Abstract
The methylation state of the paternal genome is rapidly reprogrammed shortly after fertilization. Recent studies have revealed that loss of 5-methylcytosine (5mC) in zygotes correlates with appearance of 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). This process is mediated by Tet3 and the 5mC oxidation products generated in zygotes are gradually lost during preimplantation development through a replication-dependent dilution process. Despite these findings, the biological significance of Tet3-mediated oxidation of 5mC to 5hmC/5fC/5caC in zygotes is unknown. DNA methylation plays an important role in silencing gene expression including the repression of transposable elements (TEs). Given that the activation of TEs during preimplantation development correlates with loss of DNA methylation, it is believed that paternal DNA demethylation may have an important role in TE activation. Here we examined this hypothesis and found that Tet3-mediated 5mC oxidation does not have a significant contribution to TE activation. We show that the expression of LINE-1 (long interspersed nucleotide element 1) and ERVL (endogenous retroviruses class III) are activated from both paternal and maternal genomes in zygotes. Inhibition of 5mC oxidation by siRNA-mediated depletion of Tet3 affected neither TE activation, nor global transcription in zygotes. Thus, our study provides the first evidence demonstrating that activation of both TEs and global transcription in zygotes are independent of Tet3-mediated 5mC oxidation.
Keywords: DNA demethylation, transposable element, preimplantation embryo, Tet3
Introduction
Previous studies have revealed that global reprogramming of DNA methylation in germ cells takes place at two different phases. One takes place in the paternal genome a few hours after fertilization1,2. Although this dynamic event was reported a decade ago, the molecular mechanism as well as its biological significance just begin to be revealed. The discovery that the ten-eleven translocation (Tet) family of proteins have the capacity to oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC)3,4, 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)5,6 in combination with the fact that the glycosylase Tdg can remove 5fC and 5caC5,7 suggest a mechanism by which 5mC can be removed through the coordinated action of Tet and Tdg proteins followed by DNA repair8. Consistently, loss of 5mC in paternal genome in the zygotes correlates with Tet3-mediated 5mC oxidation9,10,11. Recently, we demonstrated that loss of 5mC in the paternal genome involves two steps, including the oxidation of 5mC to 5hmC/5fC/5caC in zygotes followed by replication-dependent dilution during preimplantation development12,13. Despite the rapid progress in understanding the mechanism of paternal DNA demethylation, the biological significance of this molecular event remains unknown.
One of the most important functions of DNA methylation is to maintain genome stability by silencing the expression of transposable elements (TEs)14. TEs account for a large proportion of eukaryotic genomes. About 40% of the mouse genome is composed of TEs, including long interspersed nucleotide elements (LINEs), short interspersed nucleotide elements (SINEs), and LTR retrotransposable elements15. Since expression of TEs can result in their translocation into the host genome, leading to the disruption of genome integrity16,17, they are silenced in most cell types. A major epigenetic mechanism used in silencing TEs is DNA methylation14. For example, a strict correlation between LINE-1 silencing and DNA methylation at its 5′ regulatory region has been established in various human cell lines18. Similar studies in human embryonic fibroblasts demonstrated that 5-azadeoxycytidine-induced loss of DNA methylation resulted in derepression of LINE-145. In addition, deletion of the DNA methyltransferase 3L (Dnmt3l) or lymphoid-specific helicase (Lsh) gene results in the loss of DNA methylation on TEs and the derepression of TEs in germ cells16,17,19. Furthermore, an in vitro study suggested that DNA methylation on LINE-1 promoter is essential for inhibition of LINE-1 expression20. Thus, it is well established that DNA methylation on the regulatory regions of TEs plays an important role in silencing TE expression.
Several types of TEs are highly expressed during early stages of preimplantation development21,22. Importantly, LINE-1 expression has been shown to be important for preimplantation development23. However, how the expression of TEs in early preimplantation embryos is controlled remains unknown. Based on the observations that: (1) CpG sites in TEs are highly methylated in sperm genome and are demethylated after fertilization24,25,26; (2) the hypomethylated state of these CpG sites is maintained during preimplantation development25, it is assumed that removal of paternal DNA methylation contributes to TE activation. In this study, we tested this assumption by evaluating the role of Tet3-mediated 5mC oxidation in TE activation. Our results indicate that Tet3-mediated 5mC oxidation is not required for TE activation or general transcription in zygotes.
Results and Discussion
TE expression from both paternal and maternal alleles in 1-cell embryos
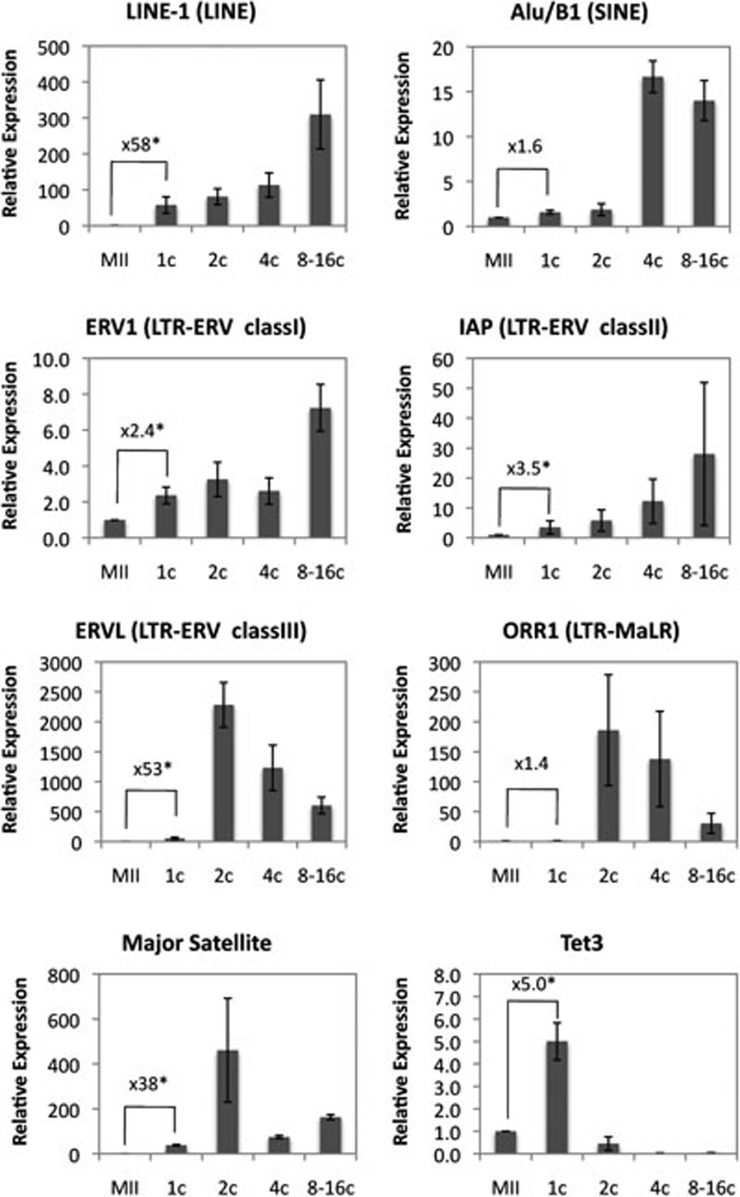
To determine which type of TEs is dynamically regulated during preimplantation development, we analyzed the expression dynamics of six different types of TEs during preimplantation development including the late 1-cell stage at 11 h post-insemination (hpi) when oxidation of 5mC in the paternal DNA is complete12. We profiled open reading frame 1 of LINE-1 (LINE family, Tf subfamily), Alu/B1 (SINE family), ERV1 (LTR retrotransposon, class I family), IAP (LTR retrotransposon, class II family), ERVL (LTR retrotransposon, class III family), and ORR1 (LTR retrotransposon, MaLR family)15 by quantitative reverse transcriptase PCR (RT-qPCR). Results shown in Figure 1 demonstrated that the expressions of TEs are dynamically regulated during preimplantation development, consistent with a previous semi-quantitative study22. Compared with MII-stage oocytes, LINE-1 and ERVL are activated more than 50-fold at the 1-cell stage embryos when oxidation of 5mC to 5hmC/5fC/5caC is complete. In contrast, ERV1 and IAP only showed a modest activation (2.4- and 3.5-fold in ERV1 and IAP, respectively), while Alu/B1 and ORR1 exhibited very little activation (1.6- and 1.4-fold, respectively). RT-qPCR analysis confirmed that the expression dynamics of major satellite DNA and Tet3 previously shown to be unregulated at the 1-cell stage are consistent with previous results10,29. This result indicates that some TEs are activated in the 1-cell embryos when global transcription is also initiated27,28.
Figure 1.
Expression dynamics of TEs during preimplantation development. Shown are RT-qPCR results of the relative expression levels of the various TEs at different preimplantation developmental stages. The results are normalized to an external control and the values of MII-stage oocytes were set as 1. The relative value of transcripts in 1-cell-stage embryos compared with that of MII oocytes was indicated in each graph. Asterisk represents statistical significance (P < 0.01). The experiments were repeated for 3-4 times. A total of 30-55 embryos were used for each experiment in a given developmental stage. Error bars represent standard deviations.
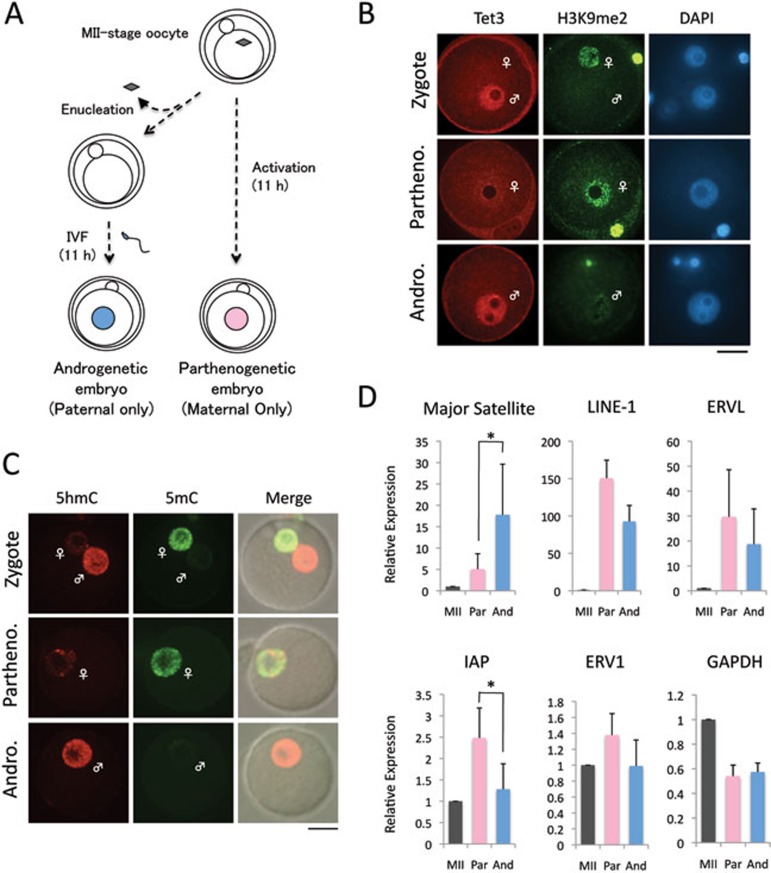
The asymmetric nature of 5mC oxidation in zygotes prompted us to ask whether activation of the TEs in zygotes exhibits allele specificity. To this end, we prepared parthenogenetic and androgenetic 1-cell embryos that contain either maternal genome or paternal genome, respectively (Figure 2A). We confirmed that the asymmetric distribution of both 5mC/5hmC and Tet3 in the parental pronuclei of normal zygotes is not altered in the parthenogenetic and androgenetic embryos (Figure 2B, 2C). Consistent with previous reports demonstrating that the major satellite DNA is mainly expressed from the paternal genome in zygotes29,30, RT-qPCR analysis demonstrated that its expression is at a much higher level in androgenetic embryos than in parthenogenetic embryos (Figure 2D), verifying the reliability of our experimental systems. We then compared the expression of four types of TEs that exhibited significant activation at the 1-cell stage (Figure 1) and found that their expression exhibited no allele specificity with the exception of IAP, whose expression is at a higher level in parthenogenetic embryos, suggesting that transcripts of IAP are mainly derived from the maternal allele at the 1-cell stage (Figure 2D). These results demonstrate that activation of LINE-1 and ERVL takes place in both alleles in zygotes.
Figure 2.
Expression of TEs in parthenogenetic and androgenetic 1-cell embryos. (A) Experimental scheme for generating parthenogenetic and androgenetic 1-cell embryos. Androgenetic embryos were produced by in vitro fertilization of enucleated MII-stage oocytes. Parthenogenetic embryos were generated by artificial activation with strontium. Embryos were collected 11 h after insemination or activation. (B) Representative images of zygotes, parthenogenetic (Partheno) and androgenetic (Andro) embryos co-stained with Tet3 (red) and H3K9me2 (green) antibodies. H3K9me2 marks the maternal pronucleus. Scale bar = 20 μm. (C) Representative images of zygotes co-stained with 5hmC (red) and 5mC (green) antibodies. More than six embryos were examined in each group. (D) RT-qPCR analysis of the expression of TEs in parthenogenetic (Partheno) and androgenetic (Andro) 1-cell embryos. The relative expression level was normalized to an external control. The values of MII-stage oocytes were set as 1. The experiments were repeated for 3 times. Error bars indicate standard deviations. * represents statistical significance (P < 0.05).
Tet3-mediated 5mC oxidation is dispensable for the activation of TEs
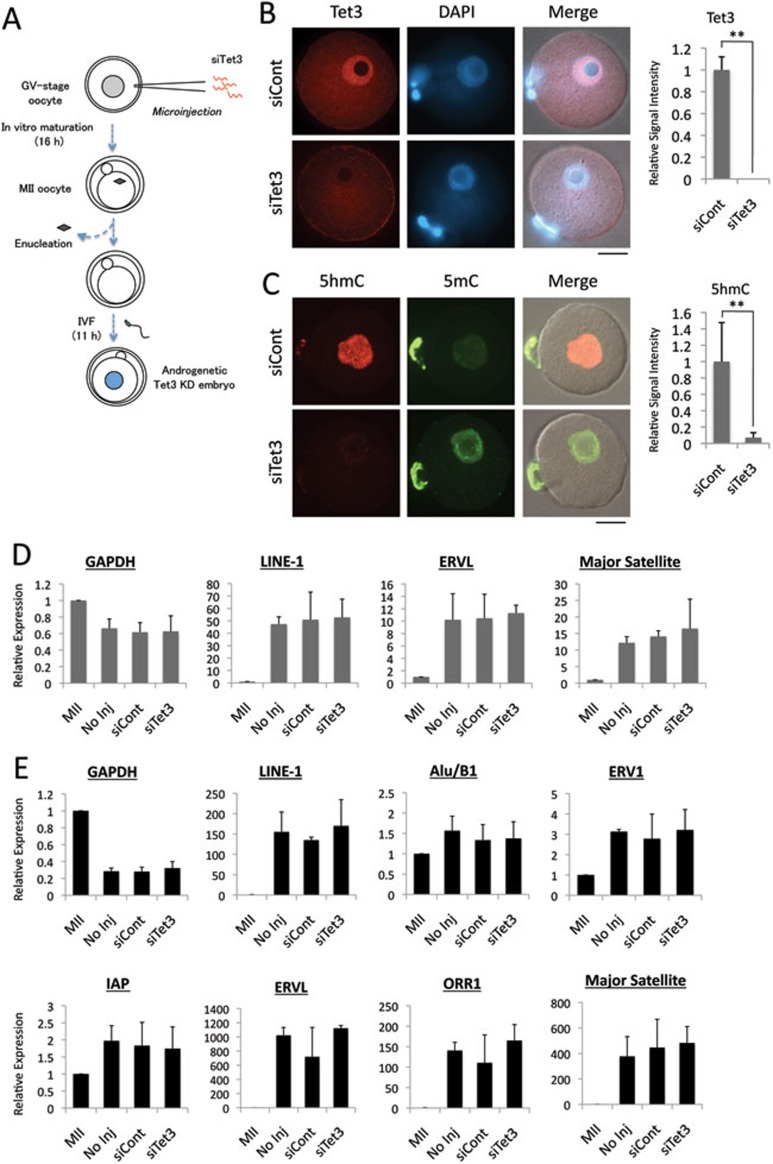
Next we examined whether activation of TEs from the paternal genome in 1-cell embryos requires Tet3-mediated 5mC oxidation. To this end, we first attempted to deplete Tet3 by injecting siRNA specific for Tet3 (siTet3) into MII-stage oocytes before fertilization. Due to the large maternal pool of Tet3 protein and the limited time window for siRNA to become functional, the depletion was not very efficient (data not shown). We then attempted to deplete Tet3 by injecting siTet3 into GV-stage oocytes (Figure 3A). Following in vitro maturation, the maternal genome was removed from the MII-stage oocytes before in vitro fertilization to generate androgenetic embryos, which enables us to evaluate TE expression from the paternal allele only. Immunostaining with a Tet3-specific antibody demonstrated that the endogenous Tet3 protein was significantly depleted in siTet3-injected embryos (Figure 3B). As expected, immunostaining revealed that the oxidation of 5mC to 5hmC is inhibited by Tet3 depletion (Figure 3C). Surprisingly, RT-qPCR analysis revealed that the expression of LINE-1, ERVL, and major satellite DNA is not altered by the depletion of Tet3 (Figure 3D). Given that the burst of transcription of some TEs including ERVL and ORR1 occurs at the 2-cell stage (Figure 1), we extended the RT-qPCR analysis to 2-cell androgenetic embryos. Results shown in Figure 3E demonstrated that neither TE nor major satellite expression is significantly affected by Tet3 depletion. Similar results were obtained when Tet3-depleted normal 1-cell embryos that contained both parental genomes were used for this analysis (data not shown). Together, these data indicate that Tet3-mediated 5mC oxidation is not required for the activation of TEs from the paternal genome in early preimplantation embryos.
Figure 3.
Activation of TEs in androgenetic embryos is independent of Tet3. (A) Scheme for generating Tet3-depleted androgenetic embryos. GV-stage oocytes were injected with siRNA specific for Tet3 (siTet3) or control siRNA (siCont). After meiotic maturation, the maternal genome was removed. The enucleated oocytes were then fertilized in vitro and collected at 11 hpi. (B) Representative images of androgenetic 1-cell embryos co-stained with Tet3 antibody (red) or DAPI (blue). Quantification of Tet3 signal intensity is shown in the right panel. The value of siCont embryos was set as 1. Five embryos in each group were quantified. Error bar indicates standard deviation. ** represents statistical significance (P <0.01). Scale bar = 20 μm. (C) Representative images of androgenetic 1-cell embryos co-stained with 5mC (green) and 5hmC (red) antibodies. Quantification of 5hmC signal intensity is shown in the right panel. The value of siCont embryos was set as 1. Six to nine embryos were quantified. Error bar indicates standard deviation. ** represents statistical significance (P <0.01). (D, E) RT-qPCR analysis of the expression of TEs in androgenetic 1-cell (D) or 2-cell (E) embryos. The relative expression level is normalized to an external control. The expression level in MII-stage oocytes was set as 1. The experiments were repeated for 3 times. Error bars represent standard deviations.
Tet3-mediated 5mC oxidation is dispensable for zygotic gene activation
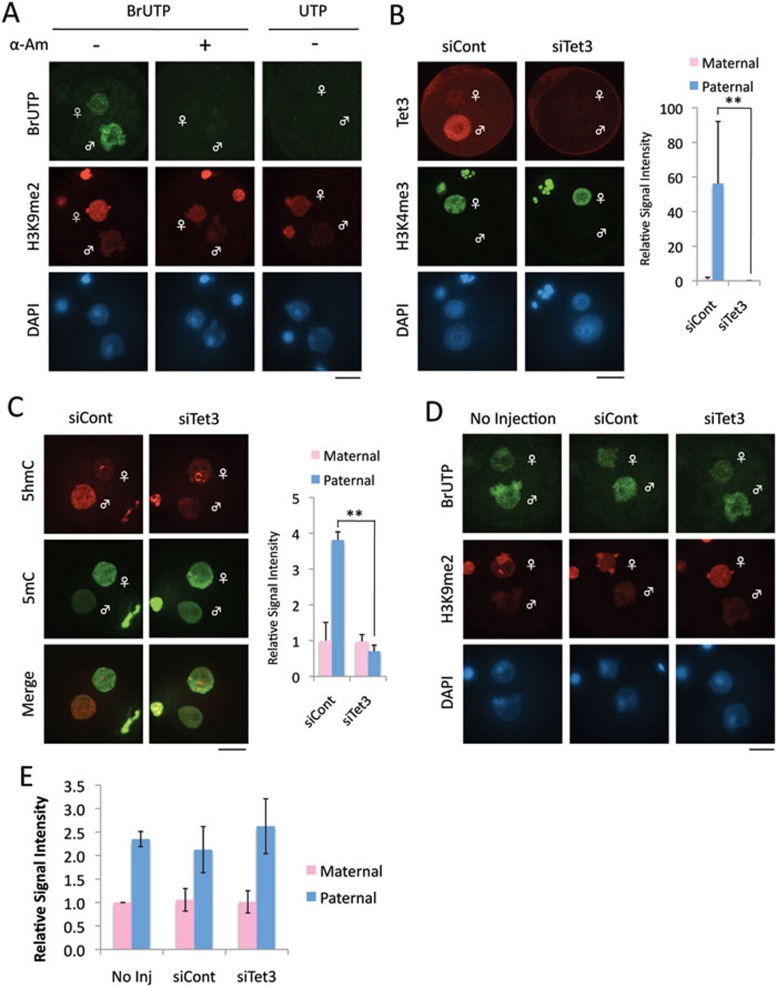
Zygotic gene activation, the earliest expression from zygote genome after fertilization, begins at the middle of 1-cell stage27,28. To test whether Tet3-mediated 5mC oxidation is involved in zygotic gene activation, we analyzed de novo RNA synthesis by measuring incorporation of 5-bromouridine-5′-triphosphate (BrUTP). We first validated the assay by demonstrating that α-amanitin treatment completely abolished the incorporation of BrUTP (Figure 4A). We also validated the BrUTP antibody specificity by substituting BrUTP with UTP (Figure 4A). Consistent with previous reports27,28, global transcriptional activity of the paternal genome, which is marked by lack of H3K9me2 staining, is significantly higher than that of the maternal genome (Figure 4A).
Figure 4.
Global transcriptional activity in zygotes is not significantly altered by Tet3 depletion. (A) Validation of BrUTP incorporation assay for measuring ongoing transcriptional activity. Representative images of zygotes co-stained with BrdU (green) and H3K9me2 (red) antibodies. DNA was stained with DAPI (blue). H3K9me2 marks the maternal pronucleus. Note that the BrUTP signal is lost in zygotes treated with α-amanitin (α-Am). Scale bar = 20 μm. (B) Representative images of zygotes co-stained with Tet3 (red) and H3K4me3 (green) antibodies. H3K4me3 marks the maternal pronucleus. Quantification of Tet3 signal intensity is shown in the right panel. The signal intensity of maternal pronucleus in siCont zygotes was set as 1. Six to nine zygotes were quantified. Error bar indicates standard deviation. ** represents statistical significance (P <0.01). (C) Representative images of zygotes co-stained with 5hmC (red) and 5mC (green) antibodies. Quantification of 5hmC signal intensity is shown in the right panel. The signal intensity of maternal pronucleus in siCont zygotes was set as 1. Fifteen to 19 zygotes were quantified. Error bar indicates standard deviation. ** represents statistical significance (P <0.01). (D) Representative images of zygotes co-stained with BrdU (green) and H3K9me2 (red) antibodies after incorporation of BrUTP. (E) Quantification of fluorescent signal intensity for BrUTP incorporation assay. The signal intensity of BrdU in maternal or paternal pronucleus was individually quantified. The signal intensity of the maternal pronucleus in non-injected zygotes was set as 1. The experiments were repeated for four times. Thirty-four to 45 zygotes were quantified in each experimental group. Error bars represent standard errors.
To deplete Tet3 protein in zygotes, GV-stage oocytes were injected with siTet3. After in vitro maturation, they were fertilized and fixed at 11 hpi. Immunostaining with Tet3 antibody demonstrated that Tet3 protein is depleted in siTet3-injected zygotes (Figure 4B). As expected, loss of 5mC and the generation of 5hmC were prevented in the paternal pronucleus of siTet3-injected zygotes (Figure 4C). Of note, 5hmC level in the paternal genome is reduced to a level similar to that in the maternal genome in Tet3-depleted zygotes, indicating effective inhibition of 5mC oxidation. BrUTP incorporation assay revealed no significant change in BrUTP incorporation upon Tet3 depletion (Figure 4D, 4E), indicating that Tet3-mediated 5mC oxidation is not required for zygotic gene activation. Given that only a limited number of genes are activated in zygotes31, this result is consistent with the notion that Tet3 is dispensable for TE activation as TEs account for a large proportion of the genome.
It is well known that one of the major functions of DNA methylation is to maintain genome integrity through suppressing the expression of TEs14,32. Given that Tet3-mediated 5mC oxidation is part of the DNA demethylation mechanism8, the results described above are surprising. Then, how is TE expression activated in early preimplantation embryos? Several studies have suggested that endogenous siRNAs against TEs play an important role in TE silencing33,34,35,36. Despite that this endogenous siRNA silencing system is functional in zygotes37, certain classes of TEs somehow find a way to escape this silencing system for their activation in zygotes.
It is possible that global chromatin remodeling in zygotes may be responsible for TE activation. Zygotic chromatin is subjected to global remodeling. For example, histone N-terminal tails are hyperacetylated in both paternal and maternal chromatins after fertilization38. In addition, asymmetric distribution of H3K36me3 and H3K79me2/3 are lost at the 1-cell stage, leading to global hypomethylation of these residues on both chromatins39,40. Furthermore, global replacement of histone variants, such as deposition of H3.3 to paternal chromatin as well as removal of H2A.Z and macroH2A from maternal chromatin41,42, takes place soon after fertilization. These chromatin remodeling events might contribute to the derepression of TEs in zygotes. Indeed, a recent study has demonstrated that TEs can be derepressed without the loss of DNA methylation or histone methylation in MORC ATPases mutant in Arabidopsis and C. elegans43. In this mutant, reactivation of TEs is associated with chromatin decondensation and disruption of higher-order chromatin structure43. Since changes in chromatin structure, including the reorganization of heterochromatin29,44, takes place in early preimplantation embryos, it is possible that these structural changes may contribute to TE activation.
The biological function of 5mC oxidation in the paternal genome in zygotes is currently unknown. Although Tet3-knockout embryos exhibit a delayed expression of Oct4 from the paternal genome, they have no problem going through preimplantation development and, importantly, the expression level of Oct4 in mutants is comparable to that in control embryos at the blastocyst stage9. Thus, it is unlikely that a slight delay in Oct4 expression can account for the decreased fertility of the mutants. Further studies are needed to understand the biological significance of 5mC to 5hmC/5fC/5caC conversion in zygotes.
Materials and Methods
Collection and culture of preimplantation embryos
All animal studies were performed in accordance with guidelines of the Institutional Animal Care & Use Committee at the University of North Carolina-Chapel Hill. MII-stage oocytes were collected from 3-week old superovulated BDF1 females by injecting 5 IU of PMSG (Harbor, UCLA) and hCG (Sigma-Aldrich) and transferred to HTF medium supplemented with 10 mg/mL bovine serum albumin (BSA; Sigma-Aldrich). Oocytes were inseminated with spermatozoa obtained from the caudal epididymides of adult BDF1 male mice. The spermatozoa had been activated for insemination by incubation for 2 h in the HTF medium before they were used. 6 h after insemination, the fertilized oocytes were washed and cultured in KSOM (Millipore) in a humidified atmosphere of 5% CO2/95% air at 37 °C. One-cell, 2-cell, 4-cell, 8-16 cell stage embryos were collected at 11, 30, 48, 72 hpi.
Generation of parthenogenetic and androgenetic embryos
For generation of parthenogenetic embryos, MII-stage oocytes were collected from oviducts and the cumulus cells were removed by short incubation in M2 media containing 0.3 mg/ml hyaluronidase (Millipore). After incubation in KSOM for 1 h, oocytes were treated with 10 mM Sr2+ for 1 h in Ca2+-free KSOM and then washed and cultured in KSOM. For generation of androgenetic embryos, the chromosomes of MII-stage oocytes were removed in M2 media containing 5 μg/ml cytochalasin B (CB; Sigma-Aldrich) by using a Piezo impact-driven micromanipulator (Prime Tech Ltd., Ibaraki, Japan). One hour after incubation in α-minimum essential medium (α-MEM; Life technologies, #12571-063) supplemented with 5% fetal bovine serum (FBS; Sigma-Aldrich) and 10 ng/ml epidermal growth factor (EGF; Sigma-Aldrich), the enucleated oocytes were inseminated in HTF medium. 6 h later, they were transferred into KSOM. Embryos that exhibit one pronucleus were used for experiments.
siRNA injection into GV-stage oocytes
Fully grown GV-stage oocytes were obtained from 8- to 12-week-old BDF1 mice 44-48 h after injection with 7.5 I.U PMSG. The ovaries were removed from the mice and transferred to M2 media (Millipore) containing 0.2 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich). The ovarian follicles were punctured with a 27-gauge needle, and the cumulus cells were gently removed from the cumulus-oocyte complexes using a narrow-bore glass pipette. After 1 h of incubation in α-MEM containing 0.2 mM IBMX, 2 μM of ON-TARGETplus siRNA specific for Tet3 (Dharmacon; the sequence was 5′-AGGCCAAGCUCUACGGGAA-3′) or ON-TARGETplus non-targeting control siRNA #1 (Dharmacon) was injected into oocytes using a Piezo impact-driven micromanipulator. 2 h after injection, the oocytes were washed in IBMX-free α-MEM medium supplemented with 5% FBS and 10 ng/ml EGF and incubated for 16-18 h to complete meiotic maturation.
Reverse transcription and real-time PCR analysis
Total RNA was purified using Trizol (Life Technologies) according to manufacturer's instruction. As an external control, an equal amount (10 pg) of Spike RNA (Dharmacon) was added into each tube that contained equal number of oocytes/embryos before RNA extraction to normalize RNA extraction and RT efficiencies. To remove genomic DNA from purified RNA samples, the samples were treated with Turbo DNase (Life Technologies) at 37 °C for 1 h. cDNA was generated with random primers (Life Technologies) and Superscript III First Strand synthesis kit (Life Technologies). Real-time quantitative PCR reactions were performed on a CFX384 real-time PCR detection system (BioRad) using SYBR Green (Applied Biosystems). Relative gene expression levels were analyzed using comparative Ct methods, where Ct is the cycle threshold number, and normalized to an external control. No contamination of the genomic DNA was verified by comparing the sample with or without reverse transcription. RT-qPCR primers are as follows: Dap (External Control)-F, 5′-CCAGACCGCGGCCTAATAATG-3′ Dap-R, 5′-CGCTTCTTCCACCAGTGCAG-3′ Tet3-F, 5′-CCGGATTGAGAAGGTCATCTAC-3′ Tet3-R, 5′-AAGATAACAATCACGGCGTTCT-3′ Gapdh-F, 5′-CATGGCCTTCCGTGTTCCTA-3′ Gapdh-R, 5′-GCCTGCTTACCACCTTCTT-3′ LINE-1 (accession number D84391)-F, 5′-GGACCAGAAAAGAAATTCCTCCCG-3′ LINE-1-R, 5′-CTCTTCTGGCTTTCATAGTCTCTGG-3′ Alu/B1-F, 5′-CGCCTTTAATCCCAGCACTC-3′ Alu/B1-R, 5′-CTGTCCTGGAACTCACTCTG-3′ ERV1-F, 5′-GCCTTTGTTGCCCAGGTAAGTCAG-3′ ERV1-R, 5′-CTCTCTACCTGTCCTGAGCTTTGAGG-3′ IAP-F, 5′-GGCTTAGTAGTCCACCCTGGAG-3′ IAP-R, 5′-CAGCAGCTGAGCTATCCTATCTCC-3′ ERVL-F, 5′-CTCTACCACTTGGACCATATGAC-3′ ERVL-R, 5′-GAGGCTCCAAACAGCATCTCTA-3′ ORR1-F, 5′-AGCCCTTAGGAGGATTCCAA-3′ ORR1-R, 5′-TGGTTCCACTCCCTGTTAGC-3′, Major satellite F, 5′-GATTTCGTCATTTTTCAAGTCGTC-3′ Major Satellite R, 5′-GCACACTGAAGGACCTGGAATATG-3′.
Whole mount immunostaining
Embryos were fixed in 3.7% paraformaldehyde (PFA) in PBS for 20 min, washed with PBS containing 10 mg/mL BSA (PBS/BSA), permeabilized with 0.5% Triton X-100 for 15 min. After blocking in PBS/BSA overnight, anti-Tet3 rabbit polyclonal (1/4 000, a gift from GL Xu) and anti-H3K4me3 mouse monoclonal (1/500, Millipore #05-1339) or anti-H3K9me2 mouse monoclonal antibodies (1/100, Millipore #05-1249) were incubated for 2 h at room temperature. When zygotes were stained with 5mC and 5hmC antibodies, the DNA was denatured with 4M HCl for 10 min and neutralized with 100 mM Tris-HCl (pH 8.5) for 20 min, and the zygotes were then incubated with PBS/BSA overnight. Anti-5mC (1/200: BI-MECY-0500, Eurogentec) and anti-5hmC (1/500: Active Motif) antibodies diluted in PBS/BSA were treated for 1 h at room temperature. After washing with PBS/BSA for 1 h, the cells were incubated with a 1:250 dilution of fluorescein isothiocyanate-conjugated anti-mouse IgG (Jackson Immuno-Research, West Grove, PA) and Alexa Flour 546 donkey anti-rabbit IgG (Life technologies) for 1 h. The oocytes were then mounted on a glass slide in Vectashield anti-bleaching solution with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Fluorescence was detected under a laser-scanning confocal microscope with a spinning disk (CSU-10, Yokogawa) and an EM-CCD camera (ImagEM, Hamamatsu). All images were acquired and analyzed using Axiovision software (Carl Zeiss). The fluorescent signal intensity was quantified with Axiovision.
BrUTP incorporation assay
BrUTP incorporation assay was performed as described previously27. Briefly, zygotes at 10 hpi were treated with a physiological buffer (PB) containing 0.05% Triton X-100 for 1-2 min at room temperature to permeabilize the ooplasm. After wash with PB, the zygotes were transferred to PB containing ATP, GTP, CTP and BrUTP (Sigma-Aldrich) and incubated for 15 min at 33 °C. For a negative control, BrUTP was replaced with UTP. Subsequently, the reaction was terminated by permeabilizing the nuclear membrane in PB containing 0.2% Triton X-100 for 3 min. The zygotes were fixed in 3.7% PFA/PB for 20 min. After washing, they were subjected to immunocytochemistry with anti-BrdU mouse monoclonal antibody (1/50, Roche Diagnostic) that crossreacts with BrU, and anti-H3K9me2 rabbit polyclonal antibody (1/200, Millipore #07-441). When zygotes were treated with α-amanitin, they were cultured in KSOM containing 0.1 mg/ml α-amanitin (Sigma-Aldrich) from 2 hpi.
Statistical analysis
Data were analyzed by student's t-test. A value of P <0.05 was considered statistically significant unless otherwise noted.
Acknowledgments
We thank Dr Guoliang Xu for a Tet3 antibody. We are grateful to Dr Shinpei Yamaguchi for reading the manuscript. This work was supported by HHMI and NIH (U01DK089565). AI is a research fellow for Research Abroad of the Japan Society for the Promotion of Science. YZ is an investigator of the HHMI.
References
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Bourc'his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol. 2004;69:381–387. doi: 10.1101/sqb.2004.69.381. [DOI] [PubMed] [Google Scholar]
- Mandal PK, Kazazian HH. SnapShot: Vertebrate transposons. Cell. 2008;135:192–192.e1. doi: 10.1016/j.cell.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- Thayer RE, Singer MF, Fanning TG. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133:273–277. doi: 10.1016/0378-1119(93)90651-i. [DOI] [PubMed] [Google Scholar]
- Huang J, Fan T, Yan Q, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic acids research. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev. 2006;73:279–287. doi: 10.1002/mrd.20423. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Arand J, Sebastiano V, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer HS, Rossi JJ. Small interfering RNAs to the rescue: blocking L1 retrotransposition. Nat Struct Mol Biol. 2006;13:758–759. doi: 10.1038/nsmb0906-758. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HH. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, et al. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010;38:5141–5151. doi: 10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Boskovic A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla ME. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics. 2012;7:747–757. doi: 10.4161/epi.20584. [DOI] [PubMed] [Google Scholar]
- Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod. 2008;78:413–424. doi: 10.1095/biolreprod.107.063453. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7:e 1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137:3785–3794. doi: 10.1242/dev.051805. [DOI] [PubMed] [Google Scholar]
- Moissiard G, Cokus SJ, Cary J, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Santos F, Reik W, Almouzni G, Dean W. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma. 2007;116:403–415. doi: 10.1007/s00412-007-0106-8. [DOI] [PubMed] [Google Scholar]
- Woodcock D, Lawler C, Linsenmeyer M, Doherty J, Warren W. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272:7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]