Abstract
Plant responses to developmental and environmental cues are often mediated by calcium (Ca2+) signals that are transmitted by diverse calcium sensors. The calcineurin B-like (CBL) protein family represents calcium sensors that decode calcium signals through specific interactions with a group of CBL-interacting protein kinases. We report functional analysis of Arabidopsis CBL2 and CBL3, two closely related CBL members that are localized to the vacuolar membrane through the N-terminal tonoplast-targeting sequence. While cbl2 or cbl3 single mutant did not show any phenotypic difference from the wild type, the cbl2 cbl3 double mutant was stunted with leaf tip necrosis, underdeveloped roots, shorter siliques and fewer seeds. These defects were reminiscent of those in the vha-a2 vha-a3 double mutant deficient in vacuolar H+-ATPase (V-ATPase). Indeed, the V-ATPase activity was reduced in the cbl2 cbl3 double mutant, connecting tonoplast CBL-type calcium sensors to the regulation of V-ATPase. Furthermore, cbl2 cbl3 double mutant was compromised in ionic tolerance and micronutrient accumulation, consistent with the defect in V-ATPase activity that has been shown to function in ion compartmentalization. Our results suggest that calcium sensors CBL2 and CBL3 serve as molecular links between calcium signaling and V-ATPase, a central regulator of intracellular ion homeostasis.
Keywords: calcium sensor, tonoplast, V-ATPase, ion homeostasis, V-ATPase
Introduction
In order to coordinate the developmental programs as well as to adapt to the environmental changes, plants have evolved sophisticated machineries for signal transduction linking perception and overall responses. The Ca2+-dependent signaling pathways participate in many aspects of plant development and environmental responses1,2,3. It is generally believed that stimulus-specific calcium signals are encoded by temporally and spatially defined patterns of Ca2+ fluctuations that result from concerted action of Ca2+ transporters4,5. To decode these cellular “Ca2+ signatures”, plant cells are equipped with a variety of calcium sensor proteins, which undergo conformational changes upon Ca2+ binding and regulate their target proteins that further orchestrate downstream responses2,6,7,8,9.
The calcineurin B-like (CBL) family proteins are related to the regulatory B subunit of calcineurin and neuronal calcium sensor proteins from animals and yeast6,10,11. These non-catalytic Ca2+ sensors play crucial roles in Ca2+-dependent processes through physically interacting with and activating a group of protein kinases designated as CBL-interacting protein kinases (CIPKs)12,13,14. In Arabidopsis, several CBLs coupled with their target CIPKs have been demonstrated to function in response to ionic stress conditions including high sodium (Na+) and low potassium (K+)15. Through forward genetic screens, the CBL Ca2+ sensor SOS3 (for Salt Overly Sensitive 3)/CBL4 and CIPK-type kinase SOS2/CIPK24 have been identified as key components in a Ca2+-signaling pathway that specifically mediates salt stress adaptation by regulating the Na+/H+ antiporter SOS1 at the plasma membrane11,16,17,18. Further studies indicated that another CBL member, CBL10, also interacts with CIPK24/SOS2 to protect Arabidopsis shoots from salt stress19,20. In response to low-K+ status, CIPK23 is activated and recruited to the plasma membrane by Ca2+ sensors CBL1 and CBL9, and the CBL-CIPK23 complexes activate the shaker-like K+ channel AKT1 for optimal K+ uptake under limiting K+ supply conditions21,22,23. Apart from controlling K+ transport in roots, the CBL1-CIPK23 and CBL9-CIPK23 complexes also regulate the dual-affinity nitrate (NO3−) transporter CHL1 by phosphorylation that serves as a molecular switch between low- and high-affinity NO3− transport modes24. In addition to their central regulation of ion fluxes in plant cells, CBLs play other roles in various abiotic stress adaptations. For instance, CBL1 integrates plant responses to cold, drought, salinity and hyperosmotic stresses25,26 and CBL9 is involved in abscisic acid (ABA) signaling and biosynthesis during seed germination27.
Distinct subcellular localizations of CBL proteins facilitate the localization of their targets including CIPKs, and thereby determine the functional specificity of each CBL28,29. While CBL1 and CBL9 harbor motifs for N-myristoylation and S-acylation that anchor them to the plasma membrane and target their function towards regulation of transporters21,22,23,30, several other CBL members, including CBL2, CBL3, CBL6 and CBL10, contain variable N-terminal extensions that target the proteins specifically to the vacuolar membranes20,28. As discussed above, substantial progress has been made in characterizing plasma membrane-targeted CBL proteins such as CBL1, CBL4 and CBL9. However, little is known about the function of tonoplast-associated CBLs despite the tempting hypothesis that these CBLs may play a central role in the regulation of intracellular ion transport across the vacuolar membrane. In this study, we investigated the function of CBL2 and CBL3 in Arabidopsis and found that the tonoplast-localized CBL2 and CBL3 are critical for plant development and ion homeostasis in plant cells, at least in part through regulation of vacuolar H+-ATPase activity in plants. In the context of previous studies on plasma membrane CBLs, this study on tonoplast CBLs thus bridges a critical gap in our understanding of CBL-CIPK network in regulating ion homeostasis in plant cells.
Results
CBL2 and CBL3 exhibit overlapping but distinct expression patterns in Arabidopsis
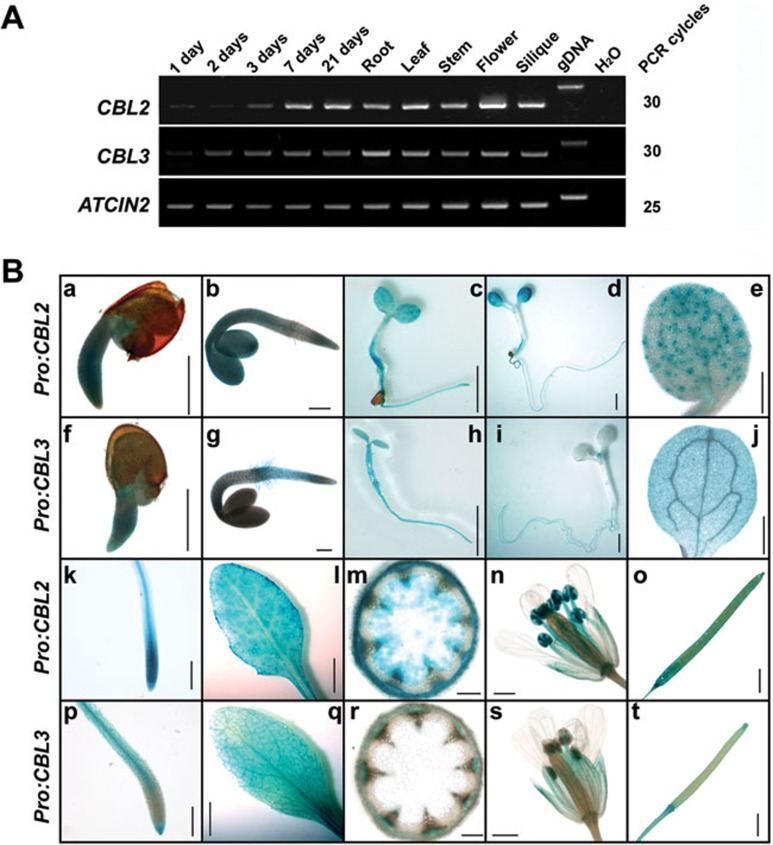
CBL2 and CBL3 share 92% identity in amino acid sequence and their genes may originate from a gene duplication event10,31. The function of these two highly similar genes may be highly redundant unless their expression pattern is distinct. As a step to dissect the functions of these two genes, we performed reverse transcription PCR (RT-PCR) analysis to determine their relative transcript abundance at different developmental stages and in various organs of Arabidopsis. The results showed that transcript levels of CBL2 and CBL3 were detectable early during germination and increased to a peak level when seedlings were established (approximately 7 days after germination). In mature plants, both CBL2 and CBL3 were ubiquitously expressed in roots, leaves, inflorescence stems, flowers and siliques, although the overall expression level of CBL2 was slightly higher than that of CBL3 (Figure 1A).
Figure 1.
Expression patterns of CBL2 and CBL3 in Arabidopsis. (A) RT-PCR analysis of CBL2 and CBL3 transcripts during seedling emergence and in different organs of Arabidopsis plants. Total RNA was isolated from germinating seeds and young seedlings (1 day, 2 days, 3 days, 7 days, 21 days after sowing) or from various tissues (root, leaf, stem, flower, silique) of wild-type Col-0 plants. RT-PCR was performed with CBL2/3-specific primers or ACTIN2-specific primers at indicated numbers of PCR cycles. Genomic DNA was used as a control template to rule out its possible contamination in each cDNA sample. PCR products were visualized by agarose gel electrophoresis followed by ethidium bromide staining. (B) CBL2 and CBL3 promoter-GUS expression in transgenic Arabidopsis plants. Histochemical GUS staining was carried out at different germinating stages and in various tissues of CBL2 (the first and third rows) or CBL3(the second and fourth rows) promoter-GUS transgenic plants. (a) and (f) one-day old germinating seed, scale bar = 0.5 mm; (b) and (g) two-day old germinating seed, scale bar = 0.5 mm; (c) and (h) three-day old seedling, scale bar = 2 mm; (d) and (i) seven-day old seedling, scale bar = 2 mm; (e) and (j) cotyledon from a five-day old seeding, scale bar = 1 mm; (k) and (p) young root, scale bar = 1 mm; (l) and (q) rosette leaf, scale bar = 5 mm; (m) and (r) section of an inflorescence stem, scale bar = 0.05 mm; (n) and (s) flower, scale bar = 0.5 mm; (o) and (t) silique, scale bar = 0.5 mm.
To analyze the expression pattern of CBL2 and CBL3 in more detail, we examined the histochemical activity of β-glucuronidase (GUS) in transgenic Arabidopsis plants expressing a CBL2 or CBL3 promoter-driven GUS reporter transgene. Such analysis revealed rather distinct patterns of expression between CBL2 and CBL3 genes. During early seedling development, expression of ProCBL2:GUS was detected in all tissues with stronger expression in aerial parts, whereas ProCBL3:GUS expression was more evident in roots (Figure 1B, a-d and f-i). In cotyledons, strong GUS staining was observed in guard cells of transgenic plants harboring ProCBL2:GUS but not in those expressing ProCBL3:GUS (Figure 1B, e, j). Moreover, ProCBL2:GUS expression was predominantly detected in the meristem and elongation zones of roots, the mesophyll cells of rosette leaves, the cortex and pith of inflorescence stems as well as anthers and stamen filaments of flowers (Figure 1B, k-n). By contrast, expression of ProCBL3:GUS was detected mainly in the root tip and root hair zone, leaf veins, vascular bundles and the vasculature of sepals (Figure 1B, p-s). Both ProCBL2:GUS and ProCBL3:GUS were also expressed in siliques, but the expression of ProCBL3:GUS was weaker and mainly restricted to the stigma and receptacle of developing siliques (Figure 1B, o and t).
Further quantitative real-time RT-PCR (qRT-PCR) analyses indicated that the expression of CBL2 or CBL3 was not obviously altered under high salt, hyperosmotic stress, or ABA treatment, but marginally induced by dehydration and low temperature (Supplementary information, Figure S1). In conclusion, CBL2 and CBL3 genes are broadly expressed in plant tissues but display both overlapping and differential expression patterns in various cell types of Arabidopsis.
CBL2 and CBL3 are anchored specifically to the vacuolar membrane via the N-terminal sequence
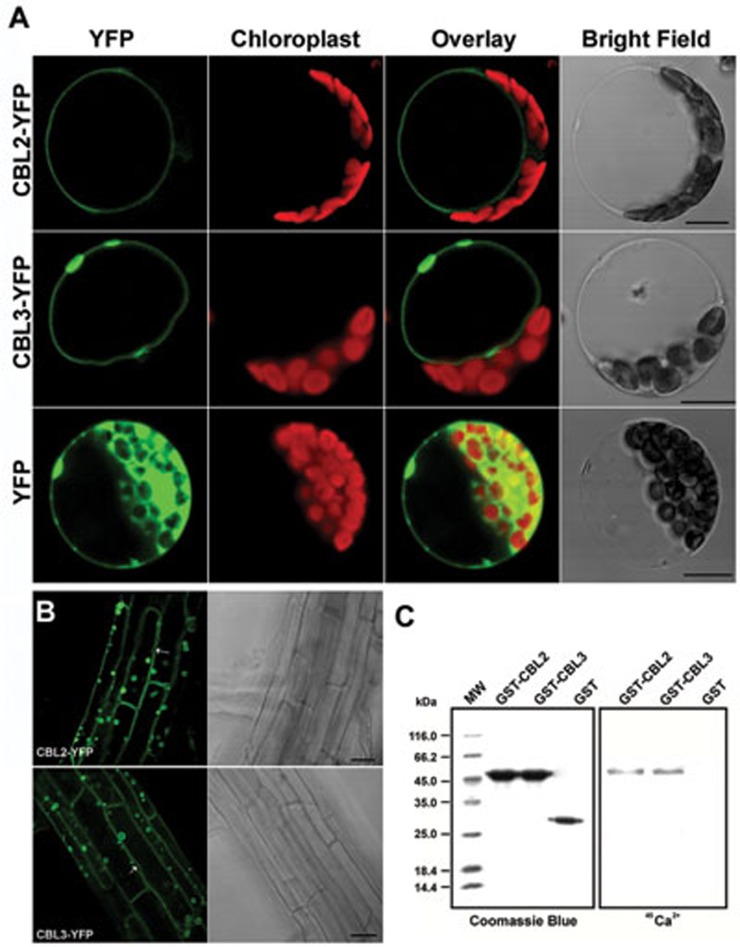
A recent study showed that Arabidopsis CBL2 and CBL3 proteins were targeted to the tonoplast by transient expression of GFP fusions in the leaf epidermal cells of Nicotiana benthamiana28. As subcellular localization is critical for functional analysis, we decided to re-examine the subcellular localization of these two proteins in Arabidopsis cells. First we performed the transient expression assay. Yellow fluorescent protein (YFP)-tagged CBL2 or CBL3 was expressed in mesophyll protoplasts isolated from Arabidopsis rosette leaves. Confocal imaging showed that fluorescence signals of CBL2-YFP or CBL3-YFP fusion protein precisely coincided with the vacuolar membrane in Arabidopsis mesophyll cells, unlike the ubiquitous distribution of free YFP (Figure 2A). Furthermore, we generated transgenic Arabidopsis lines stably expressing the CBL2-YFP or CBL3-YFP fusion protein. In these transgenic plants, the fluorescence of CBL2-YFP or CBL3-YFP in roots was also localized to the tonoplast, as indicated by membrane invaginations surrounding the nuclei (Figure 2B). In addition, fluorescence signals of CBL2-YFP and CBL3-YFP were not only concentrated on central vacuoles but also evident in the membranes of smaller lytic or storage vacuoles scattered in plant cells (Figure 2B).
Figure 2.
Subcellular localization and Ca2+ binding of CBL2 and CBL3. (A) Confocal microscopy analysis of YFP signals from Arabidopsis mesophyll protoplasts transiently expressing CBL2-YFP and CBL3-YFP fusions or YFP alone as indicated. In each panel, the YFP signals (green), chloroplast fluorescence (red), overlay (green plus red) and bright field images from the same cell are shown. Scale bar = 10 μm. (B) CBL2/3-associated YFP signals in mature root cells of transgenic Arabidopsis plants stably expressing CBL2-YFP (top panel) or CBL3-YFP (bottom panel). Arrows point to vacuolar membrane invagination away from the plasma membrane. (C) Purified GST-CBL2 and GST-CBL3 fusion proteins as well as GST control were separated on 12.5% SDS-polyacrylamide gel by electrophoresis and stained with Coomassie Blue (left panel) or electroblotted onto a nitrocellulose membrane, incubated with 45Ca2+ and autoradiographed (right panel).
Sequence analysis revealed that the tonoplast-localized CBL2 and CBL3 proteins contain an extension of 16-amino acids (16-aa) at the N-terminus compared to CBL1 that is predominantly targeted to plasma membrane (Supplementary information, Figure S2A). Because CBL2 and CBL3 share a highly conserved N-terminal sequence, we employed the N-terminus of CBL2 as a model to map the tonoplast targeting determinant. Deletion of the N-terminal 16-aa sequence aborted the tonoplast localization of CBL2-YFP, and on the other hand, adding this short peptide to YFP did not change the cytoplasmic and nuclear distribution of free YFP (Supplementary information, Figure S2B). This observation suggests that the N-terminus 16-aa sequence is required but not sufficient for targeting of CBL2 to the tonoplast. We next fused various fragments (from 17 to 28 aa) of CBL2 N-terminus to YFP N-terminus and expressed the fusion proteins in Arabidopsis mesophyll protoplasts. The results showed that a minimal length of 19-aa N-terminal region of CBL2 is sufficient to confer tonoplast localization (Supplementary information, Figure S3), which was therefore designated as “Tonoplast-Targeting Sequence (TTS)”.
We also constructed various YFP-TTS chimeras for protoplast transfection, where the TTS was in the middle or at the C-terminus of the fusion protein, respectively. Both types of YFP-TTS fusions were associated with vacuolar membrane, similar to the N-terminal fusion of CBL2-YFP, suggesting that TTS works regardless of its location in the protein (Supplementary information, Figure S4). Interestingly, we further found that TTS was capable of directing different soluble proteins to the vacuolar membrane32 (Supplementary information, Figure S5A), but it failed to change the subcellular localization of proteins with multiple transmembrane domains (Supplementary information, Figure S5B).
CBL2 and CBL3 proteins both bind Ca2+
Although deduced amino acid sequence and structural analysis suggest that all CBL proteins including CBL2 and CBL3 contain putative EF-hand motifs that contribute to calcium binding10,33, experimental evidence for the calcium-binding property of CBL2 and CBL3 is limited. To determine whether CBL2 and CBL3 are indeed capable of binding Ca2+, we performed a 45Ca-overlay assay using the recombinant CBL2 and CBL3 proteins expressed in Escherichia coli. The GST-CBL2 or GST-CBL3 fusion protein was present as an approximately 50 kDa polypeptide in the soluble fraction and purified by glutathione sepharose affinity chromatography (Supplementary information, Figure S6). Both GST-CBL2 and GST-CBL3, but not the GST control, were able to bind Ca2+, confirming that CBL2 and CBL3 are indeed Ca2+-binding proteins that can function as calcium sensors (Figure 2C).
The cbl2 cbl3 double mutant displays developmental defects
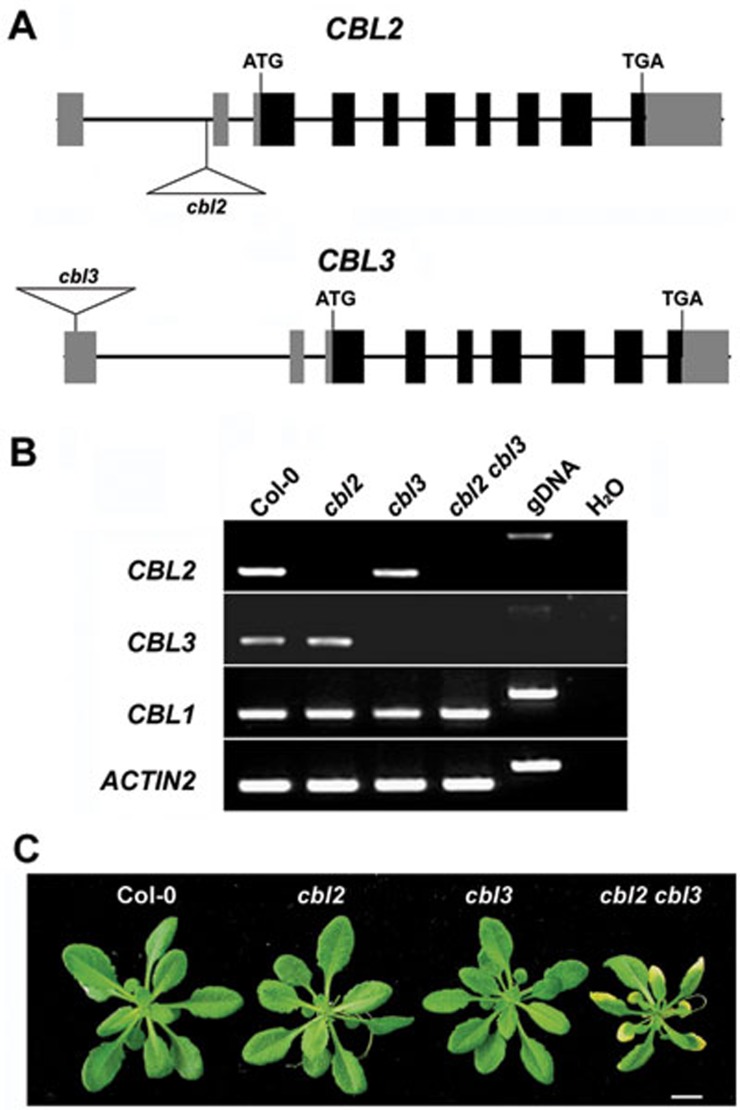
To identify the function of CBL2 and CBL3, two T-DNA insertion lines SALK_151426 (cbl2) and SAIL_785_C10 (cbl3) were characterized. The cbl2 mutant contained a T-DNA insertion in the first intron of CBL2, and the cbl3 mutant carried a T-DNA insertion in the first exon of CBL3 (Figure 3A). Homozygous cbl2 or cbl3 individuals were isolated and the cbl2 cbl3 double mutant was identified in the F2 populations derived from a cross between cbl2 and cbl3 single mutants. The cbl2 and cbl3 single mutants lacked detectable level of transcripts for CBL2 and CBL3, respectively, and cbl2 cbl3 double mutant was deficient in both transcripts. As a control, expression of CBL1 was not significantly altered in all mutants (Figure 3B).
Figure 3.
Molecular identification of the cbl2 cbl3 double mutant. (A) Schematic diagram of T-DNA insertion in cbl2 and cbl3 mutants. Exons and introns are depicted to scale by boxes and lines, respectively. The coding region of the gene is shown as black boxes while the 5′ and 3′ UTR of the cDNA is shown as light-shaded boxes. The position of the T-DNA is indicated by the triangle. (B) RT-PCR analysis of CBL2 and CBL3 gene expression in wild-type Col-0, homozygous cbl2 or cbl3 single knockout mutant and the cbl2 cbl3 double mutant. Total RNA was isolated from the rosette leaves of 40-day old plants. Expression of CBL1 and ACTIN2 was analyzed as internal controls. (C) Phenotype of the cbl2 cbl3 double mutant. Wild-type (Col-0), cbl2, cbl3 and cbl2 cbl3 Arabidopsis plants were grown in soil under 12-h light/12-h dark cycles and pictures were taken 40-day after germination. Growth of cbl2 or cbl3 is similar to that of wild-type (Col-0), whereas the cbl2 cbl3 double mutant displays smaller size and leaf tip necrosis. Bar = 10 mm.
Compared with wild-type plants (Col-0), neither of the single mutants exhibited any obvious phenotypic change during the life cycle. However, cbl2 cbl3 double mutant plants were stunted with necrosis at leaf tips (Figure 3C). In the segregation analysis of cbl2+/−/cbl3+/−, 16 out of 240 individual plants in the F2 generation were smaller in size with leaf tip necrosis (χ2 = 0.07, P = 0.79 for a 1:15 segregation ration), and all of these plants were verified to be homozygous for both cbl2 and cbl3 alleles. Further analyses of the progeny derived from a heterozygote cbl2−/−/cbl3+/− or cbl2+/−/cbl3−/− revealed a 3:1 phenotypic segregation ratio of wild-type over cbl2 cbl3, consistent with the expectation that the presence of a wild-type copy of either CBL2 or CBL3 restored wild-type phenotype. Moreover, transformation of cbl2 cbl3 with either 35S:CBL2 or 35S:CBL3 restored plant growth phenotype to the wild-type (Supplementary information, Figure S7). These genetic data demonstrate that the phenotypic changes manifested by the double mutant indeed result from disruption of both CBL2 and CBL3 genes.
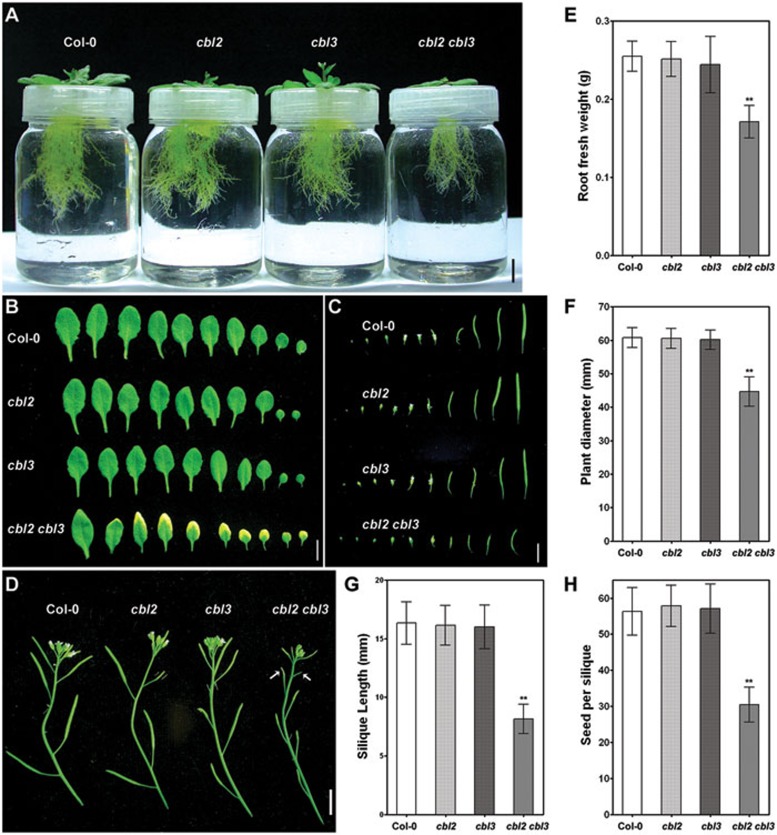
We further assessed the cbl2 cbl3 double mutant phenotype and found that simultaneous disruption in CBL2 and CBL3 had drastic effects on plant growth and development in Arabidopsis. Under hydroponic conditions, root growth of cbl2 cbl3 was severely reduced compared to that of wild-type plants or single mutants (Figure 4A), with fresh weight measured at about 60% of the wild-type roots (Figure 4E). When grown in the soil, cbl2 cbl3 double mutant had smaller rosette leaves (Figure 4B), with a diameter reduced by 25-30% in comparison to that of Col-0 or single mutants before bolting in the growth chamber (Figure 4F). Under conditions of higher light intensity, stunted growth of the double mutant was even more evident (Supplementary information, Figure S8A). Most strikingly, the cbl2 cbl3 mutant displayed obvious necrotic symptoms on the rosette leaves of various ages and on cauline leaves as well (Figure 4B and Supplementary information, Figure S8B). In the reproductive phase, the cbl2 cbl3 mutant bolted two weeks later than the wild-type (Supplementary information, Figure S8B). At harvesting time, the mutant plants displayed shorter inflorescence, smaller flowers and underdeveloped siliques (Figure 4C and 4D). The fertility and yield of the double mutant were also severely reduced, as indicated by significantly shorter siliques and fewer seeds (Figure 4G and 4H). Taken together, both vegetative growth and reproductive development were considerably impaired in the cbl2 cbl3 double knockout plants.
Figure 4.
Morphological phenotypes of the cbl2 cbl3 double mutant during different developmental stages. (A) Wild-type (Col-0), cbl2, cbl3 and cbl2 cbl3 plants were hydroponically grown in a plant growth chamber. Pictures were taken 28-day after germination to exhibit root systems of each genotype. (B) Fully expanded rosette leaves detached from 28-day old plants of wild-type (Col-0), cbl2, cbl3 and cbl2 cbl3. (C) Flower set and silique formation in wild-type (Col-0), cbl2, cbl3 and cbl2 cbl3 plants. (D) Primary inflorescence of 56-day old wild-type (Col-0), cbl2, cbl3 and cbl2 cbl3 plants. Arrows point to the abortive silique development in an early inflorescence of cbl2 cbl3 double mutant. (E) Quantification of root weight of 28-day old hydroponically grown plants. Values are mean ± SD (n = 6). (F) Quantification of plant size (average diameter of rosette) of 28-day old plants grown in soil. Values are mean ± SD (n =12). (G) Quantification of length of fully developed siliques. Values are mean ± SD (n = 20). (H) Quantification of the number of seeds per silique. Values are mean ± SD (n = 20). Scale Bars = 10 mm. Asterisks indicate statistically significant difference between wild-type Col-0 and cbl2 cbl3 double mutant (Student's t-test, **P < 0.01).
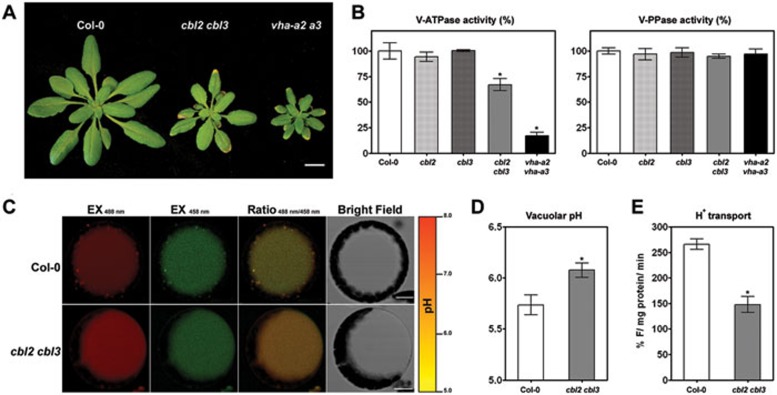
cbl2 cbl3 exhibits reduced vacuolar H+-ATPase activity and elevated vacuolar pH
The cbl2 cbl3 double mutant showed stunted growth, reduced fertility and necrotic lesions at the leaf tips. All these developmental abnormalities are reminiscent of the recently characterized vha-a2 vha-a3 double mutant lacking the tonoplast-localized membrane-integral vacuolar H+-ATPase subunit VHA-a (Figure 5A, Supplementary information, Figure S8, and34). As both CBL2 and CBL3 are tonoplast calcium sensors, we thus speculated that the phenotypes of cbl2 cbl3 could be attributed to the malfunction of vacuolar H+-ATPase (V-ATPase). To this end, we measured total V-ATPase activity as Concanamycin A and NO3−-sensitive ATP hydrolysis in microsomal membranes from rosette leaves of the wild-type, cbl2 and cbl3 single mutants, as well as cbl2 cbl3 and vha-a2 vha-a3 double mutants34,35,36. Compared to the wild-type control, similar levels of V-ATPase activity was found in both single mutants. In contrast, activity from the cbl2 cbl3 double mutant was reduced by more than 30%, whereas 85% V-ATPase activity was lost in vha-a2 vha-a3(Figure 5B). On the other hand, the activity of vacuolar H+-pyrophosphatase (V-PPase) measured as KCl-stimulated PPi hydrolysis remained unchanged in all samples (Figure 5B). These results suggest that disruption of CBL2 and CBL3 reduced tonoplast V-ATPase activity in Arabidopsis, presumably leading to growth and developmental defects that are similar to those in the vha-a2 vha-a3 mutant, specifically deficient in V-ATPase activity. These defects are milder in the cbl2 cbl3 double mutant as compared to the vha-a2 vha-a3 mutant, consistent with the observation that cbl2 cbl3 mutant plants retained higher V-ATPase activity than the latter.
Figure 5.
Reduction of tonoplast V-ATPase activity in cbl2 cbl3 double mutant results in alkalinization of vacuolar pH. (A) The cbl2 cbl3 double mutant phenocopies small size and leaf-tip necrosis of the vha2-a2 vha-a3 double mutant. Scale bar = 10 mm. (B) Vacuolar H+-ATPase (left panel) and H+-PPase (right panel) hydrolytic activity was determined from microsomal membranes of 4-week old plants of wild-type (Col-0), cbl2, cbl3, cbl2 cbl3 and vha-a2 vha-a3. Results are shown as percentage of the Col-0 control activity. Values are mean ± SE of at least three replicate experiments. (C) The images show emission intensities of mesophyll protoplast vacuoles loaded with BCECF at 488 nm (the first column, red) and 458 nm (the second column, green). The ratio images indicate an increased vacuolar pH in cbl2 cbl3 mutant than in wild-type (the third column). Pseudo-color scale on the right indicates intensity of fluorescence in which yellow and red represent minimum and maximum intensity, respectively. Scale bar = 10 μm. (D) Quantification of the luminal pH in wild-type Col-0 and cbl2 cbl3 double mutant vacuoles. Error bars represent SE of 60 measurements from 15 different intact vacuoles. (E) ATP-dependent H+ transport into purified tonoplast vesicles from Col-0 and cbl2 cbl3 plants was monitored by the quenching of quinacrine fluorescence. Results are mean ± SE of at least three replicates. Asterisks indicate statistically significant difference between wild-type Col-0 and cbl2 cbl3 mutant (Student's t-test, *P < 0.05).
Plant V-ATPase catalyzes ATP hydrolysis that is coupled to the translocation of protons (H+) into the vacuole37. To corroborate our results in an independent assay, we measured the H+-transport activity of vacuolar membrane vesicles purified from wild-type and cbl2 cbl3 microsomal membranes. The cbl2 cbl3 double mutant showed a 40% reduction in V-ATPase H+ transport activity compared to the wild-type control (Figure 5E).
Transport of H+ into vacuole contributes to acidification of vacuolar compartments and establishment of pH gradients across tonoplast38,39. To assess if the decline of V-ATPase activity in cbl2 cbl3 mutant affects vacuolar pH, we used the rationmetric fluorescent pH indicator 2′,7′-bis-(2-carboxyethyl)-5-(6)-carboxyfluorescein (BCECF) to measure vacuolar pH40. The pH values were calculated from fluorescence ratios of confocal images (Figure 5C) based on an in situ calibration curve (Supplementary information, Figure S9). In wild-type mesophyll cells, the vacuolar pH was averaged at 5.74, but that of cbl2 cbl3 mutant was shifted to 6.08 (Figure 5D). Hence a lower luminal H+ concentration in cbl2 cbl3 mutant as indicated by the higher pH further supports that CBL2 and CBL3 are required to maintain tonoplast V-ATPase activity.
cbl2 cbl3 double mutant plants are hypersensitive to excessive metal ions
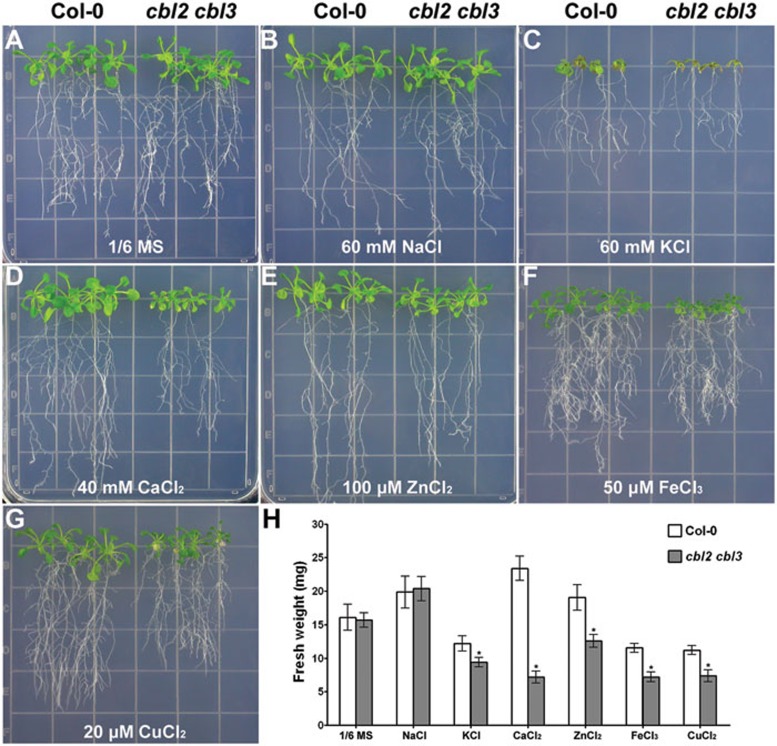
Ionic sequestration into the vacuole plays a key role in detoxification of deleterious metals from the cytoplasm. The cytosol-vacuole ion transport is often mediated by secondary transporters that are energized by the electrochemical proton gradient produced by V-ATPase and V-PPase39. In the light of reduced V-ATPase activity in cbl2 cbl3, we hypothesized that the double mutant might be compromised in its tolerance to ionic perturbations. Therefore, we compared the cbl2 cbl3 mutant and wild-type plants for their sensitivity to excessive levels of external ions. In identifying a control medium, we found that cbl2 cbl3 mutant and wild-type seedlings grew similarly on the 1/6-strength Murashige and Skoog medium (1/6 MS)41, which was subsequently used as the control medium (Figure 6A). The growth of cbl2 cbl3 seedlings were severely impaired on control medium supplemented with 40 mM Ca2+, 50 μM Fe3+ or 20 μM Cu2+(Figure 6D, 6F and 6G). A milder but still significant growth inhibition of cbl2 cbl3 plants was observed in the presence of 60 mM K+ or 100 μM Zn2+, (Figure 6C and 6E). At the end of each assay, fresh weight measurements of seedlings statistically confirmed the ion sensitivity phenotypes of cbl2 cbl3 double mutant (Figure 6H). Interestingly, despite the earlier finding that Na+ sequestration into vacuole depends on Na+/H+ antiporters42,43, cbl2 cbl3 double mutant did not show hypersensitive response to 60 mM NaCl (Figure 6B). Consistent with the results from above V-ATPase activity assays, cbl2 or cbl3 single mutants did not show any discernible difference as compared to the wild-type plants under these ionic stress conditions (Supplementary information, Figure S10).
Figure 6.
Ion sensitivity of the cbl2 cbl3 double mutant. (A-G) Growth of wild-type Col-0 and cbl2 cbl3 mutant plants under various ionic stress conditions. Five-day old Col-0 and cbl2 cbl3 seedlings were transferred onto 1/6 MS medium (A) or 1/6 MS medium supplemented with 60 mM NaCl (B), 60 mM KCl (C), 40 mM CaCl2 (D), 100 μM ZnCl2 (E), 50 μM FeCl3 (F) or 20 μM CuCl2 (G), respectively. Pictures were taken on the 21th day after transfer. (H) Measurements of seedling fresh weight of Col-0 and cbl2 cbl3 on the 21th day after transfer. Error bars represent SE of 12 independent seedlings from triplicate experiments. Asterisks indicate statistically significant difference between wild-type Col-0 and cbl2 cbl3 double mutant (Student's t-test, *P < 0.05).
cbl2 cbl3 mutant displays an altered ionic profile
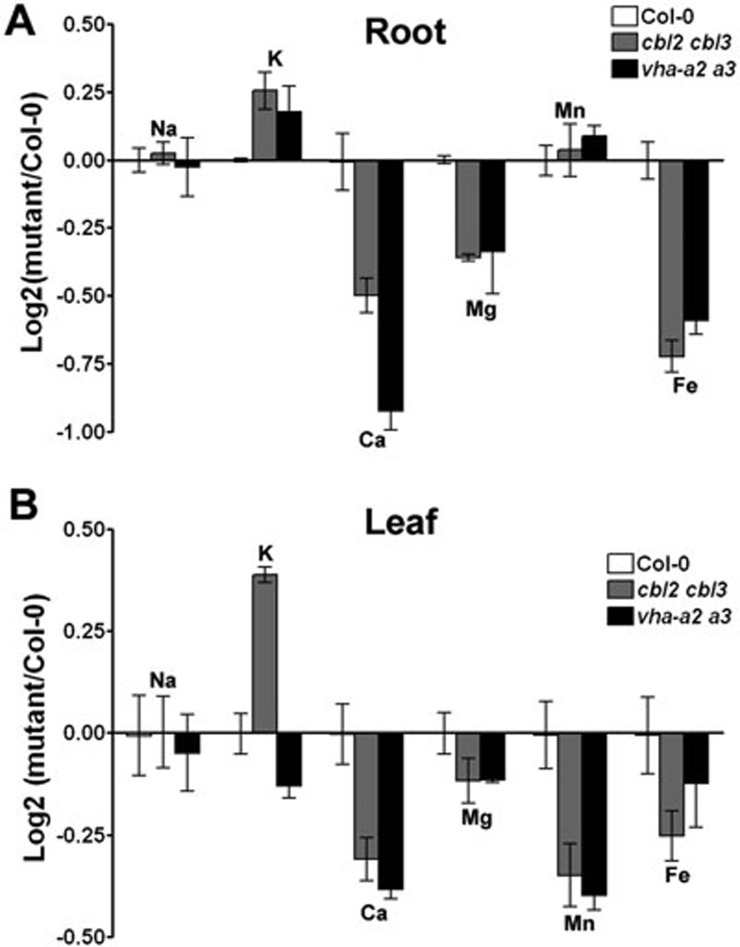
Proton-dependent transport across the tonoplast is indispensable to efficient nutrient uptake and storage in plants38. As a result of impaired V-ATPase function in cbl2 cbl3, we reasoned that accumulation of mineral ions might also be affected in mutant plants, thus serving as a model system to understand the correlation between V-ATPase activity and ion homeostasis. We therefore resorted to ionomic analysis in the cbl2 cbl3 and vha-a2 vha-a3 mutants. For this assay, roots and shoots were harvested separately from plants in hydroponic culture and analyzed for Na+, Mg2+, K+, Ca2+, Mn2+, Fe3+, Co2+, Cu2+, Zn2+ and Mo2+ using the inductively coupled plasma-mass spectrometry44. Comparison of ionic profiles of the cbl2 cbl3 mutant plants versus the wild-type controls grown under the same conditions revealed pronounced changes of several elements in the ionomes. More importantly, cbl2 cbl3 and vha-a2 vha-a3, both defective in V-ATPase, shared very similar ionic profiles (Figure 7), further supporting the conclusion that CBL2 and CBL3 are functionally linked to regulation of plant V-ATPase activity. In roots, both cbl2 cbl3 and vha-a2 vha-a3 mutants showed sharp reductions in concentrations of Ca, Mg and Fe, but significant augment in that of K, along with marginal changes in concentrations of Na and Mn (Figure 7A). In rosette leaf tissues of both mutants, the concentrations of Ca, Mg, Mn and Fe were decreased, but the K contents were differentially regulated in these two mutants (Figure 7B). The leaf K content of vha-a2 vha-a3 mutant plants was slightly reduced, but that of cbl2 cbl3 mutant was nearly 30% higher than that of wild-type (Figure 7B). Taken together, our data suggest that accumulation of micronutrients but not potassium was dramatically affected in cbl2 cbl3 as well as in vha-a2 vha-a3, consistent with the fact that these two mutants displayed lower tonoplast V-ATPase activity compared to wild-type Arabidopsis.
Figure 7.
Altered ion profiles in the cbl2 cbl3 double mutant. (A) Comparison of the root ionome of wild-type Col-0, cbl2 cbl3 and vha-a2 vha-a3 mutants. (B) Comparison of the leaf ionome of wild-type Col-0, cbl2 cbl3 and vha-a2 vha-a3 mutants. Root and leaf samples from wild-type Col-0 and cbl2 cbl3 mutant plants hydroponically grown under the same condition were analyzed for an array of ion contents using inductively coupled plasma-mass spectrometry. Ion profile data among Col-0, cbl2 cbl3 and vha-a2 vha-a3 plants were compared. For each element, the mean concentration in the wild-type Col-0 was used as the control, and log2 of the ratio of each mutant divided by the control was calculated and shown in the diagrams. Data represent mean ± SE of 12 independent plant samples in three replicate experiments.
cbl2 cbl3 mutant is tolerant to low-K+ conditions
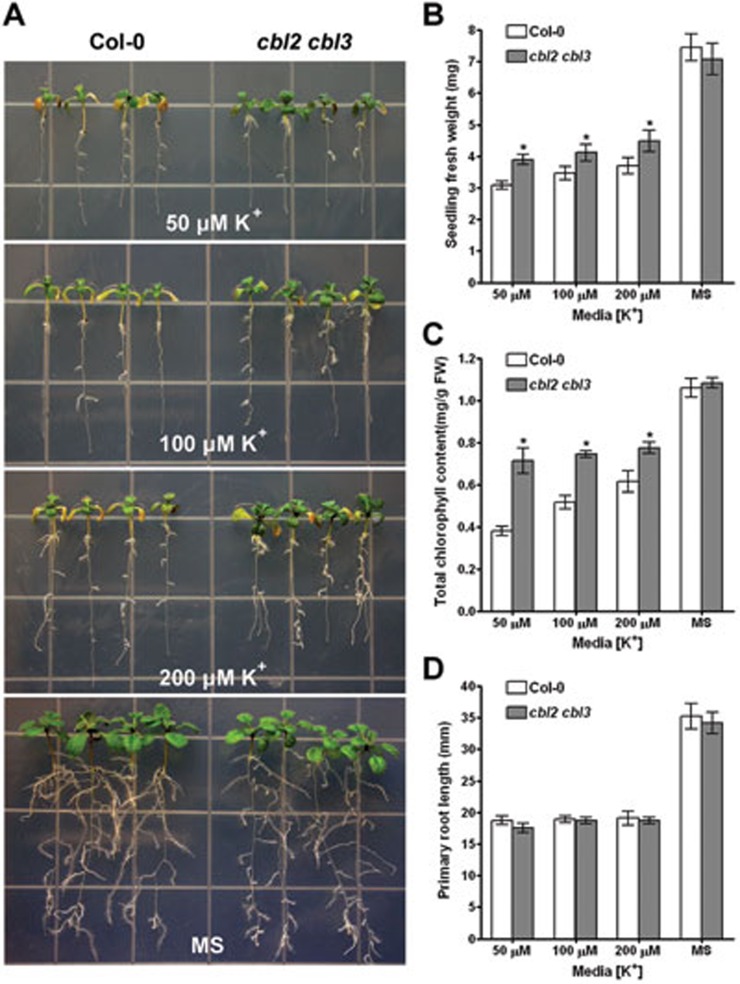
The discrepancy of leaf K concentrations in cbl2 cbl3 and vha-a2 vha-a3 mutants implies that CBL2 and CBL3 may contribute to ion homeostasis in some ways other than regulating V-ATPase. To investigate the possible role of CBL2/3 in response to low potassium, we analyzed the low-K+ stress sensitivity of the cbl2 cbl3 double mutant. After 10-day growth under K+-limiting conditions, the leaves of cbl2 cbl3 mutant were less chlorotic than those of wild-type (Figure 8A), consistent with higher leaf chlorophyll content in the double mutant (Figure 8C). Although there was no difference for root growth under K+-deficient conditions (Figure 8D), the fresh weight of cbl2 cbl3 mutant plants was higher compared to the wild-type plants, likely as a result of healthier leaves (Fgiure 8B). These results add to our understanding with regard to the role of CBL-type calcium sensors in plant responses to low-K+ stress. While CBL1 and CBL9 function at plasma membrane to regulate K+ uptake from the soil through activation of AKT121,22,45, CBL2 and CBL3 may control intracellular K+ homeostasis through regulating K+ transport across tonoplast.
Figure 8.
Low-K+ tolerance of the cbl2 cbl3 double mutant. (A) Seedlings were grown on vertical MS plates with complete nutrients for 5 days and then transferred onto K+-deficient media (modified MS with 50, 100 or 200 μM K+) or MS medium for 10 more days before taking the photographs. (B) Measurements of seedling fresh weight under various K+-deficient conditions. (C) Total chlorophyll content of wild-type (Col-0) and cbl2 cbl3 double mutant under various K+-deficient conditions. The total chlorophyll content was expressed as mg/g shoot fresh weight. (D) Measurements of primary root length under various K+-deficient conditions. Three independent experiments were performed and data represent mean ± SE (n=3). Asterisks indicate statistically significant difference between wild-type (Col-0) and the cbl2 cbl3 mutant (Student's t-test, *P < 0.05).
Discussion
Calcium plays a vital role as a ubiquitous secondary messenger in plant cells during many developmental processes and in response to environmental stimuli. Plants possess a diversity of sensor proteins that bind Ca2+ through EF-hand motifs and link the calcium changes to further cellular responses. The CBL-type calcium sensors can be divided into two major groups based on their subcellular localizations. The plasma membrane-targeted group includes CBL1, CBL4, CBL5, CBL8 and CBL9, and the tonopalst-associated group is comprised of CBL2, CBL3, CBL6 and CBL10. The fact that almost all CBLs are associated with either plasma membrane or tonoplast suggests that CBLs and their target kinases (CIPKs) build up an extensive network for the regulation of membrane transport processes. Indeed, findings of CBL1, CBL4 and CBL9 as critical regulators of plasma membrane transporters such as SOS1, AKT1, and CHL1 support this hypothesis17,18,21,22,23,24. Do tonoplast-associated CBLs coordinate with the plasma membrane CBLs in regulating cellular ionic homeostasis? Our present study answers this question by characterizing the function of CBL2 and CBL3, two highly homologous CBLs associated with vacuolar membrane.
In addition to high sequence homology, the expression patterns of CBL2 and CBL3 also largely overlap throughout plant development. Although differential expression in various cell types of the same organs may suggest cell-specific functions, genetic analysis has shown that the two genes overlap in their function as cbl2 or cbl3 single mutant closely resembles wild-type, but the double mutant displays evident abnormalities in development and ionic sensitivity (Figures 3 and 4; Supplementary information, Figure S10). We thus conclude that CBL2 and CBL3 primarily operate in a functionally redundant manner in Arabidopsis. However, we cannot rule out the possibility that either CBL2 or CBL3 may individually control yet unknown physiological processes in specific cell types.
Not only the expression pattern, but the subcellular localization of CBL-type calcium sensors is also believed to reflect spatial specification of Ca2+ signaling8,28. In this study, we confirmed that CBL2 and CBL3 are specifically associated with the tonoplast in Arabidopsis cells, consistent with an earlier study using N. benthamiana28. We also further demonstrated that the N-terminal 19-aa of the CBL2 protein defines a tonoplast targeting sequence (TTS) that is necessary and sufficient for subcellular localization to the vacuolar membrane. During the preparation of this report, Batistič et al.46 reported that S-acylation of three cysteine residues in the N-terminus is required for vacuolar targeting of CBL2 in a Brefeldin A-insensitive pathway. Particularly relevant to this study, it was also shown that the CBL6-GFP chimera with a similar TTS at the N-terminus was targeted to tonoplast independently from the typical secretory pathway47. In other words, TTS is distinct from a typical signal peptide that mediates protein translocation through the secretory pathway. Instead, TTS translocates CBL2 and CBL3 to the vacuolar membrane after protein synthesis in the cytosol. Also distinct from a typical signal peptide, TTS does not serve as a transmemrbane helix, can work in the middle or in the C-terminus of the protein, and would not be cleaved. Such sequences would be a useful tool to target other proteins to the tonoplast in genetic engineering processes32. Future work will be required to fully characterize this elusive pathway for protein trafficking in plant cells.
Due to functional redundancy, the physiological roles of CBL2 and CBL3 were approached by double mutant analysis. The most dramatic phenotypic changes in the cbl2 cbl3 double mutant include stunted growth, leaf necrosis and reduced fertility. When we correlated the developmental defects with possible functions of these two CBLs in vacuolar transport, we found that cbl2 cbl3 double mutant phenotype is largely shared by the vha-a2 vha-a3 double mutant as well, as the cax1 cax3 double mutant, both of which have defects in V-ATPase activity34,48. It is very likely that the reduced V-ATPase activity accounts for growth inhibition of all these mutants. Indeed, the cbl2 cbl3 double mutant was also compromised in its V-ATPase activity albeit to a lesser extent than the vha-a2 vha-a3 double mutant (Figure 5).
The cbl2 cbl3 phenotypic changes can be largely ascribed to the observed defect in tonoplast V-ATPase. Acidification of the vacuolar compartment by the V-ATPase is known to energize secondary ion and metabolite transport. The cbl2 cbl3 mutant is smaller than wild-type presumably because alkalized pH in the vacuoles of cbl2 cbl3 hampers cell expansion owing to inadequate turgor pressure49. As well, the reduced H+ transport across the tonoplast hinders the sequestration of excessive levels of cations into the vacuoles so that the cbl2 cbl3 mutant is hypersensitive to a variety of exogenous cations. Moreover, the symptom of necrosis at leaf tips is indicative of nutrient deficiency, consistent with ionomic profiles of cbl2 cbl3 mutants that show lower levels in nutrient cations such as Ca2+, Mg2+, Mn2+ and Fe3+.
It is intriguing to find that the cbl2 cbl3 double mutant is hypersensitive to a handful of cations but not to Na+. Previously, several mutants with reduced V-ATPase activity appeared to be hypersensitive to Na+, supporting the notion that increased V-ATPase activity is required for Na+ compartmentalization under salt stress49,50. However, consistent with our results, the vha-a2 vha-a3 double mutant does not show hypersensitivity to external Na+, suggesting that tonoplast V-ATPase activity may not be a limiting factor for Na+ sequestration and tolerance34. In contrast, reduction of V-ATPase activity in the early endosome results in increased salt sensitivity34. Therefore, different isoforms of V-ATPase may have distinct functions and CBL2 and CBL3 appear to be specifically associated with tonoplast V-ATPase activity essential for nutrient storage. Along this line, it is relevant to compare CBL2 and CBL3 with CBL10, another CBL that is targeted to tonoplast through a single transmembrane domain. The formation of CBL10-CIPK24 complex at the tonoplast mediates shoot salt tolerance by regulating unknown targets20. It will be important to understand how distinct CBL-type calcium sensors differentially regulate downstream targets and coordinate ion transport across the vacuolar membrane in plant cells.
Another interesting observation is that the cbl2 cbl3 mutant has a higher K+ content than the wild type. A simple explanation is that more K+ accumulation occurs in the vacuole as a compensation for loss of other mineral ions. However, it may be more interesting to take the other results into account, that is, cbl2 cbl3 mutant was more sensitive to high external K+ and more tolerant to low-K+ conditions as compared to the wild-type. These phenotypes are probably independent of mis-regulation of V-ATPase because vha-a2 vha-a3 did not show obvious phenotypes in response to external K+ perturbations34. K+ translocation into the lumen can be mediated by vacuolar NHX-type K+/H+ antiporters such as NHX151,52,53. Relevant to this finding, the activity of NHX1 is shown to be regulated by another calcium sensor, calmodulin54. It would be interesting to determine how K+/H+ exchange at the tonoplast was altered in the cbl2 cbl3 mutant and how these CBL-type calcium sensors coordinate with calmodulin in linking calcium and vacuolar K+transport in plant cells.
It has been long suggested that the activity of plant V-ATPase is regulated by developmental and environmental factors55,56, but the molecular mechanism remains unclear. Here we provide several lines of evidence to support that CBL2 and CBL3 may mediate a Ca2+-dependent pathway in regulation of V-ATPase activity. While the precise mechanism for this CBL2/3-mediated pathway is still lacking at present, we propose several possible scenarios. A simple possibility is that CBL2 and CBL3 may directly interact with one or several subunits of V-ATPase thereby controlling its activity or assembly. However, preliminary yeast two-hybrid assays failed to identify strong interactions between CBL2/3 and any of the known subunits (Supplementary information, Figure S11). Thus, a more inviting scenario is that the calcium sensors CBL2 and CBL3, like other CBLs, activate downstream CIPKs and recruit these kinases to the vacuole membrane8,28, followed by phosphorylation of particular V-ATPase subunits and activation of the enzyme complex. Through yeast two-hybrid assays, we found that CBL2 and CBL3 shared several interacting CIPKs (Supplementary information, Figure S12), but none of these cipk single mutants showed similar phenotypes as the cbl2 cbl3 double mutant according to our preliminary experiments. One possible reason might be functional redundancy and combination of multiple mutations among these cipk mutants would help to solve this problem. Finally, we cannot exclude the possibility that CBL2 and CBL3 modulate some secondary transporters at the tonoplast, which influence the primary H+ pump indirectly. Further work will be directed to identify other key components in this novel Ca2+-signaling pathway in regulation of plant V-ATPase activity.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Col-0 was used in this study. T-DNA insertion mutants were searched in the SALK T-DNA Express database (http://signal.salk.edu), and two lines cbl2 (SALK_151426) and cbl3 (SAIL_785_C10) were obtained from Arabidopsis Biological Resource Center (http://abrc.osu.edu/). The homozygous cbl2 or cbl3 mutant was identified using gene-specific primers in combination with T-DNA left border primers (Supplementary information, Table S1). Confirmation for T-DNA flanking sequences were presented in Supplementary information, Data S1. The cbl2 cbl3 double mutant was generated by crossing cbl3 to cbl2.
Wild-type (Col-0) and mutant Arabidopsis plants were grown in soil at 22 °C under 12-h-light/12-h-dark conditions in the green house. Hydroponically grown plants were kept in a plant growth chamber with a 14-h-light/10-h-dark photoperiod under 60-90 μmol/m2/s light and 60%-80% relative humidity. For the ion sensitivity assays, wild-type and mutant seeds were sterilized with 0.5% sodium hypochlorite for 5 min, washed four times with sterilized water and sown on MS medium41 solidified with 0.8% phytoagar. The plates were placed at 4 °C for 2 days, and then the seeds were germinated vertically at 24 °C under normal conditions. Five-day old seedlings were transferred onto 1/6-strength MS medium (containing 1% sucrose, pH = 5.8, solidified with 1% agarose) or the 1/6 MS medium supplemented with various metal ions at the indicated concentrations. For low-K+ phenotypic assay, 5-day old seedlings were transferred onto LK medium that was modified based on MS medium: KNO3 and KH2PO4 were replaced by NH4NO3 and NH4H2PO4, respectively; the final concentrations of K+ were adjusted by KNO3; and 0.8% UltraPure Agarose (Invitrogen) was added for solidification. We performed the ionic sensitivity assays three times each in two independent laboratories. The responses between wild-type and mutant plants followed the same pattern although some quantitative differences might be observed due to altered growth conditions. Data from three independent experiments performed in one laboratory were presented.
RT-PCR and qRT-PCR analyses
Total RNA was isolated from Arabidopsis seedlings of different ages as well as various organs of mature plants, or from rosette leaves of wild-type, single- and double-mutant plants, using TRIZOL Reagent (Invitrogen). RNA samples were also obtained from 10-day old seedlings grown on MS medium under various stress treatments. After being treated with DNase I (Promega), 2 μg of total RNA was subjected to reverse transcription reaction using the reverse transcriptase ReverTra Ace (TOYOBO) at 42 °C for 1 h. The resulting cDNA was used for PCR amplification with the gene-specific primers listed in Supplementary information, Table S1. qRT-PCR analysis was performed on the RotorGene 3000 system (Corbett Research) with the SYBR Green Realtime PCR Master Mix (TOYOBO) to monitor double-stranded DNA products. Data analysis was performed with Rotor-Gene software version 6.0 and relative amounts of mRNA were calculated based on the comparative threshold cycle method. The relative expression of CBL2 or CBL3 gene was double-normalized using the housekeeping gene ACTIN2 and using the control expression values measured at 0 h.
Promoter-GUS fusion analysis and histochemical assay of GUS activity
For construction of ProCBL2:GUS, an approximately 1.5 kb promoter region right upstream of ATG start codon of CBL2 gene was amplified from Col-0 genomic DNA by PCR and verified by sequencing. The PCR fragment was cloned into the Bam HI/Hin dIII site of binary vector pCAMBIA 1381Z to obtain a transcriptional fusion of the CBL2 promoter with the GUS coding sequence. To generate the ProCBL3:GUS construct, 1.6 kb of the CBL3 promoter region was amplified using Col-0 genomic DNA and the PCR product was cloned into the Bam HI/Pst I site of pCAMBIA 1381Z vector. The constructs were introduced into the Agrobaterium tumefaciens GV3101 strain, which was thereafter used to transform Arabidopsis thaliana by the floral dipping method57. At least 10 independent transgenic lines for each construct were selected on MS medium containing 30 mg/l hygromycin and seedlings of T2 generation were subjected to GUS assays. For histochemical analysis to detect GUS expression, plant materials were incubated at 37 °C for 4 h in the staining buffer (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6], 0.1% Triton X-100) supplemented with 0.5 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc). To clear chlorophyll from the green tissues, the stained materials were incubated in 75% ethanol overnight and then kept in 95% ethanol58.
Protein expression, purification and calcium binding assay
The coding region of CBL2 and CBL3 was amplified from Col-0 cDNA samples. PCR products were cloned into the pGEX-4T-1 vector through Bam HI/Sal I site and verified by sequencing. The resulting GST fusion constructs were transformed into E. coli strain BL21 (DE3). Recombinant protein expression was induced at OD600 = 0.8 with 0.5 mM IPTG. Bacterial cells were harvested by centrifugation and lysed by sonication. GST-CBL2 and GST-CBL3 proteins were affinity purified from bacterial lysates with Glutathione-Sepharose 4B (GE Healthcare).
For Ca2+-binding assay, proteins were separated by SDS-PAGE and transferred onto nitrocellulose by eletroblotting. Thereafter, the membrane was washed four times in a buffer solution containing 10 mM imidazole-HCl, pH 6.8, 60 mM KCl and 5 mM MgCl2. The membrane was then incubated in the same buffer plus 37 kBq m/l 45Ca2+ (Perkin-Elmer) for 10 min, rinsed with distilled water for 40 min, air-dried and exposed to X-ray film for 6 days.
Subcellular localization of CBL2 and CBL3
To determine the subcellular localization of CBL2, CBL3 and chimeras modified by TTS, the coding region of each target protein was in-frame fused to YFP in the transient expression vector pA7-YFP. The fusion constructs were introduced into Arabidopsis mesophyll protoplasts prepared from rosette leaves by the polyethylene glycol-mediated transformation procedure59, respectively. For stable expression, the CBL2-YFP or CBL3-YFP fusion was cloned downstream of a plant super promoter in the pCAMBIA 1300 binary vector60, and the resulting constructs were transferred into Agrobaterium tumefaciens GV3101 strain. Arabidopsis plants were transformed using the floral dipping method57. Twenty independent transgenic lines were used for localization of CBL2-YFP and CBL3-YFP. YFP fluorescence in transfected protoplasts or in the roots of transgenic plants was imaged using the LSM510 META confocal laser scanning microscope (Carl Zeiss). The filter settings are Ex 514 nm/Em BP 535-600 nm for YFP, and Ex 488 nm/Em LP 650 nm for chlorophyll.
Enzyme activity and proton (H+) transport measurements
Leaf microsomal fractions were prepared from 4-week old hydroponically grown plants. Plant materials were ground with cold homogenization buffer containing 350 mM sucrose, 70 mM Tris-HCl (pH 8.0), 3 mM Na2EDTA, 0.2% (w/v) BSA, 1.5% (w/v) PVP-40, 5 mM DTT, 10% (v/v) glycerol, 1 mM PMSF and 1× protease inhibitor mixture (Roche). The homogenate was filtered through four layers of cheesecloth and centrifuged at 4 000× g for 20 min at 4 °C. The supernatant was filtered through cheesecloth again and then centrifuged at 100 000× g for 1 h. The resulting microsomal pellet was resuspended in 350 mM sucrose, 10 mM Tris-Mes (pH 7.0), 2 mM DTT and 1× protease inhibitor mixture. V-ATPase and PPase activity of 10 μg microsomal membranes was determined as phosphate (Pi) release after 40 min incubation at 28 °C. The V-ATPase assay solution contained 25 mM Tris-Mes (pH 7.0), 4 mM MgSO4, 50 mM KCl, 1 mM NaN3, 200 μM Na2MoO4, 500 μM NaVO4, 0.1% Brij 35 and 2 mM Mg-ATP. V-ATPase activity was calculated as the value difference between the measurements in the absence and the presence of 200 nM Concanamycin A plus 50 mM NaNO3. The PPase activity was assayed in a reaction solution containing 25 mM Tris-Mes (pH 7.5), 2 mM MgSO4, 200 μM Na2MoO4, 0.1% Brij 58, and 200 μM Na4P2O7. PPase activity was expressed as the difference measured in the absence and the presence of 50 mM KCl. For measuring the amount of inorganic Pi, reactions were terminated by adding 40 mM citric acid. Freshly prepared AAM solution (50% (v/v) acetone, 2.5 mM ammonium molybdate, 1.25 M H2SO4) was then added to the reaction, vortexed and colorimetrically examined at 355 nm. 10 μg boiled membranes instead of fresh microsomes were used for the blank value.
For H+ transport assay, tonoplast vesicles were isolated and purified as previously described61. The H+ transport activity of V-ATPase was measured as a quenching of the pH-sensitive fluorescent probe quinacrine. Proton translocation was initiated in purified tonoplast vesicles (100 μg) by rapid addition of 1 mL reaction buffer containing 10 mM Tris-Mes (pH 8.0), 250 mM mannitol, 6 mM MgSO4, 50 mM (CH3)4NCl, 3 mM ATP and 3 μM quinacrine. Fluorescence quenching was monitored in a thermostated cell at 25 °C using a fluorescence spectrometer (Hitachi F-2700) at 427 nm excitation wavelength and 495 nm emission wavelength.
Measurement of vacuolar pH
Vacuolar pH was monitored with the cell-permeant and pH-sensitive fluorescent dye BCECF-AM in Arabidopsis mesophyll protoplasts. Freshly prepared protoplasts were loaded with the fluorescent dye in MMG solution (0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7) supplemented with 10 μM BCECF-AM. After 1 h staining at 23 °C in the darkness, the protoplasts were washed twice with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7), and then incubated in WI soluation (0.4 M mannitol, 20 mM KCl, 4 mM MES, pH 5.7). For in situ calibration of BCECF, the protoplasts were incubated in serial pH equilibration buffers containing 0.4 M mannitol, 20 mM KCl, 50 mM ammonium acetate supplemented with 50 mM MES-BTP (pH 5.2-6.4) or 50 mM HEPES-BTP (pH 6.8-7.6), respectively. The BCECF fluorescence was detected and imaged using the Zeiss LSM510 META confocal laser-scanning microscope. The fluorophore was excited at 488 nm and 458 nm, respectively, and the emission was detected between 505 nm and 550 nm. Vacuolar pH value was quantified by ratio analysis of the pH-dependent (488 nm) and pH-independent (458 nm) excitation wavelengths from a calibration curve (Supplementary information, Figure S9), and ratio images were produced using the ion concentration tool of Zeiss LSM confocal software.
Ionomic analysis
Ion profiling was performed essentially as described44. Wild-type and mutant plants were fed with the 1/6-strength MS solution twice a week and harvested when they were 5-week old. Plants were carefully subsampled in roots and rosette leaves, and washed with 18 MΩ water for three to five times. Sample digestion was carried out in concentrated HNO3 overnight followed by gentle oscillation in a 99 °C water bath for 1-2 h. Each sample was then diluted to 14 ml and elemental analysis was performed with an ICP-mass spectrometer (Elan DRCe; Perkin-Elmer). The experiments were repeated for at least three times and 12-18 individual plants were used in each replicate.
Acknowledgments
We thank Prof Karin Schumacher and Dr Melanie Krebs (University of Heidelberg, Germany) for providing the vha-a2 vha-a3 mutant seeds. Special thanks would go to Chen Zhong (Shanghai Institutes for Biological Sciences, CAS) for the assistance in ionomic analysis. This work is supported by the grants from the National Natural Science Foundation of China (31171169, 30872044), the National mega project of GMO crops (2011ZX08001-003, 2011ZX08004-002 and 2011ZX08010-004) to HXZ and National Science Foundation to SL.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Expression of CBL2 and CBL3 in response to various abiotic stresses.
The N-terminal 16-aa of CBL2 is required but not sufficient for its tonoplast targeting.
The first 19-aa in the N-terminus of CBL2 determines the tonoplast localization.
The efficacy of CBL2 Tonoplast Targeting Sequence (TTS) is independent of its N-terminal location.
TTS targets several soluble proteins but not transmembrane proteins to the tonoplast.
Purification of recombinant CBL2 and CBL3 proteins from E-Coli cells.
Complementation of the cbl2 cbl3 double mutant by CBL2 or CBL3.
Phenotype of cbl2 cbl3 and vha-a2 vha-a3 plants grown in the soil.
In situ calibration of BCECF-AM in protoplast vacuoles.
Growth phenotype of cbl2, cbl3 single mutants and cbl2 cbl3 double mutant under various ionic stress conditions.
Yeast two-hybrid assays on CBL2/3 in combination with VHA subunits.
CBL2 and CBL3 interact with an array of CIPK proteins.
Primers used in this study.
The flanking sequence of the T-DNA insertion lines in this study.
References
- Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistič O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Brownlee C. The generation of Ca2+ signals in plants. Annu Rev Plant Biol. 2004;55:401–427. doi: 10.1146/annurev.arplant.55.031903.141624. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. Shaping the calcium signature. New Phytol. 2009;181:275–294. doi: 10.1111/j.1469-8137.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell. 2002;14:S389–S400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem J. 2010;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- Kudla J, Xu Q, Harter K, Gruissem W, Luan S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA. 1999;96:4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KN, Ritz O, et al. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell. 1999;11:2393–2405. doi: 10.1105/tpc.11.12.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–1400. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001;20:1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Lan W, Lee SC. Potassium nutrition, sodium toxicity and calcium signaling: connections through the CBL-CIPK network. Curr Opin Plant Biol. 2009;12:339–346. doi: 10.1016/j.pbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BG, Waadt R, Cheong YH, et al. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007;52:473–484. doi: 10.1111/j.1365-313X.2007.03249.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, et al. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin S, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Albrecht V, Weinl S, Blazevic D, et al. The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 2003;36:457–470. doi: 10.1046/j.1365-313x.2003.01892.x. [DOI] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell. 2003;15:1833–1845. doi: 10.1105/tpc.012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, et al. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Waadt R, Steinhorst L, Held K, Kudla J. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 2010;61:211–222. doi: 10.1111/j.1365-313X.2009.04045.x. [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008;56:505–516. doi: 10.1111/j.1365-313X.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- Batistič O, Sorek N, Schultke S, Yalovsky S, Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell. 2008;20:1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu Ü, Weinl S, Blazevic D, Batistič O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M, Held K, Binder A, et al. FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 2012;69:181–192. doi: 10.1111/j.1365-313X.2011.04780.x. [DOI] [PubMed] [Google Scholar]
- Nagae M, Nozawa A, Koizumi N, et al. The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J Biol Chem. 2003;278:42240–42246. doi: 10.1074/jbc.M303630200. [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, et al. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA. 2010;107:3251–3256. doi: 10.1073/pnas.0913035107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüx A, Liu TY, Krebs M, et al. Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell. 2008;20:1088–1100. doi: 10.1105/tpc.108.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Schumacher K, Müller ML, Padmanaban S, Taiz L. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci. 2002;7:157–161. doi: 10.1016/s1360-1385(02)02240-9. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Krebs M. The V-ATPase: small cargo, large effects. Curr Opin Plant Biol. 2010;13:724–730. doi: 10.1016/j.pbi.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Palmgren MG, Schumacher K. Plant proton pumps. FEBS Lett. 2007;581:2204–2214. doi: 10.1016/j.febslet.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Swanson SJ, Jones RL. Gibberellic acid induces vacuolar acidification in barley aleurone. Plant Cell. 1996;8:2211–2221. doi: 10.1105/tpc.8.12.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–495. [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, et al. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002;30:529–539. doi: 10.1046/j.1365-313x.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol. 2003;21:1215–1221. doi: 10.1038/nbt865. [DOI] [PubMed] [Google Scholar]
- Geiger D, Becker D, Vosloh D, et al. Heteromeric AtKC1·AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem. 2009;284:21288–21295. doi: 10.1074/jbc.M109.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O, Rehers M, Akerman A, et al. S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 2012;22:1155–1168. doi: 10.1038/cr.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottanelli F, Foresti O, Hanton S, Denecke J. Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. Plant Cell. 2011;23:3007–3025. doi: 10.1105/tpc.111.085480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Shigaki T, et al. Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 2005;138:2048–2060. doi: 10.1104/pp.105.061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban S, Lin X, Perera I, Kawamura Y, Sze H. Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol. 2004;134:1514–1526. doi: 10.1104/pp.103.034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli G, Verslues PE, Agius F, et al. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol. 2007;27:7781–7790. doi: 10.1128/MCB.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010;61:495–506. doi: 10.1111/j.1365-313X.2009.04073.x. [DOI] [PubMed] [Google Scholar]
- Bassil E, Tajima H, Liang YC, et al. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011;23:3482–3497. doi: 10.1105/tpc.111.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell. 2012;24:1127–1142. doi: 10.1105/tpc.111.095273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc Natl Acad Sci USA. 2005;102:16107–16112. doi: 10.1073/pnas.0504437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla RJ, Vera-Estrella R, Maldonado-Gama M, Pantoja O. Abscisic acid induction of vacuolar H+-ATPase activity in Mesembryanthemum crystallinum is developmentally regulated. Plant Physiol. 1999;120:811–820. doi: 10.1104/pp.120.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge C, Lahr J, Hanitzsch M, Bolte S, Golldack D, Dietz KJ. New insight into the structure and regulation of the plant vacuolar H+-ATPase. J Bioenerg Biomembr. 2003;35:377–388. doi: 10.1023/a:1025737117382. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-Glucurodinase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yang Q, Chen ZZ, Zhou XF, et al. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant. 2009;2:22–31. doi: 10.1093/mp/ssn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell. 2003;15:347–364. doi: 10.1105/tpc.007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of CBL2 and CBL3 in response to various abiotic stresses.
The N-terminal 16-aa of CBL2 is required but not sufficient for its tonoplast targeting.
The first 19-aa in the N-terminus of CBL2 determines the tonoplast localization.
The efficacy of CBL2 Tonoplast Targeting Sequence (TTS) is independent of its N-terminal location.
TTS targets several soluble proteins but not transmembrane proteins to the tonoplast.
Purification of recombinant CBL2 and CBL3 proteins from E-Coli cells.
Complementation of the cbl2 cbl3 double mutant by CBL2 or CBL3.
Phenotype of cbl2 cbl3 and vha-a2 vha-a3 plants grown in the soil.
In situ calibration of BCECF-AM in protoplast vacuoles.
Growth phenotype of cbl2, cbl3 single mutants and cbl2 cbl3 double mutant under various ionic stress conditions.
Yeast two-hybrid assays on CBL2/3 in combination with VHA subunits.
CBL2 and CBL3 interact with an array of CIPK proteins.
Primers used in this study.
The flanking sequence of the T-DNA insertion lines in this study.