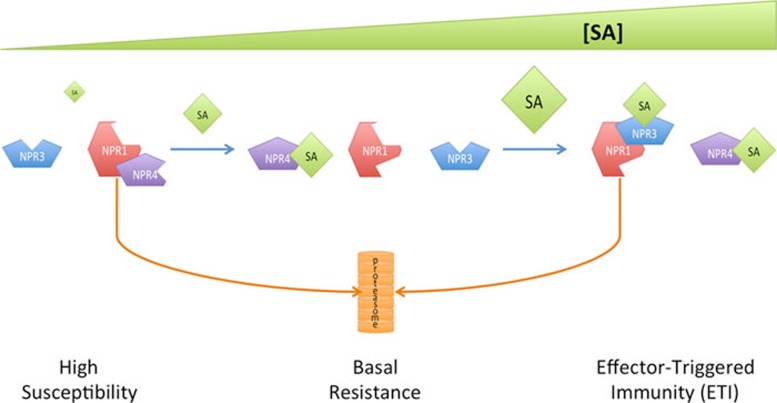
In this context, the Nature paper from Xinnian Dong's group6 represents a major step forward in our understanding of SA signaling mechanisms during plant-pathogen interactions. Dong's group has been instrumental in characterizing the function of NPR1 (Nonexpresser of Pathogenesis-Related genes 1) in plant defense7. While NPR1 is a key player in one of the SA-mediated defense signaling pathways, it does not appear to be an SA receptor as it does not directly bind SA6. Instead, SA regulates the conversion of NPR1 from an oligomeric to a monomeric form, which leads to its nuclear translocation8. SA also regulates NPR1 phosphorylation, which facilitates NPR1's recruitment to a Cullin3 (CUL3) E3 ligase and subsequently proteasome-mediated degradation9. Now Dong's group has demonstrated that the NPR1 paralogues NPR3 and NPR4 are adaptor proteins for the CUL3 E3 ligase that specifically target NPR1 for degradation in an SA concentration-dependent manner6. Supporting their conclusion, NPR3 and NPR4 contain domains typically found in CUL3 substrate adaptors, and npr3/4 single and double mutants contain elevated levels of NPR1. Furthermore, NPR3 and NPR4 directly interact with NPR1. Strikingly, SA disrupts the NPR1-NPR4 interaction, thereby making NPR1 less susceptible to degradation, whereas SA promotes the NPR1-NPR3 interaction, which makes NPR1 more accessible for degradation (Figure 1). Since NPR4 has high affinity for SA (nanomolar range) while NPR3 has low affinity for SA (micromolar range), low SA levels should reduce NPR1 degradation, whereas high SA levels should enhance it.
Figure 1.
NPR1 homeostasis is controlled by SA binding to NPR3/NPR4 in a concentration-dependent manner. At low SA levels (High Susceptibility, left), NPR1 is unavailable to induce defense gene since it is targeted through its binding to NPR4 for degradation in proteasomes. As SA concentration increases after infection (Basal Resistance, middle), SA binds to NPR4 disrupting its interaction with NPR1. Free NPR1 can now play its role in defense gene activation. At very high concentrations (ETI, right), SA levels are sufficient to bind to NPR3 and promote its interaction with NPR1, leading to NPR1 turnover.
At the biological level, nuclear accumulation of NPR1 is required for basal defense gene expression, whereas proteasome-mediated turnover is required for ETI, and a combination of NPR1 accumulation and turnover is necessary for SAR development6,9. The results presented by Fu et al.6 suggest that the interplay between NPR1, NPR3/4, and an SA concentration gradient finetunes NPR1 homeostasis and thus helps specify disease resistance. According to their working model, the enhanced susceptibility exhibited by SA-deficient plants is due to unrestricted NPR4 binding to NPR1, which depletes NPR1 due to CUL3NPR4-mediated degradation6. In wild-type plants, low basal SA levels may bind to NPR4, thereby allowing some NPR1 to accumulate to confer basal resistance. Following pathogen infection, recognition of pathogen effectors by plant resistance proteins induces a high level of SA in local infected tissues; in this case, CUL3NPR3-mediated degradation would allow fast NPR1 turnover, leading to ETI. In systemic tissues, an intermediate level of SA would enable both NPR1 accumulation and turnover, leading to SAR.
Clearly, the study by Fu et al.6 represents a major step towards elucidating the mechanism(s) of SA perception in programming defense gene expression. However, NPR3 and NPR4 may not be SA receptors in a traditional sense. An increasing body of evidence indicates the existence of SA-dependent, but NPR1-independent defense signal transduction pathways10, in which NPR3/4 may not participate. In addition, it is unknown whether NPR3/NPR4-mediated SA perception is involved in the diverse roles that this hormone plays in growth and development, or in abiotic stress. Even for NPR1-dependent defense signal transduction, it is unclear whether NPR3/NPR4 are involved in SA's ability to induce nuclear translocation of NPR1 and/or promote NPR1 phosphorylation to facilitate the proteasome-mediated turnover. Moreover, since SA binding did not affect the gel filtration elution profile of NPR46, the mechanism through which SA binding influences the ability of NPR4 (or NPR3) to bind NPR1 is currently unknown. Thus, many aspects of SA-mediated signaling remain to be explored.
References
- Duthie GG, Wood AD. Natural salicylates: foods, functions and disease prevention. Food Funct. 2011;2:515–520. doi: 10.1039/c1fo10128e. [DOI] [PubMed] [Google Scholar]
- Hart RG, Harrison MJ. Aspirin wars: the optimal dose of aspirin prevent stroke. Stroke. 1996;27:585–587. doi: 10.1161/01.str.27.4.585. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Fullerton MD, Ross FA, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot. 2011;62:3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, et al. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C, Mou Z. Salicylic acid and its function in plant immunity. J Integr Plant Biol. 2011;53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]