Abstract
MicroRNAs (miRNAs) are a class of short endogenous non-coding RNAs regulating gene expression in many biological processes, including proliferation, apoptosis and differentiation. The deregulation of miRNA expression is believed to be an important regulator of tumor development and progression of thyroid cancer. In this review, we discussed important roles and expression profiles of miRNA in differentiated thyroid cancer (DTC) as well as the promising implication in clinical practice.
Keywords: Differentiated thyroid cancer, microRNA, mutations, fine needle aspiration biopsy
Introduction
MicroRNAs (miRNAs) represent a class of short endogenous non-coding RNAs regulating gene expression at mRNA post-transcriptional level in many biological processes, including proliferation, apoptosis, and differentiation [1]. These 19-23 nucleotide (nt) RNAs are able to block translation or degradation of target mRNA through complementary binding to the 3’-untranslated region (UTR) of mRNAs. Mi-RNA genes were firstly transcribed by RNA polymerase II (PloII) into a long hairpin structure known as primary miRNA (pri-miRNA). Subsequently, pri-miRNA is processed by Drosha and DGCR8 to form about 70-nucleotide-long precursor miRNA (pre-miRNA). After exported into cytoplasm, pre-miRNA is cleaved by endonuclease enzyme Dicer into mature miRNA. Mature miRNA is loaded into RNA-induced silencing complex (RISC), which results in cleavage or translational repression of target mRNA. (See Figure 1).
Figure 1.
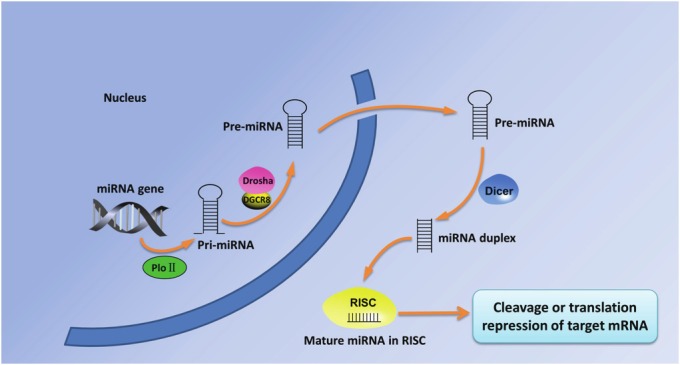
The biogenesis of miRNAs. Mi-RNA genes were transcribed by RNA polymerase II (PloII) into primary miRNA (pri-miRNA). Subsequently processed by Drosha and DGCR8, pri-miRNA forms precursor miRNA (pre-miRNA) and is exported into cytoplasm. In the cytoplasm, pre-miRNA is cleaved by endonuclease enzyme Dicer into mature miRNA. Mature miRNA is loaded into RNA-induced silencing complex (RISC), which results in cleavage or translational repression of target mRNA.
Increasing evidence has revealed the involvement of mi-RNA in human malignancies. The deregulation of miRNA expression is believed to be an important regulator of tumor development and progression. Due to its repression effect, deregulation of specific mi-RNA could lead to the repression of tumor suppressor gene and/or increase of oncogene expression. Consequently, these molecular changes favor cell proliferation, differentiation and apoptosis. MicroRNA expression profiling of human tumors has identified signatures associated with diagnosis, staging, prognosis, and response to treatment [2]. MiRNA expression profiles resulted in being different not only between tumors and normal tissues but also between different subtypes of tumors and between primary tumors and metastatic tumors. In this review, we discussed important roles and different expression profiles of miRNA in differentiated thyroid cancer as well as the promising implication in clinical practice.
MicroRNA expression profiles in thyroid cancer
Differentiated thyroid carcinomas (DTCs) comprise two most common histologic types, papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC). PTCs and FTCs, making up about 94% of total thyroid cancer cases, have a favorable prognosis with an 85% 10-year survival [3]. However, both PTCs and FTCs may progress to poorly differentiated thyroid carcinomas (PDTCs) or may completely loose differentiation and transform to anaplastic thyroid carcinoma (ATC).
Outcomes regarding mi-RNA expression profiles in DTCs based on a number of independent studies were collected in Table 1. These studies strongly suggested a vital role for specific miRNAs as key factors in the development and progression of thyroid cancer. Moreover, functional studies suggested that miRNA deregulation may play a critical role in thyroid carcinogenesis. A comprehensive analysis by microarray found that an aberrant miRNA expression profile that clearly differentiates PTCs from normal thyroid tissues, especially significant upregulation of miR-221, -222 and -181b in PTCS [4]. Further data demonstrated that miR-221 and miR-222 are endogenous regulators of P27Kip1 protein expression, which represents a very important regulator of cell cycle [5]. Another study showed that three microRNAs (miR-221, -222, and -146) are transcriptionally upregulated in PTC tumors in comparison with normal thyroid tissue [6]. Concomitant with upregulation of the three miRNAs was dramatic loss of KIT transcript and Kit protein, both of which involves in the pathogenesis of thyroid cancer. Microarray analysis of PTCs showed numerous genes were directly and indirectly regulated by miR-221 and further studies in vitro or in vivo using the bioluminescence imaging system confirmed the downregulation of HOXB5 by endogenous or exogenous miR-221 [7]. Significant downregulation of miRNA-1 was detected in a panel of thyroid tumors compared with normal thyroid tissues. Functional studies identified miRNA-1 as a tumor suppressor targeting CCND2, CXCR4, and SDF-1 genes, suggesting its ability to inhibit thyroid carcinoma cell proliferation and migration [8].
Table 1.
Deregulation of mi-RNA expression profile in differentiated thyroid cancer
| Reference | Tumor type | Sample sources | Profiling method | Upregulated mi-RNAs | Downregulated mi-RNAs |
|---|---|---|---|---|---|
| He et al.[6] | PTC | Frozen tissue | Microarray, q RT-PCR and Northern blots | miR-146b, miR-221, and miR-222 | — |
| Pallante et al. [4] | PTC | Frozen tissue | Microarray, q RT-PCR and Northern blots | miR-181b, miR-221, and miR-222 | — |
| Tetzlaff et al. [9] | PTC | FFPE | Microarray, q RT-PCR and Northern blots | miR-21, miR-31, miR-34a, miR-172, miR-181a, miR-181b, miR-213, miR-221, miR-222, miR-223, and miR-224 | miR-19b-1,2, miR-30a-5p, miR-30c, miR-130b, miR-145sh, miR-218, miR-292-as, miR-300, and miR-345 |
| Chen et al. [10] | PTC vs non-PTC | FFPE | q RT-PCR | miR-146b, miR-221, and miR-222 | — |
| Chou et al. [11] | PTC | Frozen tissue | q RT-PCR | miR-146b, miR-221, and miR-222 | — |
| Nikiforova et al.[12] | PTC and FTC | Frozen tissue | q RT-PCR | PTC: miR-187, miR-221, miR-222, miR-181b, miR-146b, miR-155 | — |
| Conventional FTC: miR-146b, miR-155, miR-187, miR-221, miR-222, and miR-224 | |||||
| Oncocytic FTC: miR-183, miR-187, miR-197, miR-221, miR-222, and miR-339 | |||||
| Weber et al. [13] | FTC | Frozen tissue | Microarray, q RT-PCR | miR-192, miR-197, miR-328, and miR-346 | — |
| Rossing et al. [14] | FTC vs. FTA/NT | Frozen tissue | Microarray | FTC vs. NT: 37 mi-RNAs FTC vs. FTA: 12 mi-RNAs | FTC vs. NT: 113 mi-RNAs FTC vs. FTA: 44 mi-RNAs |
Footnotes: PTC, papillary thyroid cancer; FTC, follicular thyroid cancer; FTA, follicular thyroid adenoma; NT, normal thyroid tissues; FFPE, formalin-fixed paraffin-embedded samples; q RT-PCR, quantitative RT-PCR; miR, mi-RNA.
In contrast to PTCs, mi-RNA expression profile in FTCs comparatively lack comprehensive studies especially with regard to distinct subtypes like oncocytic vs. conventional FTCs. One observation by miRNA microarray analysis showed that two miRNAs (miR-197 and miR-346) were differentially expressed between malignant and benign follicular thyroid tumors and both contributed to FTC carcinogenesis [13]. Rossing et al. [14] reported that 150 and 107 miRNAs were differentially expressed in follicular thyroid carcinoma and adenoma respectively compared with normal thyroid tissues. Among the most downregulated miRNAs, miR-199b-5p and miR-144 were found essentially lost in the carcinomas, indicating their potential to promote malignant transformation and a useful diagnostic tool.
Correlation of MicroRNA expression pattern with genetic background in thyroid cancer
Similar to other cancer types, development and progression of differentiated thyroid cancer involves a number of genetic alterations including two distinct molecular mechanisms: point mutation or chromosomal rearrangement. The most common genetic alterations in PTCs include BRAF mutation, RAS mutation, and RET/PTC rearrangement which mainly involve in the RAS/BRAF/MAPK signal pathway. Interesting, these molecular alterations are exclusive in PTC patients, suggesting that each of them alone is sufficient for malignant transformation of thyroid cells. Two major genetic alternations (RAS mutation and PAX8/PPARγ rearrangement) that have been found in FTC are rarely overlap in the same tumor, suggesting two independent pathways involved in the development of FTC [15]. In contrast to RAS mutation that is not restricted to a particular histological subtype of thyroid tumor, PAX8/PPARγ rearrangement mainly presented with follicular neoplasms and a small portion of follicular variant of papillary thyroid cancers [15-21].
By the unsupervised hierarchical clustering analysis, Nikiforova et al. [12] found that BRAF, RET/PTC and PAX8/PPAR positive tumors can form individual clusters, suggesting a strong correlation between miRNA expression pattern and mutational status. Further analysis demonstrated that five specific miRNAs (miR-187, miR-221, miR-222, miR-146b, and miR-155) presented with significantly expressed differences between the groups according to the mutational status. MiR-146b expression in PTCs harboring BRAF mutation was significantly higher than those without BRAF mutation and has a close association with high risk clinicopathological feature such as extrathyroidal invasion [11]. Expression profiling of mi-RNA were found to be able to distinguish tumors containing the BRAF mutation from the other tumor types as well as make a distinction between the more aggressive insular & anaplastic tumors and the classic variant [22]. Cahill et al. [23] found that that 21 miRNAs were significantly upregulated and 14 miRNAs were significantly downregulated in two human PTC cell lines harboring RET mutation compared with normal thyroid cell lines. These differentially expressed miRNAs potentially regulate genes involved in thyroid functions, and their deregulation could be implicated in thyroid carcinoma progression. Another study regarding PTC cell lines carrying BRAF mutation showed a unique miRNA expression signature in comparison with a normal thyroid cell line [24]. However, a study that compared 28 BRAF mutated and 26 wild type BRAF PTC tissues showed no difference between the mutated and nonmutated tumors [25].
Promising implication of MiRNAs in clinical practice
In contrast to mRNAs, mature miRNAs are comparatively stable and remain largely intact in routinely collected, formalin-fixed paraffin-embedded (FFPE) clinical tissues [9]. The ability to detect miRNA profiles in FFPE tissues implicated a great opportunity to perform the large retrospective analyses necessary to confirm the diagnostic role and investigate the prognostic significance of miRNA profiles. Furthermore, miRNA detection presents important molecular information that may determine tumor clinicopathological characteristics and will unlock a rich resource for future tumor researches. Fine-needle aspiration biopsy (FNAB) is currently the most widely used tool for the preoperative diagnosis of thyroid lesions with limitation for up to 30% indeterminate cases [26]. Investigation of miRNA expression pattern for differential diagnosis of thyroid neoplasms in fine needle aspiration biopsy samples is feasible and may improve the accuracy of FNAB cytology.
Numerous studies have demonstrated that the potential diagnostic value of mi-RNA expression signatures in thyroid cancer, especially for indeterminate results on FNAB samples. Pallante et al. [4] investigated that expression of miR-221, -222 and -181b had 5- to 35-fold differential in FNAB samples of PTCs compared with other thyroid nodules. Overexpression of four miRNAs (miR-100, miR-125b, miR-138, and miR-768-3p) was detected in malignant samples of follicular origin and only miR-125b was significantly overexpressed in FTC samples [27]. Seven mi-RNAs Chen et al. [10] found that four mi-RNAs (miRNA-146b, miRNA-221, and miRNA-222) from tested six miRNAs might potentially be adjunct markers to distinguish between PTCs and benign lesions. These findings suggested that specific miRNAs can be potential diagnostic tools with high accuracy in both surgical and preoperative FNA samples. Mazeh et al [28] found that miR-221 was the most favorable miRNA in differentiating benign from malignant thyroid pathology with specificity (100%), negative (96%) and positive (100%) predictive value, and accuracy (98%) respectively. For FNAB with indeterminate results, studies have showed that miR-126 and miR-7 may be useful adjunct diagnostic indicators [29,30]. Given the high negative predictive value of miR-7 (100%), patients may benefit from the result based on the predictor and avoid an immediate diagnostic thyroidectomy. Another study showed that a set of four miRNAs (miR-146b, -221, -187, and -30d) was identified that could differentiate malignant from benign lesions especially for the atypia cases in preoperative patients with an accuracy of 93.3% for the training sample set and an accuracy of 85.3% for the validation sample set. However, this particular miRNA panel is subject to low accuracy in classifying follicular lesions [31].
Mi-RNA expression profiles have a close association with clinicopathological features which help determine optimum management of thyroid cancer. A recent study suggested that miRNA signature can distinguishes the degree of PTC aggressiveness [32]. The results showed that four miRNAs (miR-146b, miR-222, miR-34b, miR-130b) were differentially expressed in aggressive in comparison with nonaggressive PTCs. MiR-146b was demonstrated to have a close association with aggressive behavior of PTC among BRAF-positive tumors, which further refine the prognostic importance of BRAF. Similar correlation was observed between downregulated miRNAs (miR-34b and miR-1) and higher MET expression in aggressive PTC. Chou et al. [11] investigated that overexpression of miR-146b, miR-221, and miR-222 were significantly associated with extra-thyroidal invasion in PTCS. MiRNA-100 was observed to have a significantly expression level between T1 and T4 tumors [33]. Schwertheim et al. [34] reported that poorly differentiated thyroid carcinoma had a distinct mi-RNA expression profile in comparison with PTC and ATC, suggesting that deregulation of some miRNAs may take part in selecting a subset of PTC progressing to PDTC.
Up to date, detection of mi-RNAs was only performed in specialized laboratories and could not be introduced into clinical practice before standardization of the extraction techniques and the testing methods. Due to its diverse roles in regulating cell proliferation, differentiation and apoptosis, mi-RNA could be potential target of therapeutic genetic strategies in human tumors. Although still in experimental phase, mi-RNAs provide a perspective for a RNA-based therapy by either upregulating or inhibiting the expression of specific miRNAs [35,36].
Acknowledgments
The author wishes to acknowledge funding from the National Natural Science Foundation of China (NO.30600601), China, and funds from the Tulane Cancer Center (TCC) and Louisiana Cancer Research Consortium (LCRC).
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Busaidy NL, Cabanillas ME. Differentiated thyroid cancer: management of patients with radioiodine nonresponsive disease. J Thyroid Res. 2012;2012:618985. doi: 10.1155/2012/618985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 5.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 6.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HJ, Kim YH, Lee DS, Chung JK, Kim S. In vivo imaging of functional targeting of miR-221 in papillary thyroid carcinoma. J Nucl Med. 2008;49:1686–1693. doi: 10.2967/jnumed.108.052894. [DOI] [PubMed] [Google Scholar]
- 8.Leone V, D'Angelo D, Rubio I, de Freitas PM, Federico A, Colamaio M, Pallante P, Medeiros-Neto G, Fusco A. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1alpha. J Clin Endocrinol Metab. 2011;96:E1388–1398. doi: 10.1210/jc.2011-0345. [DOI] [PubMed] [Google Scholar]
- 9.Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, Livolsi VA, Baloch ZW. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18:163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 10.Chen YT, Kitabayashi N, Zhou XK, Fahey TJ 3rd, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 11.Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, Yang KD, Cheng JT, Huang CC, Liu RT. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20:489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 12.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance an diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 14.Rossing M, Borup R, Henao R, Winther O, Vikesaa J, Niazi O, Godballe C, Krogdahl A, Glud M, Hjort-Sorensen C, Kiss K, Bennedbaek FN, Nielsen FC. Down-regulation of microRNAs controlling tumourigenic factors in follicular thyroid carcinoma. J Mol Endocrinol. 2012;48:11–23. doi: 10.1530/JME-11-0039. [DOI] [PubMed] [Google Scholar]
- 15.Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW 2nd, Tallini G, Kroll TG, Nikiforov YE. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88:2318–2326. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- 16.Dwight T, Thoppe SR, Foukakis T, Lui WO, Wallin G, Hoog A, Frisk T, Larsson C, Zedenius J. Involvement of the PAX8/peroxisome proliferator-activated receptor gamma rearrangement in follicular thyroid tumors. J Clin Endocrinol Metab. 2003;88:4440–4445. doi: 10.1210/jc.2002-021690. [DOI] [PubMed] [Google Scholar]
- 17.Kroll TG. Molecular events in follicular thyroid tumors. Cancer Treat Res. 2004;122:85–105. doi: 10.1007/1-4020-8107-3_4. [DOI] [PubMed] [Google Scholar]
- 18.Lacroix L, Mian C, Barrier T, Talbot M, Caillou B, Schlumberger M, Bidart JM. PAX8 and peroxisome proliferator-activated receptor gamma 1 gene expression status in benign and malignant thyroid tissues. Eur J Endocrinol. 2004;151:367–374. doi: 10.1530/eje.0.1510367. [DOI] [PubMed] [Google Scholar]
- 19.Lacroix L, Lazar V, Michiels S, Ripoche H, Dessen P, Talbot M, Caillou B, Levillain JP, Schlumberger M, Bidart JM. Follicular thyroid tumors with the PAX8-PPARgamma1 rearrangement display characteristic genetic alterations. Am J Pathol. 2005;167:223–231. doi: 10.1016/s0002-9440(10)62967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7:569–580. doi: 10.1038/nrendo.2011.142. [DOI] [PubMed] [Google Scholar]
- 21.Koenig RJ. Detection of the PAX8-PPARgamma fusion protein in thyroid tumors. Clin Chem. 2010;56:331–333. doi: 10.1373/clinchem.2009.137679. [DOI] [PubMed] [Google Scholar]
- 22.Aherne ST, Smyth PC, Flavin RJ, Russell SM, Denning KM, Li JH, Guenther SM, O'Leary JJ, Sheils OM. Geographical mapping of a multifocal thyroid tumour using genetic alteration analysis & miRNA profiling. Mol Cancer. 2008;7:89. doi: 10.1186/1476-4598-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahill S, Smyth P, Finn SP, Denning K, Flavin R, O'Regan EM, Li J, Potratz A, Guenther SM, Henfrey R, O'Leary JJ, Sheils O. Effect of ret/PTC 1 rearrangement on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer. 2006;5:70. doi: 10.1186/1476-4598-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahill S, Smyth P, Denning K, Flavin R, Li J, Potratz A, Guenther SM, Henfrey R, O'Leary JJ, Sheils O. Effect of BRAFV600E mutation on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer. 2007;6:21. doi: 10.1186/1476-4598-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheu SY, Grabellus F, Schwertheim S, Handke S, Worm K, Schmid KW. Lack of correlation between BRAF V600E mutational status and the expression profile of a distinct set of miRNAs in papillary thyroid carcinoma. Horm Metab Res. 2009;41:482–487. doi: 10.1055/s-0029-1215558. [DOI] [PubMed] [Google Scholar]
- 26.Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL, Mandel SJ, Mazzaferri EL, McIver B, Pacini F, Schlumberger M, Sherman SI, Steward DL, Tuttle RM. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19:1167–1214. doi: 10.1089/thy.2009.0110. [DOI] [PubMed] [Google Scholar]
- 27.Vriens MR, Weng J, Suh I, Huynh N, Guerrero MA, Shen WT, Duh QY, Clark OH, Kebebew E. MicroRNA expression profiling is a potential diagnostic tool for thyroid cancer. Cancer. 2012;118:3426–3432. doi: 10.1002/cncr.26587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazeh H, Mizrahi I, Halle D, Ilyayev N, Stojadinovic A, Trink B, Mitrani-Rosenbaum S, Roistacher M, Ariel I, Eid A, Freund HR, Nissan A. Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid. 2011;21:111–118. doi: 10.1089/thy.2010.0356. [DOI] [PubMed] [Google Scholar]
- 29.Kitano M, Rahbari R, Patterson EE, Steinberg SM, Prasad NB, Wang Y, Zeiger MA, Kebebew E. Evaluation of candidate diagnostic micrornas in thyroid fine-needle aspiration biopsy samples. Thyroid. 2012;22:285–291. doi: 10.1089/thy.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitano M, Rahbari R, Patterson EE, Xiong Y, Prasad NB, Wang Y, Zeiger MA, Kebebew E. Expression profiling of difficult-to-diagnose thyroid histologic subtypes shows distinct expression profiles and identify candidate diagnostic microRNAs. Ann Surg Oncol. 2011;18:3443–3452. doi: 10.1245/s10434-011-1766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen R, Liyanarachchi S, Li W, Wakely PE Jr, Saji M, Huang J, Nagy R, Farrell T, Ringel MD, de la Chapelle A, Kloos RT, He H. MicroRNA signature in thyroid fine needle aspiration cytology applied to "atypia of undetermined significance" cases. Thyroid. 2012;22:9–16. doi: 10.1089/thy.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE, Nikiforova MN. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. 2011;18:2035–2041. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vriens MR, Weng J, Suh I, Huynh N, Guerrero MA, Shen WT, Duh QY, Clark OH, Kebebew E. MicroRNA expression profiling is a potential diagnostic tool for thyroid cancer. Cancer. 2012;118:3426–3432. doi: 10.1002/cncr.26587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwertheim S, Sheu SY, Worm K, Grabellus F, Schmid KW. Analysis of deregulated miRNAs is helpful to distinguish poorly differentiated thyroid carcinoma from papillary thyroid carcinoma. Horm Metab Res. 2009;41:475–481. doi: 10.1055/s-0029-1215593. [DOI] [PubMed] [Google Scholar]
- 35.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 36.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]


