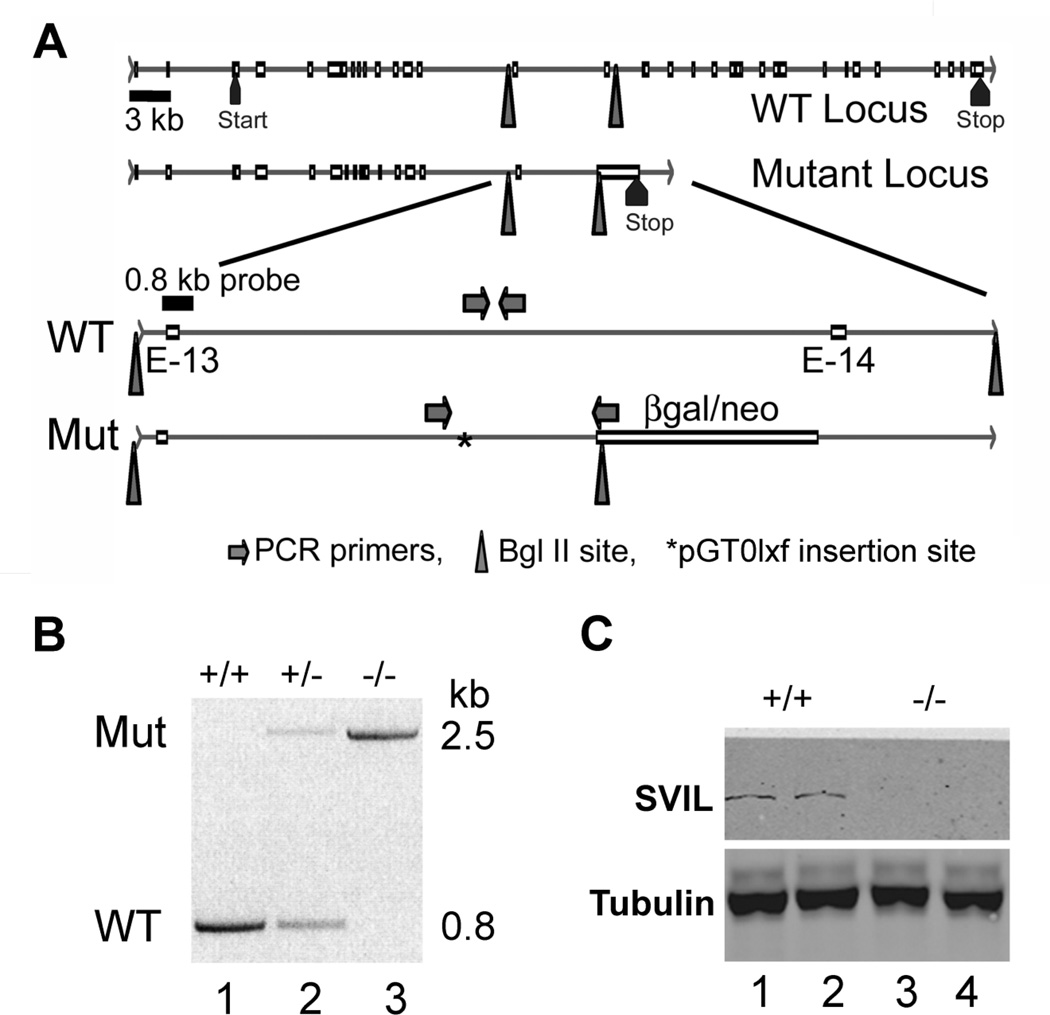
Figure 5.
Disruption of the Svil gene and loss of Svil expression in platelets. (A) Diagram of the wild-type (WT) and mutated (Mut) Svil loci (top 2 lines) with enlargements (bottom 2 lines) showing the insertion site (*) of the pGT0lxf vector sequence within the intron between coding exons 13 (E-13) and 14 (E-14). Also shown are the locations of PCR primer sets diagnostic for the wild-type and mutant loci (arrows), the 0.8-kb probe used for Southern analyses, and the Bgl II sites (triangles) associated with endogenous genomic DNA and with the inserted β-galactosidase-neomycin (β-gal/neo) coding sequence. (B) PCR of genomic DNA from mouse tails using the primer sets shown in Panel A. Primers specific for the insertion identify a ~2.5-kb product (Mut, top) while primers specific for the wild-type allele generate a 0.8-kb product (WT, below). (C) Immunoblot of mouse platelets with anti-supervillin antibody (H340) and anti-tubulin as a loading control.