Abstract
Tongue diagnosis is a unique method in traditional Chinese medicine (TCM). This is the first investigation on the association between traditional tongue diagnosis and the tongue coating microbiome using next-generation sequencing. The study included 19 gastritis patients with a typical white-greasy or yellow-dense tongue coating corresponding to TCM Cold or Hot Syndrome respectively, as well as eight healthy volunteers. An Illumina paired-end, double-barcode 16S rRNA sequencing protocol was designed to profile the tongue-coating microbiome, from which approximately 3.7 million V6 tags for each sample were obtained. We identified 123 and 258 species-level OTUs that were enriched in patients with Cold/Hot Syndromes, respectively, representing "Cold Microbiota" and "Hot Microbiota". We further constructed the tongue microbiota-imbalanced networks associated with Cold/Hot Syndromes. The results reveal an important connection between the tongue-coating microbiome and traditional tongue diagnosis, and illustrate the potential of the tongue-coating microbiome as a novel holistic biomarker for characterizing patient subtypes.
Tongue diagnosis is a non-invasive, simple and valuable diagnostic tool that has been repeatedly testified by clinical practitioners of traditional Chinese medicine (TCM) for at least 3,000 years (Fig. 1A). TCM treats tongue appearance as an outer manifestation of the status of the human body. TCM practitioners differentiate conditions of individual patients according to the TCM Syndromes (“ZHENG” in Chinese), which are used as a holistic summary of the patient's status and are determined mainly by the colour and texture of the coating of the patient's tongue in addition to other symptoms1. The Cold Syndrome and Hot Syndrome are the two most common and representative syndromes that represent two opposite but interrelated conditions of the human body and are identified by a “white-greasy” and “yellow-dense” coating of the tongue, respectively. These syndromes have been used to characterise patient status in many diseases or conditions, including inflammation, infection, and stress, as well as immune and endocrine disorders2,3. Cold and Hot Syndromes have also been a focus in our previously studies4,5. In addition to the extensive practice of tongue diagnosis in TCM, modern technologies, especially tongue imaging analyses, have been introduced for tongue diagnosis6,7. The relationships between tongue diagnosis and diseases including cancer8, gastritis and precancerous lesions9,10 have been explored. However, the biological bases of different tongue-coating appearances are still poorly understood and lack systematic investigation at the molecular level.
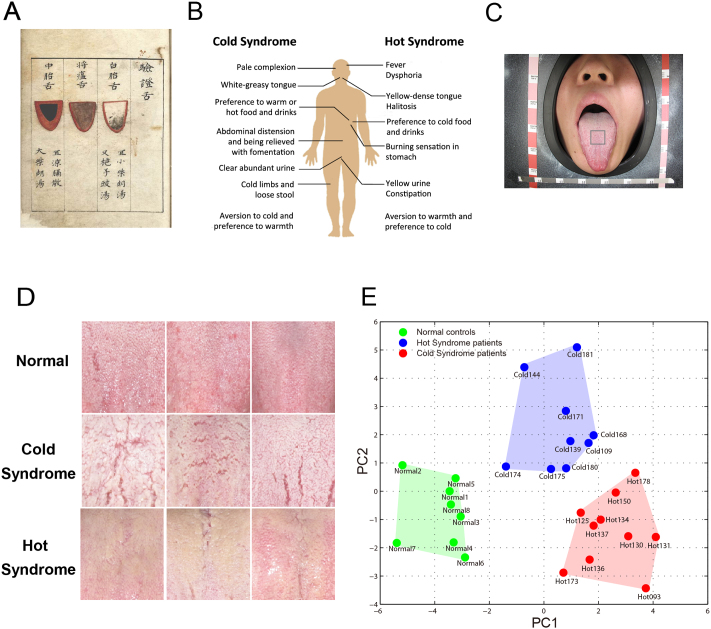
Figure 1. TCM tongue diagnosis and tongue-coating appearance classification.
(A) An ancient instruction for tongue diagnosis recorded in Shang-han-dian-dian-jin, a TCM book compiled in the Ming Dynasty of China (1368–1644 AD). (B) Symptoms all over human body used as features for the TCM classification of Cold and Hot Syndromes; the tongue-coating feature is highly important in clinical practice. (C) Sampling images of tongue coating from the centre of the tongue, an area regarded as reflecting conditions of the “spleen-stomach” in the traditional tongue diagnosis. (D) Typical tongue-coating appearances: normal tongue coating of healthy control examples (upper), white-greasy tongue coating of Cold Syndrome examples (middle), and yellow-dense tongue coating of Hot Syndrome examples (lower). (E) Principal Component Analysis (PCA) of the tongue-coating images. The healthy controls, Cold Syndrome patients and Hot Syndrome patients are distributed in separate regions, indicating that the three classes are distinguishable based on their tongue-coating images.
In modern Western medicine, the tongue has increasingly been viewed as an extension of the upper gastrointestinal tract that can provide important clues to the body's current condition and global information about the body's health status. Some Western medicine literature has used the term “geographic tongue” to refer to tongues with discoloured regions or cracks that accompany illness or environmental sensitivity11,12. Some researchers have also considered TCM tongue diagnosis to be a helpful approach for clinical decision making in Western medicine13. For example, tongue coating can be a risk indicator for aspiration pneumonia in edentate patients, as it is associated with a number of viable salivary bacteria14. It has also been reported that amyloidosis of the tongue may be a diagnostic manifestation of plasmacytoma15. These examples indicate the potential for inferring systemic disorders from the tongue in clinical diagnoses. Compared with other diagnostic regimens, such as blood tests or tissue biopsies, tongue diagnosis is more amenable to and convenient for patients as well as doctors. Therefore, studying the molecular bases of tongue diagnosis is important for understanding this long-standing medical practice and might be a promising contribution to personalized medicine.
Recent studies have indicated that the microbial community in the human body is associated with human physiology and pathology16. The microbiome on the tongue coating is one of the main microbiomes of the human body17 and is at the forefront of the human digestive system18. It was reported that the tongue shares considerable similarity with the gut not only in mucosal epithelia but also microbial diversity19,20. Densely populated by a variety of microorganism communities, the human gut microbiome is considered essential to maintaining natural host-environment interactions involved in nutrient absorption, epithelium regeneration, energy metabolism and immune response21. The characteristics and structures of microbiomes are closely associated with the health of the gastrointestinal tract22,23,24. Patients with inflammatory bowel disease, obesity, diabetes and colorectal cancer have been found to exhibit different gastrointestinal microbial structural patterns25,26,27,28.
The characterization and differentiation of Cold and Hot Syndromes has played an important role in the clinical practice of TCM for gastrointestinal diseases, and the appearance of the tongue coating has long been a major parameter in discriminating the Cold and Hot Syndromes. We speculated that the microbiome composition is a key factor that affects the appearance of the tongue coating and reasoned that the microbiome composition on the tongue coating could be associated with characteristics of stomach diseases. If this is true, then the underlying association of the Cold and Hot Syndromes defined by tongue coating features may be rooted in the association of microbiome features with the health status of the human body. Therefore, we investigated the possible relationship between the tongue microbial community and the health status of the patient as described in TCM practice to begin to bridge the gap between traditional tongue diagnosis and molecular systems biology.
The development of next-generation sequencing (NGS) technology for characterising microbial communities provides an opportunity to deepen our understanding of the tongue-coating microbiome; thus, we used NGS to profile tongue-coating microbiomes. We collected white-greasy and yellow-dense tongue-coating samples from Cold and Hot Syndrome patients with chronic atrophic gastritis (CAG), respectively, along with normal tongue-coating samples from healthy controls. The tongue-coating microbiomes were sequenced using an accurate Illumina paired-end with double-barcode (Ipedb for short) sequencing protocol that we developed. The protocol takes advantage of the sequencer's high-throughput capability and uses a double-barcode, paired-end strategy to ensure the accuracy of measuring 16S rRNA hyper-variable regions with short-read sequencing. To the best of our knowledge, this is the first analysis of tongue-coating microbiomes and their relation with traditional tongue diagnosis. As a result, we identified 715 differentially abundant, species-level Operational Taxonomic Units (OTUs) on tongue coatings of the enrolled patients compared to healthy controls. We further identified two subtypes of tongue-coating microbiomes and detected 123 “Cold OTUs” and 258 “Hot OTUs” that are enriched in patients with Cold Syndrome and Hot Syndrome representing the “Cold Microbiota” and “Hot Microbiota”, respectively. These observations revealed that the biological bases underlying traditional tongue diagnosis may root in the relationship between the tongue-coating microbiome and the body's health status.
Results
Tongue-coating sample collection
We collected samples from 19 CAG patients who were independently diagnosed by Western medical gastroenterologists according to “the Updated Sydney System”29,30 and by two TCM doctors according to the “National TCM Diagnosis Principle”2. According to Western medicine, these patients are considered to have the same disease, whereas in the view of TCM, they are further categorised into two subtypes, the Cold Syndrome (n = 9) and Hot Syndrome (n = 10), based on an overall medical examination of symptoms (Fig. 1B). Despite the common manifestations experienced by the patients, such as abdominal discomfort or pain, nausea, vomiting and diarrhoea, patients with the Cold and Hot Syndromes exhibited characteristic differences. Tongue-coating appearance is one of the most important features in Syndrome classification. Fig. 1C shows the method we used to capture tongue-coating images according to instructions by TCM practitioners. We also recruited 8 volunteers who had reported no stomach discomforts in the past five years and showed normal tongue coating; these volunteers were the healthy controls in the study. For the selection of samples in this pilot study, we focused on the comparability of characteristic tongue appearances diagnosed by traditional Chinese medical doctors. The healthy volunteers with typical healthy white-thin tongue coatings were found to be younger than the study patients and matched the genders of the study groups, resulting in a group of sex-matched but age-unmatched normal controls. The genders and ages of gastritis patients with Cold Syndrome and Hot Syndrome were well matched (Table 1). Representative tongue-coating images from patients with Cold and Hot Syndromes and from normal controls are shown in Fig. 1D. Tongue-coating images of all the collected samples are provided in the Supplementary Materials (Fig. S1). Consistent with the traditional medical findings, the Cold Syndrome patients presented with the typical white-greasy tongue coatings, and Hot Syndrome patients exhibited the typical yellow-dense tongue coatings. In addition to the tongue-coating image features, the 9 Cold Syndrome patients presented with other typical features of Cold Syndrome according to their clinical records as follows: all patients reported that they preferred warm or hot food and drinks, and 7 reported abdominal distension that was relieved by using a warm compress or consuming hot drinks. Similarly, the 10 Hot Syndrome patients showed representative features in addition to the tongue-coating appearance as follows: 7 patients reported halitosis, and 6 reported yellow urine.
Table 1. General information of patients and healthy controls.
| Normal controls | Gastritis patients with Cold Syndrome | Gastritis patients with Hot Syndrome | |
|---|---|---|---|
| Number of Cases | 8 | 9 | 10 |
| Age (Mean±SD) | 26±1.2 | 60±4.3 | 53±4.2 |
| Male (%) | 3 (37.5%) | 4 (44.4%) | 4 (40%) |
| Female (%) | 5 (62.5%) | 5 (55.6%) | 6 (60%) |
We applied the Principal Component Analysis (PCA) to digital features of the tongue-coating images (see Methods). Fig. 1E shows the distribution of samples in the space of the first two principal components of the PCA of the tongue-coating images. The first two principal components accounted for 67.5% of the total variations. The samples were distributed in three regions corresponding to a white-greasy tongue coating in Cold Syndrome patients, a yellow-dense tongue coating in Hot Syndrome patients and a normal (white-thin) tongue coating in healthy controls (Fig. 1E). This finding confirms the empirical judgments by TCM doctors using their naked eyes. We then collected microbiome samples from the tongue coatings of the study patients and normal controls (see Methods).
16S rRNA sequencing analysis and taxonomy of the tongue-coating microbiome
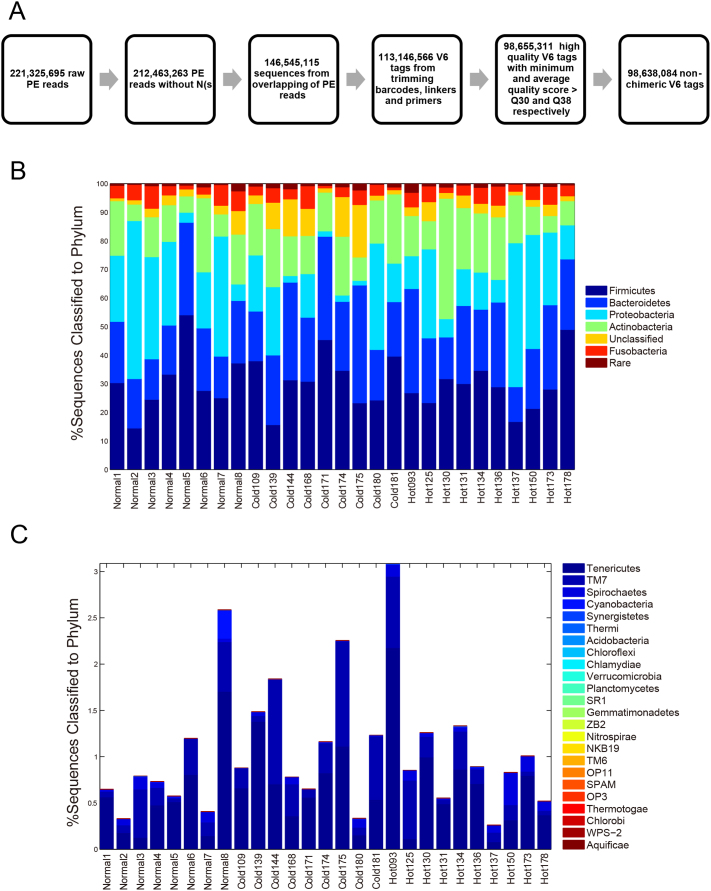
We characterized the diversity of the tongue-coating microbiome via targeted 16S rRNA gene sequencing. We developed an Illumina paired-end with double barcodes (called Ipedb for short) sequencing protocol for this purpose (Fig. S2) and obtained 3.65±0.52 million high-quality V6 (the 6th hyper-variable region in the 16S rRNA gene) sequences per sample from 221.33 million paired-end (PE) raw reads (Table S1). The details of the protocol are described in the Methods section. In brief, bacterial V6 regions were PCR-amplified with designed primers (Table S2), and PCR amplicons (120~150 bp) were barcoded at both ends and sequenced using the Illumina GAIIx with 120×2-bp PE reads. High-quality V6 sequences were yielded after overlapping each pair of raw PE reads; trimming barcodes, linkers and primers; and filtering low-quality or chimeric sequences (Fig. 2A). In the Ipedb protocol, the criterion for removing reads with mismatch(s) within the 90~120-bp overlapping region was crucial for reducing sequencing errors because singled-end reads demonstrated a high sequencing error rate in this region. The average sequencing error rate in the resulting V6 sequences was estimated at ~0.22%, which was much less than the rate of 0.6~1.0% reported for Illumina GA31 and the rate of 0.50% reported for pyrosequencing32. Additionally, our unique “double-barcode” strategy of adding barcodes at both ends enabled the identification of approximately 0.20% of the sequences as chimeras generated during Illumina image processing, further increasing the sequencing accuracy.
Figure 2. 16S rRNA sequencing analysis and taxonomy classification of the tongue-coating microbiome at the phylum level.
(A) The flowchart of sequence preprocessing, quality control and chimera detection. (B) Relative abundances of the dominant phyla in samples. (C) Relative abundances of the rare phyla in samples.
These sequences were first assigned to specific taxonomies using the GAST (Global Alignment for Sequencing Taxonomy) rRNA classifier33, which uses a reference database of SSU sequences to determine the taxonomy of hyper-variable-region tags. As shown in Fig. 2B and Fig. 2C, 95.23% of the sequences were classified into 29 phyla in the Bacteria domain. The dominant phyla (relative abundance >1%) across all 27 samples were Firmicutes (30.40%±9.47%), Bacteroidetes (23.57%±7.57%), Proteobacteria (20.07%±15.51%), Actinobacteria (15.79%±7.84%) and Fusobacteria (4.37%±2.02%). The rare phyla (relative abundance < 1%) were Acidobacteria, Aquificae, Acidobacteria, Chlamydiae, Chlorobi, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Nitrospira, Planctomycetes, Spirochaetes, Synergistetes, Tenericutes, Thermi, Thermotogae, Verrucomicrobia and nine candidate divisions (NKB19, OP11, OP3, SPAM, SR1, TM6, TM7, WPS-2, ZB2).
A comparison of the relative abundance of the dominant phyla between the normal controls and all gastritis patients showed no significant difference between the two groups. The phylum that showed the largest difference was Bacteroidetes, which was more abundant in the tongue-coating microbiomes of patients, although the difference was not significant (controls: 20.08%±5.86%; patients: 25.03%±7.86%; two-tailed t-test, p = 0.44 after a Bonferroni correction). The proportions of unclassifiable sequences also showed no significant differences between patients and controls (controls: 2.99%±2.30%; patients: 5.50%±4.97%; two-tailed t-test, p = 0.085). Among the patient samples, Cold Syndrome patients had slightly more sequences that were not classified to specific phyla than Hot Syndrome patients (Cold Syndrome patients: 7.95%±6.31%; Hot Syndrome patients: 3.30%±1.58%; two-tailed t-test, p = 0.061), whereas no dominant phyla showed significant differences in relative abundance between the patients with the two Syndromes.
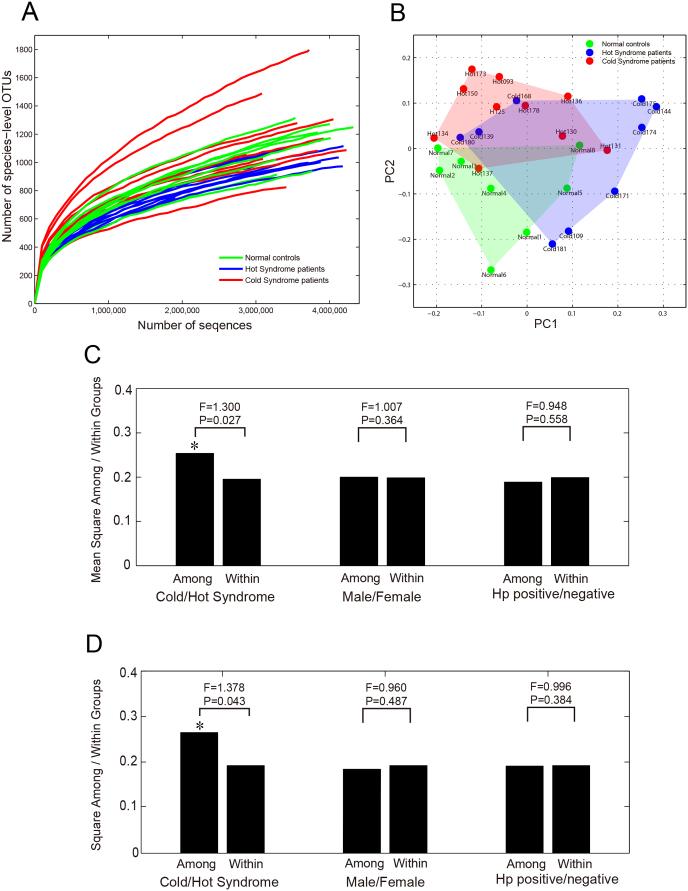
We further conducted alpha diversity and beta diversity analyses to identify features of the tongue-coating microbiome composition that might be associated with the condition of patients with chronic atrophic gastritis, categorised as Hot or Cold Syndromes. V6 tag sequences were clustered into OTUs, which shared different levels of genetic similarities (unique sequence, 100%; species level, 97%; genus level, 95%; family level, 90%), using the CROP method34, an unsupervised Bayesian clustering algorithm. For each sample, a rarefaction curve was drawn at a specific OTU level as the function of the observed number of OTUs on sequence counts by re-sampling at different sequencing depths (Figs. 3A and S3). Richness, evenness and phylogenetic diversity estimators were also calculated (see Figs. S4, S5 and S6 in the Supplementary Materials). The alpha diversities were consistent among tongue-coating microbial communities of different samples.
Figure 3. 16S rRNA diversity analysis detected significant differences in the tongue-coating microbiome between gastritis patients with Cold and Hot Syndromes.
(A) Alpha diversity rarefaction curves of all 27 samples based on species-level OTUs. (B) PCoA plot of the between-sample Jaccard matrix of all 27 samples. (C) AMOVA results of the between-sample Jaccard distance matrix of the 19 gastritis patients. The F-statistic is the ratio of the mean square among groups within the groups. (D) AMOVA results of the between-sample unweighted Unifrac distance matrix of the 19 gastritis patients.
We then compared the microbial community composition (based on presence / absence information of OTUs) and structure (based on relative abundances of OTUs) between samples by calculating the Jaccard distance35 and the Bray-Curtis distance36, respectively. We applied the AMOVA (Analysis of Molecular Variances) method37 (see details in the Supplementary Materials) to determine whether there were stratifications in the samples. With the distances based on species-level OTUs, AMOVA showed that microbial communities on the tongue coatings of study patients and normal controls exhibited significantly different community memberships (P = 0.005), but their community structures did not show significant difference (P = 0.364). We also used the Unifrac distance based on the comparison of phylogenetic trees38 in the AMOVA analysis. With the unweighted Unifrac distance, a noticeable difference was observed between the two groups, although it was not statistically significant (P = 0.063). This finding indicates that there can be differences in the OTU compositions of tongue-coating microbiomes between different types of samples. We used the Principal Coordination Analysis (PCoA) method39 to visualise such differences. PCoA is a variation of the MDS (Multi-Dimensional Scaling) method that visualises a set of samples on a two-dimensional plot by keeping the relative dissimilarity relations of the samples defined by a matrix of dissimilarity measures or distances between all pairs of samples. Fig. 3B shows the PCoA plot of the 27 samples when the Jaccard distance measure was used. Although there was not a clear boundary between the patients and normal control samples, there was a tendency for the normal controls, Hot Syndrome patients and Cold Syndrome patients to be locally distributed as three groups with some overlap on the plot.
We were more interested in whether there were features of the tongue-coating microbiomes that were significantly different between patients with the two Syndromes. We therefore focused on the 19 gastritis patient samples for further analysis. We used four types of distance measures – Jaccard, Bary-Curtis, unweighted Unifrac and weighted Unifrac – to obtained the dissimilarity matrices of the samples and applied AMOVA to these matrices to check whether there was significant stratification between the Cold and Hot Syndrome patients. As a result, we found significant differences between 9 Cold Syndrome samples and 10 Hot Syndrome samples within the Jaccard and unweighted Unifrac distance matrices (P = 0.027, Fig. 3C; P = 0.043, Fig. 3D). This finding indicates that there are systemic differences in the tongue-coating microbial composition between the two TCM-defined groups. We also checked other factors that could group the patients, including gender and Helicobacter pylori positivity, using the AMOVA method. In contrast to the significant differences found between the two Syndromes, no significant difference was detected between male (n = 8) and female (n = 10) cases or between Hp-positive (n = 6) and -negative cases (n = 13) (Figs. 3C and 3D).
Cold and hot microbiota of the tongue-coating microbiome
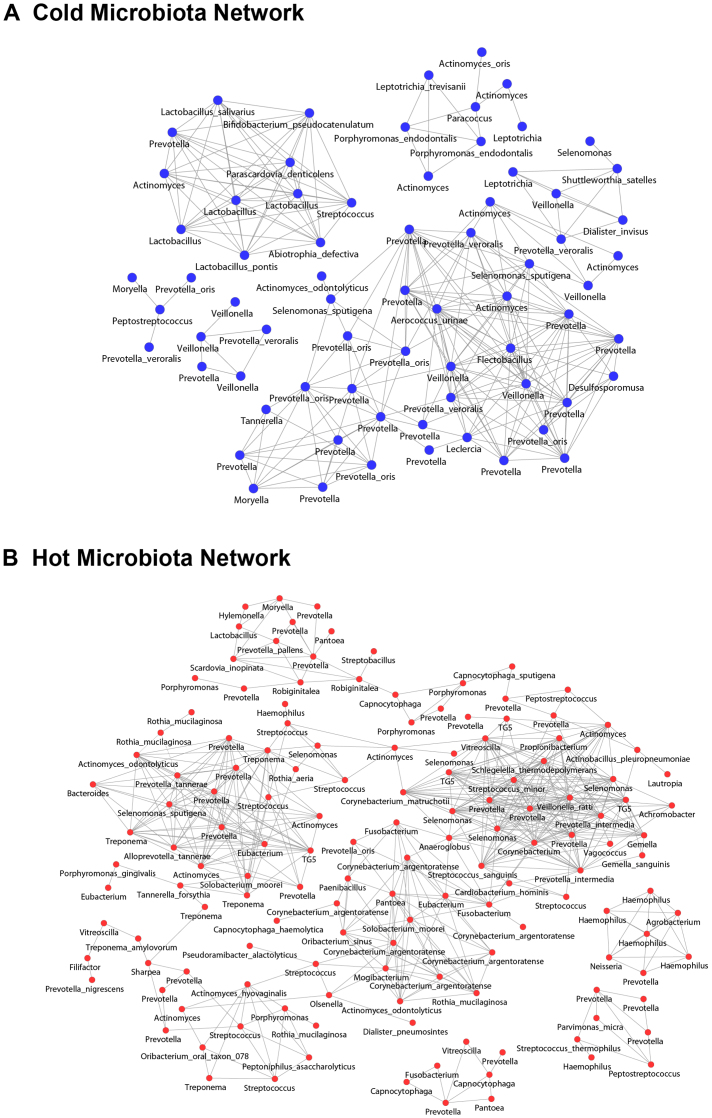
The above analyses showed that there is information in the microbial composition that can discriminate Hot Syndrome from Cold Syndrome. We therefore adopted a supervised analysis approach to further study the phylotypes that could distinguish the tongue-coating microbiomes of the Cold and Hot Syndrome patients. We employed the DEGseq tool40, which was originally designed to find differentially expressed genes with RNA-seq read counts. First, we compared the sequence abundances of all of the 6,103 species-level OTUs between the normal controls and gastritis patients. A total of 755 species-level OTUs showed significant differential abundance between the two groups, with p-value<0.001 (FDR<0.005). The OTUs that were represented in fewer than three samples were then screened, which included 40 OTUs. Among the remaining 715 OTUs, 483 were abundant in study patients, and 232 were abundant in controls. Among the 483 abundant OTUs in study patients, we identified 381 that were significantly different between patients with Cold and Hot Syndromes by comparing their abundances in the two patient groups. A total of 123 OTUs were highly enriched in Cold Syndrome patients, and 258 OTUs were highly enriched in Hot Syndrome patients. These “Cold OTUs” and “Hot OTUs represented the “Cold Microbiota” and “Hot Microbiota”, respectively. The other 102 OTUs that distinguished patients from controls did not show significant differences between patients with Cold and Hot Syndromes; these are referred to as “Shared OTUs”. The 123 Cold OTUs were classified to taxonomy by the GAST classifier33. Among them, 83 Cold OTUs were classified into 24 genera, including 28 Cold OTUs more precisely classified into 16 species. Similarly, among the 258 Hot OTUs, 170 were classified into 50 genera, including 57 Hot OTUs classified into 37 species. Some of the genera and species identified in the Cold and Hot OTUs overlapped. As shown in Fig. 4, we constructed two co-occurrence networks with a Pearson Correlation coefficient > 0.85 of 83 genus-classifiable OTUs in the Cold Microbiota and 170 genus-classifiable OTUs in the Hot Microbiota, illustrating potential relationships among the two groups of microbiota presented in patients with Cold and Hot Syndromes, respectively. In total, the 251 Cold or Hot OTUs were classified into 61 genera, including 85 Cold or Hot OTUs more precisely classified into 49 species (Table S3).
Figure 4. Tongue microbiota-imbalanced networks of genus-classifiable OTUs in (A) the Cold Microbiota and (B) the Hot Microbiota.
OTUs as nodes are marked with the genus names (some with more precise species names), and edges with Pearson's correlation coefficient (PCC) > 0.85 are indicated.
Discussion
In the long history of traditional clinical practice in China and other Eastern countries, TCM practitioners have typically classified patients of the same disease into sub-groups as different Syndromes from a holistic perspective on patients' overall status. Many features are used in the discrimination of Syndromes, but the tongue-coating appearance is one of the major factors. It has been understood that the human microbiome is related to human health status and many diseases41,42. We hypothesized that the tongue-coating appearance may reflect characteristics of the tongue microbiome, which is associated with the status of the human body. To test this hypothesis, we conducted a next-generation sequencing analysis of tongue-coating microbiome samples from 9 Cold Syndrome patients with a white-greasy tongue coating, 10 Hot Syndrome patients with a yellow-dense tongue coating, and 8 healthy cases with a normal tongue coating. We designed an Illumina paired-end with double-barcode (Ipedb) protocol to sequence the 16S rRNA V6 regions of the microbiome, which was more accurate and had deeper coverage than existing methods.
Compared with the available healthy tongue dorsa microbiomes43,44,45,46, we found that although different 16S rRNA library construction techniques and data analysis methods were used, the results from our normal control samples and those of previous studies43,44,45,46 showed consistent phyla catalogues, namely Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria, as the main parts of the tongue microbiome. Moreover, both our study and others reported abundance information44,46 showing that Firmicutes, Proteobacteria and Bacteroidetes were the most abundant phyla, followed by Fusobacteria and Actinobacteria in healthy tongue dorsa microbiomes. The only difference is the increased level of Actinobacteria and decreased level of Bacteroidetes in three samples44 compared with our results. The comparison results demonstrate the reliability and consistency of our study in terms of healthy tongue microbiome.
Statistical analysis of the obtained sequences demonstrated the potential connection between the tongue-coating microbiome and traditional tongue diagnosis. Two subtypes of tongue-coating microbiomes were identified in gastritis patients who correspond well to the Cold and Hot Syndromes. We identified 715 species-level OTUs in the tongue-coating microbiome of chronic atrophic gastritis patients who were different in abundance compared to the controls and were further classified into Cold, Hot or Shared OTUs. A total of 251 of these OTUs were assigned to 61 genera and 49 species, implying the potential of using these 61 genera and 49 species as biomarkers for the status of patients (Table S4). Because the oral cavity is at the forefront of the alimentary and respiratory tracts, it is reasonable that the tongue-coating microbiome might reflect the status of the gastrointestinal and respiratory ecosystems.
We further explored the function of the OTUs and the possible relationship between the Cold and Hot OTUs and body symptoms in human diseases by studying these genera or species in the literature; the functions of Cold OTUs are summarised in Table 2, and those of Hot OTUs are in Table 3. Interestingly, the functional role of Hot OTUs can clearly explain Hot Syndrome. Hot OTUs are closely associated with Hot Syndrome, namely, the well-accepted concept “Shang-Huo” in Chinese, a condition manifested as inflammation or irritability in some parts of body. For example, 4 Hot OTUs identified as Alloprevotella tannerae and Solobacterium moorei were responsible for halitosis or bad breath in the oral cavity79,80. Six Hot OTUs identified as Corynebacterium argentoratense were regarded as a pathogen of tonsillitis81. Twenty Hot OTUs were identified as Prevotella nigrescens, Capnocytophaga sputigena, and others, which are pathogens of periodontal diseases or highly abundant in patients suffering from periodontal disease (see references in Table 3). In contrast, 1 Cold OTU was identified as Actinomyces oris, which was not highly abundant in periodontitis patients53. Only 2 Cold OTUs identified as Porphyromonas endodontalis have been reported as possibly pathogenic in periodontitis52. Therefore, tongue-microbiome analysis provides new findings that effectively explain Hot Syndrome in traditional Chinese medicine.
Table 2. Summary of functions of Cold OTUs.
| #OTUs | Taxonomy Assignment | Function | References |
|---|---|---|---|
| 3 | Lactobacillus sp. | lactic acid bacteria | 47 |
| 1 | Lactobacillus pontis | ||
| 1 | Lactobacillus salivarius | ||
| 1 | Streptococcus sp. | ||
| 1 | Streptococcus anginosus | ||
| 1 | Lactobacillus salivarius | probiotics | 48 |
| 1 | Bifidobacterium pseudocatenulatum | 49 | |
| 8 | Veillonella sp. | metabolizing lactic acid | 50 |
| 1 | Parascardovia denticolens | dental caries | 51 |
| 2 | Porphyromonas endodontalis | periodontical diseases (gingivitis or periodontitis) | 52 |
| 1 | Actinomyces oris (lowly-abundant) | 53 | |
| 1 | Dialister invisus | endodontic infections | 54 |
| 1 | Shuttleworthia satelles | 55 | |
| 6 | Prevotella oris | bacteremia and endocarditis | 56 |
| 1 | Leptotrichia trevisanii | 57 | |
| 1 | Abiotrophia defectiva | 58 | |
| 1 | Shuttleworthia satelles | 59 | |
| 2 | Actinomyces odontolyticus | 60 | |
| 1 | Peptostreptococcus sp. | infection in/on all body sites | 61 |
| 1 | Aerococcus urinae | urinary tract infection | 62 |
Table 3. Summary of functions of Hot OTUs.
| #OTUs | Taxonomy Assignment | Function | References |
|---|---|---|---|
| 1 | Lactobacillus sp. | lactic acid bacteria | 47 |
| 7 | Streptococcus sp. | ||
| 1 | Streptococcus thermophilus | ||
| 1 | Streptococcus minor | ||
| 1 | Vagococcus sp. | ||
| 1 | Anaeroglobus sp. | dental caries | 63 |
| 1 | Pseudoramibacter alactolyticus | 64 | |
| 1 | Scardovia inopinata | 65 | |
| 1 | Treponema amylovorum | 66 | |
| 1 | Prevotella nigrescens | periodontical diseases (gingivitis or periodontitis) | 67 |
| 2 | Capnocytophaga sputigena | 68 | |
| 1 | Desulfomicrobium orale | 69 | |
| 1 | Treponema socranskii | 70 | |
| 3 | Fusobacterium sp. | 71 | |
| 2 | Porphyromonas gingivalis | 72,73 | |
| 1 | Streptococcus sanguinis | ||
| 1 | Tannerella forsythia | ||
| 2 | Prevotella intermedia | ||
| 2 | Mogibacterium sp. | 74 | |
| 1 | Olsenella sp. | ||
| 1 | Dialister pneumosintes | 75 | |
| 2 | Parvimonas micra | 76 | |
| 3 | Capnocytophaga sp. | dental plaque | 77 |
| 2 | Capnocytophaga sputigena | ||
| 1 | Capnocytophaga haemolytica | ||
| 1 | Rothia aeria | degrading gluten | 78 |
| 6 | Rothia mucilaginosa | ||
| 2 | Alloprevotella tannerae | halitosis | 79 |
| 2 | Solobacterium moorei | 80 | |
| 6 | Corynebacterium argentoratense | tonsillitis | 81 |
| 2 | Prevotella oris | bacteremia and endocarditis | 56 |
| 1 | Cardiobacterium hominis | 82 | |
| 1 | Gemella sanguinis | 83 | |
| 1 | Capnocytophaga haemolytica | 84 | |
| 2 | Actinomyces odontolyticus | 60 | |
| 2 | Peptostreptococcus sp. | infection in/on all body sites | 61 |
| 1 | Agrobacterium sp. | opportunistic infections | 85 |
As shown in Table 2 and Table 3, the potential functions of the Cold and Hot Microbiota show some consistency with the metabolism-immune imbalance underlying Cold and Hot Syndrome as revealed in our previous studies4,5. For example, three dominant genera in the gastric microbiome of gastric cancer patients, namely, Lactobacillus, Veillonella and Streptococcus, exhibited different patterns in Cold and Hot OTUs. In addition, some Hot OTUs were assigned to Solobacterium moorei, Corynebacterium argentoratense, Prevotella nigrescens, and other species. Pathological studies indicate that these bacteria are associated with TCM Hot Syndrome-related inflammation or irritability such as halitosis, tonsil infection, and periodontal inflammation (Table 3). Each group of Cold or Hot OTUs also has a potential symbiotic relationship and forms a microbiota-imbalanced network (Fig. 4). Taken together, we conclude that the abnormal appearances of the tongue coating as observed by TCM tongue diagnosis actually reflect the imbalanced tongue-coating microbiome in the entire human ecosystem. Therefore, this is the first study to suggest that the TCM diagnosis of Cold and Hot Syndromes based on tongue-coating appearance successfully detects signals in the tongue-coating microbiomes that are indicators of the entire body's status.
Other available factors, such as gender and Helicobacter pylori infection, did not significantly affect the microbiome features. Although it had been well established that the bacterium Helicobacter pylori is closely related to gastritis and gastric ulcers86 and that Helicobacter pylori could be detected in the saliva and supragingival and subgingival plaque of periodontitis patients87, we did not detect the bacterium on tongue coatings of either Helicobacter pylori-positive or -negative samples. This result is consistent with a recent study that found that the oral cavity is not a reservoir for Helicobacter pylori in patients with epigastric pain syndrome88.
We acknowledge that there is a limitation in the present work in that the age of normal controls did not match that of study patients, and more investigation into the impact of age on the tongue microbiome is required in future work. Nevertheless, we believe that the major conclusions of the current study are still valid because of the following: first, youth are more likely to form a healthy tongue coating and thus serve as a good control against abnormal tongue appearances; second, our results for normal controls are in accordance with a healthy tongue dorsa microbiome for people between 18 and 40 years of age46; and third, both the sex and the ages of patients with Cold and Hot Syndromes were well matched in this study.
In summary, by studying the tongue-coating microbiomes of chronic atrophic gastritis patients and healthy controls using next-generation sequencing and bioinformatics analysis, we revealed the possible mechanism underlying the traditional tongue diagnosis and highlighted the potential for using tongue coating microbiome features as biomarkers to reflect the holistic status of the human body, especially in gastrointestinal diseases. The current sample size of this study is still small, and further investigations with more samples and complete metagenome sequencing are needed for a comprehensive understanding of the system. The observations described here might provide a new direction for understanding the biological basis of TCM tongue diagnosis and the link between the “geographic tongue” and the human body's health status observed in modern Western medicine.
Methods
Tongue-coating samples
This study was approved by the Medical Ethical Committee of the Beijing Dongzhimen TCM Hospital. All patients and healthy volunteers signed informed consent forms. The 19 gastritis patients were enrolled from Beijing Dongzhimen TCM Hospital and Xiyuan TCM Hospital; 9 of them were diagnosed with Cold Syndrome, and 10 of them were diagnosed with Hot Syndrome. Nine normal control samples were drawn from healthy volunteers with no stomach discomfort at Tsinghua University. All healthy volunteers reported no stomach discomfort in the past 5 years and exhibited a normal tongue coating as judged by TCM doctors. Samples with medical histories that included the use of glucocorticoids and antibiotics and a history of smoking within the last 3 months were excluded. All patients were tested for Helicobacter pylori infection using the fast urea test and histology, and all healthy volunteers were tested using the 13C urea breath test. All tongue coatings were sampled and photographed in the morning prior to breakfast, and then the patients were examined by gastric endoscopy. All participants were required to brush their teeth before the sampling to rinse possible food contamination that might influence the tongue microbiome. One of the nine normal control samples was withdrawn from the study due to an abnormal passing rate (~30%) in data preprocessing, resulting in a final study group of 8 controls and 19 study patients.
Tongue-imaging analysis
We used the DS01-B tongue manifestation acquisition instrument by DAOSH Co., Shanghai, China, to photograph the tongue coatings of patients and healthy volunteers (Fig. 1B). A 400×400-pixel image was cut from the centre of each tongue coating image, where the “spleen-stomach” conditions is reflected in the traditional tongue diagnosis, by a certified and experienced TCM doctor. The area is regarded to be associated with conditions of “spleen-stomach” in the TCM tongue diagnosis. A total of 18 digital features were extracted from every 400×400-pixel image in two steps. First, we listed nine colour variables in three common three-dimensional colour spaces for each pixel. These variables are hue, saturation and brightness in the HSB colour space; x, y and Y in the CIExyY colour space; and L, a and b in the CIELab colour space. Second, we summarised the mean and the variance of nine colour variables among 400×400 pixels, resulting in 18 digital features of the image. Finally, Principal Component Analysis (PCA) was performed on the 18 digital features of the 27 images.
16S rRNA sequencing
DNA samples were extracted using Qiagen extraction kits (Qiagen, Hilden, Germany). Isolated DNA was used as a template for the amplification of the V6 region of 16S rRNA. All samples were amplified in 50 µl PrimerStar HS Premix (Takara, Dalian, China) that contained 2.5 µl template DNA and a final primer concentration of 0.4 µM. In the first round of PCR amplification for extracting V6 regions, the following parameters were used: 94°C for 5 min; then 16 cycles of 94°C for 45 sec, 52°C for 45 sec, 72°C for 60 sec, and, finally, 72°C for 10 min. In the second round of PCR amplification to introduce Illumina sequencing adapters, the following parameters were used: 94°C for 5 min; then 16 cycles of 94°C for 45 sec, 56°C for 45 sec, 72°C for 60 sec; and, finally, 72°C for 10 min. Products were cleaned up with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Eight healthy samples, 9 Cold Syndrome samples and 10 Hot Syndrome samples were then sequenced on one flow cell (seven lanes and one phiX control lane) with a read length of 2×120 bp using Illumina's Genome Analyzer IIx and the Ipedb protocol that we developed. Image analysis and base calling were completed using Real Time Analysis (RTA) v1.8. The details of the Ipedb protocol are described in the Supplementary Materials.
Data preprocessing
Data preprocessing was carried out in a six-step pipeline. (1) Raw paired-end reads were removed if they contained ambiguous base calling N(s). (2) The remaining paired-end reads were assembled into sequences by overlapping their forward and reverse ends. (3) We trimmed barcodes from assembled sequences and used them to assign sequences to different samples. In this step, sequences survived if their barcodes at both ends indicated the same samples. (4) We trimmed linkers and primers from sequences to obtain V6 tags. (5) V6 tags with low-quality scores were screened. The quality score of each base in a sequence was the larger that in the forward end and the reverse end. The minimum score of all bases in the V6 tags was >Q30, and the average score of all bases in the V6 tags was >Q38. (6) UCHIME89 was used to detect chimeras from high-quality V6 tags. Finally, we acquired high-quality, non-chimeric V6 tags from raw paired-end reads.
Sequencing data analysis and statistical approaches
These high-quality, non-chimeric sequences were assigned to taxonomy by the GAST rRNA classifier33 and were clustered into OTUs at different genetic distances (0%, 3%, 5% and 10%) by CROP34, an unsupervised Bayesian clustering algorithm. We then used Mothur90 to implement a series of OTU-based analyses of the alpha and beta diversities of the microbial communities. In the alpha diversity analysis, three richness estimators – the observed OTU number, Chao191 and ACE92 – and two evenness estimators – the Shannon Index93 and Simpson Index94 – were calculated (Fig. S4). Rarefaction curves of observed OTU numbers on sampling depths were drawn with 10,000 sequences per step (Fig. 2). In the beta diversity analysis, a Jaccard membership dissimilarity estimator35 and a Bray-Curtis structure dissimilarity estimator36 were calculated. The phylogenetic-based Unifrac distance38 was also analyzed. Principle Coordinate Analysis, or PCoA3,9 was used to visualise the beta diversity. In addition, AMOVA37 was carried out by Mothur90 to test the hypothesis that dissimilarities between samples from the same groups are significantly different compared to those from different groups. For each species-level OTU, a negative binomial test was performed using DEGSeq40 to test whether the OTU was differentially abundant between healthy controls and chronic atrophic gastritis patients.
Author Contributions
BJ, JL, RJ and TC conducted data analysis and wrote manuscript. XL, YC, TM and LL conducted sample collection, microbiome experiments and wrote manuscript. SL and XZ conceived and designed the studies, analyzed the data and wrote manuscript.
Supplementary Material
SUPPLEMENTARY
Acknowledgments
We thank Drs. Yongan Ye and Zhihong Li at Dongzhimen Hospital, Beijing University of Chinese Medicine and Dr. Zhenhua Li at Xiyuan hospital, China Academy of Traditional Chinese Medicine for their helps in patient sample collection. We thank volunteers in our lab for their contributions in normal sample collection. We thank Dr. Jin Gu and Fuqiu Wang for their help in the imaging analysis. We thank Xiaolin Hao for helping to ran CROP on the data set, and thank Profs. Steven Brenner and Fengzhu Sun for their useful suggestions. This work is supported in part by the National Basic Research Program of China (2012CB316504), NSFC grants 81225025, 90709013, 61021063 and 60928007, Basic Research Funds of TNLIST and NSF 1043075.
References
- Chiu C. C. A novel approach based on computerized image analysis for traditional Chinese medical diagnosis of the tongue. Comput. Methods Programs Biomed. 61, 77–89 (2000). [DOI] [PubMed] [Google Scholar]
- State Administration of Traditional Chinese Medicine (SATCM) of The People's Republic of China. The Criteria of Diagnosis and Therapeutic Effect of Diseases and Zhengs in TCM. Nanjing University Press (1994) (in Chinese). [Google Scholar]
- Hsu C. H., Yu M. C., Lee C. H., Lee T. C. & Yang S. Y. High eosinophil cationic protein level in asthmatic patients with Heat Zheng. Am. J. Chin. Med. 31, 277–283 (2003). [DOI] [PubMed] [Google Scholar]
- Li S. et al. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst. Biol. 1, 51–60 (2007). [DOI] [PubMed] [Google Scholar]
- Ma T. et al. Bridging the gap between traditional Chinese medicine and systems biology: the connection of Cold Syndrome and NEI network. Molecular BioSystems 6, 613–619 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang D., Pang B., Li N., Wang K. & Zhang H. Computerized diagnosis from tongue appearance using quantitative feature classification. Am. J. Chinese Med. 33, 859–866 (2005). [DOI] [PubMed] [Google Scholar]
- Dong H. et al. Quantitative analysis on tongue inspection in traditional Chinese medicine using optical coherence tomography. J. Biomed. Opt. 13, 11004 (2008). [DOI] [PubMed] [Google Scholar]
- Liu Q., Yue X. Q., Deng W. Z. & Ren R. Z. Quantitative study on tongue color in primary liver cancer patients by analysis system for comprehensive information of tongue diagnosis. Journal of Chinese Integrative Medicine 1, 180–183 (2003) (In Chinese). [DOI] [PubMed] [Google Scholar]
- Xu J. T. et al. Preliminary Observation of Relationship between Feature of Tongue Manifestations and Syndrome Patterns in 166 Cases of Chronic Gastritis. Journal of Shanghai University of Traditional Chinese Medicine 17, 34–37 (2003) (In Chinese). [Google Scholar]
- Li C. D., Lan Q. F. & Zhang J. J. Study on relationship between tongue picture and cell apoptosis in patients with chronic gastritis. Journal of Chinese Integrative Medicine 23, 433–435 (2003) (In Chinese). [PubMed] [Google Scholar]
- Ship J. A., Phelan J. & Kerr R. A. Biology and Pathology of the Oral Mucosa. In Fitzpatrick's Dermatology in General Medicine, Chapter 112. McGraw-Hill Professional Publishing (2003). [Google Scholar]
- Zadik Y., Drucker S. & Pallmon S. Migratory stomatitis (ectopic geographic tongue) on the floor of the mouth. J. Am. Acad. Dermatol. 65, 459–460 (2011). [DOI] [PubMed] [Google Scholar]
- Anastasi J. K., Currie L. M. & Kim G. H. Understanding diagnostic reasoning in TCM practice: tongue diagnosis. Alternative Therapies in Health and Medicine 15, 18–28 (2009). [PubMed] [Google Scholar]
- Abe S., Ishihara K., Adachi M. & Okuda K. Tongue-coating as risk indicator for aspiration pneumonia in edentate elderly. Arch. Gerontol. Geriatr. 47, 267–275 (2008). [DOI] [PubMed] [Google Scholar]
- Hoefert S., Schilling E., Philippou S. & Eufinger H. Amyloidosis of the tongue as a possible diagnostic manifestation of plasmacytoma. Mund Kiefer Gesichtschir 3, 46–49 (1999) (in German). [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Consortium. . A framework for human microbiome research. Nature 484, 215–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N. et al. Composition of the adult digestive tract microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biology 13, R42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik E. M. et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Nat. Acad. Sci. 103, 732–737 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson H. E. et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5, e9836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. & Gordon J. I. The core gut microbiome, energy balance and obesity. J. Physiol. 587, 4153–4158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H. F. & Lamont R. J. Oral microbial communities in sickness and in health. Trends. Microbiol. 13, 589–595 (2005). [DOI] [PubMed] [Google Scholar]
- Avila M., Ojcius D. M. & Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28, 405–411 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F. & Malagelada J. R. Role of bacteria in experimental colitis. Best Pract. Res. Clin. Gastroenterol. 17, 793–804 (2003). [DOI] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S. & Gordon J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006). [DOI] [PubMed] [Google Scholar]
- Cani P. D. & Delzenne N. M. Gut microflora as a target for energy and metabolic homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 10, 729–734 (2007). [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J. & Gordon J. I. An invitation to the marriage of metagenomics and metabolomics. Cell 134, 708–713 (2008). [DOI] [PubMed] [Google Scholar]
- Dixon M. F., Genta R. M., Yardley J. H. & Correa P. Classification and grading of gastritis. Am. J. Surg. Pathol. 20, 1161–1181 (1996). [DOI] [PubMed] [Google Scholar]
- Stolte M. & Meining A. The updated Sydney system: classification and grading of gastritis as the basis of diagnosis and treatment. Can. J. Gastroenterol. 15, 591–598 (2001). [DOI] [PubMed] [Google Scholar]
- Dohm J. C., Lottaz C., Borodina T. & Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic. Acids Res. 36, e105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S. M., Huber J., Morrison H., Sogin M. & Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8, R143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S. M. et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4, e1000255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X. L., Jiang R. & Chen T. Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering. Bioinformatics 27, 611–618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. Comparative study of the distribution in the floral portion of the Alps and the Jur. Bulletin de la Societe Vaudoise des Sciences Naturelles 37, 547–579 (1901). [Google Scholar]
- Bray J. R. & Curtis J. T. An ordination of upland forest communities of southern Wisconsin. Ecological Monographs 27, 325–349 (1957). [Google Scholar]
- Martin A. P. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68, 3673–3682 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Lozupone C. & Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower J. C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338 (1966). [Google Scholar]
- Wang L. K., Feng Z. X., Wang X., Wang X. & Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- Nagalingam N. A. & Lynch S. V. Role of the microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 18, 968–984 (2012). [DOI] [PubMed] [Google Scholar]
- McGarr S. E., Ridlon J. M. & Hylemon P. B. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J. Clin. Gastroenterol. 39, 98–109 (2005). [PubMed] [Google Scholar]
- Mitchell P. M. et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 41, 558–63 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaura E., Keijser B. J., Huse S. M. & Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 9, 259 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S. M., Ye Y., Zhou Y. & Fodor A. A. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One. 7, e34242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N. et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 13, R42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K. H. & Ludwig W. Phylogenetic relationships of lactic acid bacteria. In The Lactic Acid Bacteria: The Genera of Lactic Acid Bacteria, Chapter 2. Springer (1995). [Google Scholar]
- Mayanagi G. et al. Probiotic effects of orally administered Lactobacillus salivarius WB21-containing tablets on periodontopathic bacteria: a double-blinded, placebo-controlled, randomized clinical trial. J. Clin. Periodontol. 36, 506–513 (2009). [DOI] [PubMed] [Google Scholar]
- Mayo B., & van Sinderen D. (editors) Bifidobacteria: Genomics and Molecular Aspects. Caister Academic Press (2010). [Google Scholar]
- Periasamy S. & Kolenbrander P. E. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J. Bacteriol. 192, 2965–2972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton D. et al. Isolation and identification of bifidobacteriaceae from human saliva. Appl. Environ. Microbiol. 74, 6457–6460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo Bedranm T. B. et al. Porphyromonas endodontalis in chronic periodontitis: a clinical and microbiological cross-sectional study. J. Oral. Microbiol. 4, 3402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López R., Dahlén G., Retamales C. & Baelum V. Clustering of subgingival microbial species in adolescents with periodontitis. European Journal of Oral Sciences. 119, 141–150 (2011). [DOI] [PubMed] [Google Scholar]
- Siqueira J. F. Jr. & Rôças I. N. Uncultivated phylotypes and newly named species associated with primary and persistent endodontic infections. J. Clin. Microbiol. 43, 3314–3319 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto R. C., Gomes B. P., Desai M., Rajendram D. & Shah H. N. Bacterial examination of endodontic infections by clonal analysis in concert with denaturing high-performance liquid chromatography. Oral Microbiol. Immunol. 22, 403–410 (2007). [DOI] [PubMed] [Google Scholar]
- Bein T., Brem J. & Schüsselbauer T. Bacteremia and sepsis due to Prevotella oris from dentoalveolar abscesses. Intensive Care Med. 29, 856 (2003). [DOI] [PubMed] [Google Scholar]
- Cooreman S. et al. Bacteraemia caused by Leptotrichia trevisanii in a neutropenic patient. Anaerobe 17, 1–3 (2011). [DOI] [PubMed] [Google Scholar]
- Kalavakunta J. K. et al. Destructive Abiotrophia defectiva endocarditis. J. Heart Valve Dis. 20, 111–112 (2011). [PubMed] [Google Scholar]
- Shah N. B. et al. Shuttleworthia satelles endocarditis: evidence of non-dental human disease. J. Infect. 60, 491–493 (2010). [DOI] [PubMed] [Google Scholar]
- Cone L. A., Leung M. M. & Hirschberg J. Actinomyces odontolyticus bacteremia. Emerg. Infect. Dis. 9, 1629–1632 (2003). [DOI] [PubMed] [Google Scholar]
- Finegold S. M. Anaerobic Bacteria in Human Disease. Academic Press (1977). [Google Scholar]
- Sierra-Hoffmana M., Watkinsa K., Jinadathaa C., Faderb R. & Carpentera J. L. Clinical significance of Aerococcus urinae: a retrospective review. Diagn. Microbiol. Infect. Dis. 53, 289–292 (2005). [DOI] [PubMed] [Google Scholar]
- Chugal N. et al. Molecular characterization of the microbial flora residing at the apical portion of infected root canals of human teeth. J. Endod. 37, 1359–1364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preza D. et al. Bacterial profiles of root caries in elderly patients. J. Clin. Microbiol. 46, 2015–2021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesto M., Biavati B. & Mattarelli P. Occurrence of the family bifidobacteriaceae in human dental caries and plaque. Caries. Res. 40, 271–276 (2006). [DOI] [PubMed] [Google Scholar]
- Montagner F., Jacinto R. C., Signoretti F. G. & Gomes B. P. Treponema species detected in infected root canals and acute apical abscess exudates. J. Endod. 36, 1796–1799 (2010). [DOI] [PubMed] [Google Scholar]
- Stingu C. S. Association of periodontitis with increased colonization by Prevotella nigrescens. J. Investig. Clin. Dent. 10.1111/j.2041–1626.2012.00129.x (2012). [DOI] [PubMed] [Google Scholar]
- Teles F. R. et al. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J. Periodontal. Res. 47, 95–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langendijk P. S., Kulik E. M., Sandmeier H., Meyer J. & van der Hoeven J. S. Isolation of Desulfomicrobium orale sp. nov. and Desulfovibrio strain NY682, oral sulfate-reducing bacteria involved in human periodontal disease. Int. J. Syst. Evol. Microbiol. 51, 1035–1044 (2001). [DOI] [PubMed] [Google Scholar]
- Takeuchi Y. et al. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J. Periodontol. 72, 1354–1363 (2001). [DOI] [PubMed] [Google Scholar]
- Ohkusa T. et al. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut 52, 79–83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaranayake L. P. Microbiology of periodontal disease. In Essential Microbiology for Dentistry, 275–285. Elsevier (2006a). [Google Scholar]
- Samaranayake L. P. Normal oral flora, the oral ecosystem and plaque biofilm. In Essential Microbiology for Dentistry, 255–266. Elsevier (2006b). [Google Scholar]
- Sato T. et al. Cultivable anaerobic microbiota of infected root canals. Int. J. Dent. 609689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima C. T. F., Gornic C., Sampaio A. B., Marques R. J. P. & Vieira A. P. C. Detection of Dialister pneumosintes in the subgingival biofilm of subjects with periodontal disease. Anaerobe 13, 244–248 (2007). [DOI] [PubMed] [Google Scholar]
- Moore H. L. V., Johnson J. L. & Moore W. E. C. The genus Peptostreptococcus. Bergey's Manual of Systematic Bacteriology 2, 1090–1091 (1986). [Google Scholar]
- Yamamoto T., Kajiura S., Hirai Y. & Watanabe T. Capnocytophaga haemolytica sp. nov. and Capnocytophaga granulosa sp. nov., from human dental plaque. Int. J. Syst. Bacteriol. 44, 324–329 (1994). [DOI] [PubMed] [Google Scholar]
- Zamakhchari M. et al. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS One 6, e24455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell K. L., Citron D. M., Warren Y. A., Nachnani S. & Goldstein E. J. Anaerobic bacteria cultured from the tongue dorsum of subjects with oral malodor. Anaerobe 9, 243–246 (2003). [DOI] [PubMed] [Google Scholar]
- Kazor C. E. et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41, 558–563 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegel P. et al. Corynebacterium argentoratense sp. nov., from the human throat. Int. J. Syst. Bacteriol. 45, 533–537 (1995). [DOI] [PubMed] [Google Scholar]
- Malani A. N., Aronoff D. M., Bradley S. F. & Kauffman C. A. Cardiobacterium hominis endocarditis: two cases and a review of the literature. Eur. J. Clin. Microbiol. 25, 587–595 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. K., Tak T., Haselby R. C., McCauley C. S. Jr. & Reed K. D. Second case of infective endocarditis caused by Gemella sanguinis. WMJ. 101, 37–39 (2002). [PubMed] [Google Scholar]
- Gutierrez-Martin M. A. et al. Aortic valve endocarditis by Capnocytophaga haemolytica. Ann. Thorac. Surg. 84, 1008–1010 (2007). [DOI] [PubMed] [Google Scholar]
- Hulse M., Johnson S. & Ferrieri P. Agrobacterium infections in humans: experience at one hospital and review. Clin. Infect. Dis. 16, 112–117 (1993). [DOI] [PubMed] [Google Scholar]
- Marshall B. J. The discovery that Helicobacter pylori, a spiral bacterium, caused peptic ulcer disease. In Helicobacter Pioneers: Firsthand Accounts From The Scientists Who Discovered Helicobacters, 165–202. Blackwell (2002). [Google Scholar]
- Gebara E. C. et al. Prevalence of Helicobacter pylori detected by polymerase chain reaction in the oral cavity of periodontitis patients. Oral Microbiology and Immunology 19, 277–280 (2004). [DOI] [PubMed] [Google Scholar]
- Rossi-Aguiar S. V. P. et al. Oral cavity is not a reservoir for Helicobacter pylori in infected patients with functional dyspepsia. Oral Microbiology and Immunology. 24, 255–259 (2009). [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270 (1984). [Google Scholar]
- Chao A. & Lee S. M. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87, 210–217 (1992). [Google Scholar]
- Charles K. Ecological Methodology. HarperCollins (1989). [Google Scholar]
- Simpson E. H. Measurement of diversity. Nature 163, 688 (1949). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY