Abstract
Entomologic investigations were conducted during an intense outbreak of West Nile virus (WNV) disease in Maricopa County, Arizona during July 31–August 9, 2010. The investigations compared the East Valley outbreak area, and a demographically similar control area in northwestern metropolitan Phoenix where no human cases were reported. Five mosquito species were identified in each area, and species composition was similar in both areas. Significantly more Culex quinquefasciatus females were collected by gravid traps at Outbreak sites (22.2 per trap night) than at control sites (8.9 per trap night), indicating higher Cx. quinquefasciatus abundance in the outbreak area. Twenty-eight WNV TaqMan reverse transcription-polymerase chain reaction–positive mosquito pools were identified, including 24 of Cx. quinquefasciatus, 3 of Psorophora columbiae, and 1 of Culex sp. However, Cx. quinquefasciatus WNV infection rates did not differ between outbreak and control sites. At outbreak sites, 30 of 39 engorged Cx. quinquefasciatus had fed on birds, 8 of 39 on humans, and 1 of 39 on a lizard. At control sites, 20 of 20 identified blood meals were from birds. Data suggest that Cx. quinquefasciatus was the primary enzootic and epidemic vector of this outbreak. The most important parameters in the outbreak were vector abundance and blood meal analysis, which suggested more frequent contact between Cx. quinquefasciatus and human hosts in the outbreak area compared with the control area.
Introduction
After its initial recognition in the United States in 1999 during an outbreak of encephalitis in Queens, New York City, West Nile virus (WNV; Flavivirus:Flaviviridae) spread across the country, reaching the West Coast in 2003.1 The first appearance of WNV in Arizona also occurred in 2003, when 13 human cases were reported, 10 of whom were residents of Maricopa County, which comprises the greater Phoenix metropolitan area (www.maricopa.gov/publichealth/Services/EPI/Reports/wnv.aspx). The number of cases increased dramatically in 2004, when 391 cases, and 16 deaths, were reported statewide, with 91% (355) of those cases in Maricopa County (www.westnileaz.com/data.htm). In subsequent years case numbers were lower, ranging from 19 cases in 2009 to 91 cases in 2008.
In 2010, confirmed or suspect cases of WNV disease in Maricopa County equaled the 2009 total by mid-July, with 19 cases and 1 death. The focus of the outbreak was the East Valley area of southeastern metropolitan Phoenix, encompassing the jurisdictions of Apache Junction, Chandler, Gilbert, Mesa, Queen Creek, Tempe, and portions of Phoenix, San Tan Valley, and other unincorporated areas. On July 21, the Maricopa County Department of Public Health and the Arizona Department of Health Services requested assistance from the Centers for Disease Control and Prevention (CDC) in the outbreak investigation. The CDC response consisted of two components, 1) an epidemiologic component to assist the Maricopa County Department of Public Health with case finding and follow-up, and to conduct a case-control study to identify modifiable risk factors for WNV infection;2 and 2) an entomologic/ecologic component to conduct investigations in the outbreak focus, and in a demographically similar control area having little or no documented WNV activity, and also to collaborate with the epidemiologic team assessing case-patient and control-subject residences for environmental risk factors of infection. In this report, we present the results of the entomologic investigations, specifically a comparison of entomologic parameters within and outside the outbreak area.
Materials and Methods
Study areas.
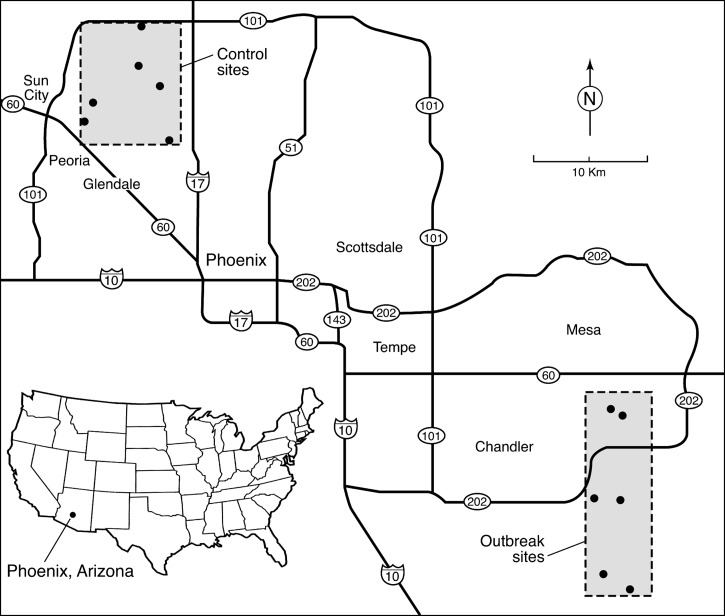
Six mosquito collection sites (outbreak sites) were selected in the Gilbert/Queen Creek area of the East Valley where human WNV disease cases were occurring and WNV-positive mosquito pools were being detected by the Maricopa County Department of Environmental Services, Vector Control surveillance (Figure 1). Three of these sites were at case-patient residences, and three sites were non-case residences or small, park-like green spaces within residential neighborhoods near Maricopa County Department of Environmental Services, Vector Control surveillance trap sites, or in one case, a ranch with horses present. An additional 6 control sites were chosen in the northwestern metropolitan area of Phoenix bounded by the Agua Fria Freeway on the north and west, Interstate 17 on the east, and US route 60 on the south. These sites were all at single-family residences, or at green spaces within residential neighborhoods. This area had no identified human cases of WNV disease, and only one virus-positive mosquito pool had been detected before our study began. Selected demographic data for the census tract containing each of the 12 sites was gathered from currently available census data (http://factfinder2.census.gov/faces/nav/jsf/pages/index.xhtml).
Figure 1.
Locations of six outbreak sites in the southeastern Phoenix, Arizona, metropolitan area (East Valley), the focus of a West Nile virus disease outbreak in 2010, and six control sites in the northwestern metropolitan area, where no human cases occurred.
Mosquito collection and virus testing.
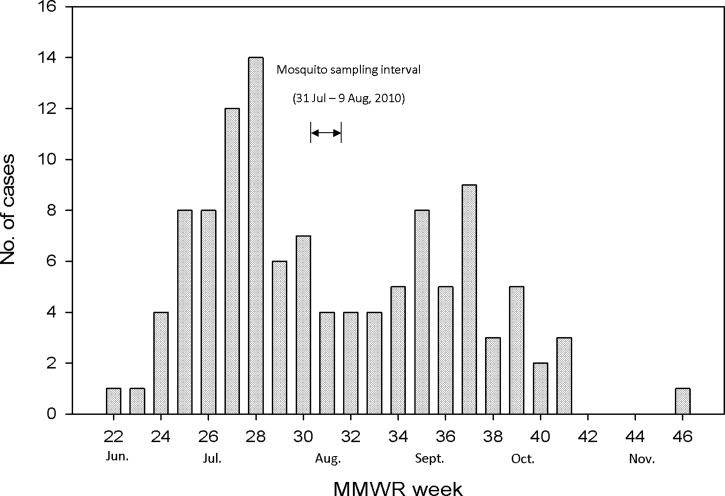
Mosquito trapping was done for 10 consecutive nights at each site during July 31–August 9. Retrospectively, this was approximately 2–3 weeks after the peak of the outbreak, on the basis of week of onset of illness (Figure 2). One dry ice-baited CDC light trap and one CDC gravid trap containing a grass infusion were located at each site. Traps were set in the late afternoon or early evening, and retrieved the next morning. Collections were transferred to 2.0-mL cryovials (Nunc, Roskilde, Denmark), frozen on dry ice until they could be returned to the laboratory, then stored at –80°C until processed. Mosquitoes were identified on a refrigerated chill table and separated into pools of approximately 50 by species, sex, collection site, and collection date. Mosquito pools were triturated in 1.75 mL of BA-1 diluent (Hanks M-199 salts, 0.05 M Tris, pH 7.5, 1% bovine serum albumin, 2 mM l-glutamine, 0.35 g/liter sodium bicarbonate, 100 U/mL penicillin, 100 μg/mL streptomycin, and 1 μg/mL Fungizone) in a Mixer Mill 300 (QIAGEN, Valencia, CA) and centrifuged at 4°C.3 A subset of 353 Culex quinquefasciatus males and females from outbreak and control sites were pooled individually and triturated in 0.5 mL of diluent for population genetic analysis in addition to virus testing. Nucleic acids were extracted from a 100-μL aliquot of each clarified supernatant by using a Biorobot 9604 (QIAGEN). All pools were tested for WNV RNA by using a TaqMan reverse transcription-polymerase chain reaction (RT-PCR) with WNV-specific primers.3,4
Figure 2.
Epidemic curve of the West Nile virus outbreak in Maricopa County, Arizona, in 2010, with the mosquito sampling interval denoted. Cases are listed by week of disease onset. MMWR = Morbidity and Mortality Weekly Report.
Identification of blood meals from engorged mosquitoes.
Identification of mosquito blood meals was performed by sequencing vertebrate mitochondrial genes, as described.5 In brief, engorged abdomens were individually triturated in 200 μL of phosphate-buffered saline diluent, and nucleic acids were extracted as described above. DNA was amplified by PCR, followed by sequencing of vertebrate mitochondrial cytochrome c oxidase 1 (COI) or cytochrome b (cytb) gene fragments. The identity of gene sequences was determined by using the DNA Barcode database (www.barcodinglife.org) for COI and GenBank for cytb.
Statistical analysis.
Abundance of Cx. quinquefasciatus in gravid traps versus light traps, and abundance in gravid traps in outbreak versus control sites, was analyzed by using the Mann-Whitney rank sum test; P < 0.05 was considered significant. Mosquito infection rates (IRs) per 1,000 mosquitoes were calculated as the maximum-likelihood estimate with 95% confidence intervals (CIs) by using a Microsoft (Redmond, WA) Excel add-in.6 The IRs of outbreak and control sites were compared by calculating the 95% CI for the difference in IRs and for the ratio of the outbreak site IR to the control site IR. In the former analysis, if the difference in IR = 0, then the IRs are the same, and if the 95% CI includes 0, then the difference in IR is not significant. Likewise, a ratio of 1 = identity, and if the 95% CI includes 1, then there is no significant difference in IRs between sites. A vector index (VI) was calculated to estimate the average number of infected mosquitoes per trap night in an area.7 The VI is the product of the IR/1,000 mosquitoes and the density of the mosquito population (expressed as mosquitoes per trap night). The distribution of Cx. quinquefasciatus blood meal sources at outbreak and control sites was assessed by using the Fisher exact test (two-tailed); P < 0.05 was considered significant. The proportion of unidentified blood meals at outbreak and control sites was compared by using the chi-square test; P < 0.05 was considered significant. Data from the 6 outbreak sites and 6 control sites were combined for host species comparisons.
Results
Demographic data.
Census information for the tracts containing each trap site was gathered to qualitatively compare outbreak and control areas for population age structure, median home value, and age. The population in the outbreak area was generally younger than in the control area; 36.5% of outbreak residents were ≤ 20 years old and 17.8% were ≥ 50 years old compared with 27.7% and 32.4%, respectively, in the control area. Residences in the outbreak area tended to be newer; > 91% were built since 1990 compared with > 25% built since 1990 in the control area. Median home prices were higher ($380,983) in the outbreak area than in the control area ($235,767).
Mosquito collections.
A total of 9,298 female mosquitoes, representing at least 6 species, was collected during 10 nights of trapping (Table 1); 6,665 from outbreak sites and 2,633 from control sites. Mosquito species composition was similar at outbreak and control sites with the following exceptions. Aedes aegypti was approximately 34-fold more abundant at control sites than at outbeak sites, Ae. vexans was collected only at outbreak sites, and Anopheles franciscanus was found only at control sites. The latter two species comprised only 0.3% of the total mosquitoes collected, and are probably not an important component of the mosquito fauna in outbreak or control areas.
Table 1.
Female mosquitoes collected by using CDC light traps and CDC gravid traps during an outbreak of West Nile virus disease, Maricopa County, Arizona, July 31–August 9, 2010*
| Species | Outbreak sites | Control sites | Grand total | ||||
|---|---|---|---|---|---|---|---|
| Light traps | Gravid traps | Total | Light traps | Gravid traps | Total | ||
| Psorophora columbiae | 5,087 | 5 | 5,092 (76.4) | 1,832 | 2 | 1,834 (69.7) | 6,926 (74.5) |
| Culex quinquefasciatus | 112 | 1,308 | 1,420 (21.3) | 17 | 524 | 541 (20.6) | 1961 (21.1) |
| Aedes aegypti | 5 | 0 | 5 (< 0.1) | 161 | 7 | 168 (6.4) | 173 (2.0) |
| Cx. tarsalis | 61 | 54 | 115 (1.7) | 33 | 44 | 77 (2.9) | 192 (2.0) |
| Ae. vexans | 19 | 0 | 19 (0.3) | 0 | 0 | 0 (0) | 19 (0.2) |
| Culex sp. | 2 | 7 | 9 (0.1) | 0 | 5 | 5 (0.2) | 14 (0.2) |
| Anopheles franciscanus | 0 | 0 | 0 (0) | 5 | 0 | 5 (0.2) | 5 (< 0.1) |
| Aedes sp. | 4 | 0 | 4 (< 0.1) | 1 | 0 | 1 (< 0.1) | 5 (< 0.1) |
| Anopheles sp. | 1 | 0 | 1 (< 0.1) | 2 | 0 | 2 (< 0.1) | 3 (< 0.1) |
| Total | 5,291 | 1,374 | 6,665 (100) | 2,051 | 582 | 2,633 (100) | 9,298 (100) |
Values are no. (%) collected. CDC = Centers for Disease Control.
Psorophora columbiae was the most abundant species, comprising 74.5% of the total, and its abundance at outbreak sites (5,092) was approximately three-fold greater than at control sites (1,834). Culex quinquefasciatus was the second most abundant species, with 1,961 collected, and also was most abundant at outbreak sites (72.3%) compared with control sites (27.7%). Aedes aegypti was collected in small numbers (2% of total mosquitoes trapped) at outbreak and control sites, but 97% were from control sites. Culex tarsalis, the other WNV vector of interest in this study, was not abundant at our sites. Of 194 collected, 59% were from outbreak sites and 41% were from control sites. In addition, many male mosquitoes were also collected, primarily in gravid traps, including 1,188 Cx. quinquefasciatus, 93 Cx. tarsalis, 81 Ps. columbiae, 25 Ae. aegypti, and 9 Culex sp.
The abundance of Cx. quinquefasciatus captured at outbreak and control sites was compared. Significantly more Cx. quinquefasciatus females were collected in gravid traps than in light traps at outbreak sites (22.2/trap night [TN] versus 1.8/TN; Mann Whitney U = 2380.0, P < 0.001) and control sites (8.9/TN versus 0.3/TN; U = 440.0, P = 0.002). Significantly more Cx. quinquefasciatus were collected in gravid traps at outbreak sites combined (22.2/TN) than at control sites combined (8.9/TN) (Mann Whitney U = 681.0, P = 0.037).
West Nile virus detection.
West Nile virus RNA was detected by TaqMan® RT-PCR in 28 mosquito pools consisting of 24 pools of Cx. quinquefasciatus, 3 of Ps. columbiae, and 1 of Culex sp. Virus was not detected in Cx. tarsalis, or in males of any species. All but one of the Cx. quinquefasciatus pools, and the single Culex sp. pool, were from gravid trap collections, and all Ps. columbiae pools were from light traps.
We calculated IRs and VIs for female Cx. quinquefasciatus to analyze differences between outbreak and control sites. The IR for all outbreak sites combined was 11.4 (95% CI = 6.5–18.8), and IR for all control sites combined was 21.8 (95% CI = 11.2, 39.1). The IRs for outbreak and control sites was compared by calculating 95% CIs for the difference in IR (95% CI = –0.029 to 0.002) and the ratio of the IRs (95% CI = 0.23–1.21). By both analyses, there was no statistically significant difference in IRs between outbreak and control sites. Vector indices were similar at both locations; combined outbreak sites VI = 0.3 and combined control sites VI = 0.2. In this calculation, the higher IR at the control area was offset by the significantly lower abundance of Cx. quinquefasciatus at control sites compared with outbreak sites.
Mosquito blood meal identification.
A total of 95 engorged females were tested for vertebrate host determination by sequencing of COI or cytb gene fragments, including 88 Cx. quinquefasciatus (87 from gravid traps, 1 from a light trap), 4 Cx. tarsalis (all from gravid traps), and 3 Ps. columbiae (all from light traps). Of these mosquitoes, blood meal identification was obtained for 63 specimens, including 59 Cx. quinquefasciatus, 3 Cx. tarsalis, and 1 Ps. columbiae. Overall, 50 (85%) of 59 Cx. quinquefasciatus blood feeds were from birds, 8 (13%) were from humans, and 1 (2%) was from a lizard (Table 2). No blood meals were identified from companion animals such as dogs and cats, or from livestock or wild mammals. Of the 59 identified Cx. quinquefasciatus blood meals 39 were from outbreak sites, and 20 were from control sites. The proportion of identified feeds taken from avian hosts was 77% (30 of 39) at outbreak sites and 100% at control sites. Thus, all of the 8 Cx. quinquefasciatus blood meals taken from humans were from outbreak sites versus 0 from control sites, which showed a significant difference (P = 0.042, by Fisher exact test).
Table 2.
Sources of Culex quinquefasciatus blood meals at outbreak and control sites for West Nile virus disease in Maricopa County, Arizona, 2010*
| Host species | Outbreak sites | Control sites |
|---|---|---|
| Avian | ||
| Mourning dove (Zenaida macroura) | 13 (33)a | 0 (0)b |
| House sparrow (Passer domesticus) | 8 (21)a | 4 (20)a |
| Curve-billed thrasher (Toxostoma curvirostre) | 1 (3)a | 6 (30)b |
| Chicken (Gallus gallus) | 1 (3)a | 5 (25)b |
| White-winged dove (Zenaida asiatica) | 2 (5)a | 1 (5)a |
| Northern mockingbird (Mimus polyglottos) | 1 (3)a | 2 (10)a |
| House finch (Carpodacus mexicanus) | 2 (5)a | 0 (0)a |
| Abert's towhee (Pipilo aberti) | 0 (0)a | 1 (5)a |
| Rock dove (Columba livia) | 0 (0)a | 1 (5)a |
| Eurasian collared dove (Streptopelia decaocto) | 1 (3)a | 0 (0)a |
| Greater roadrunner (Geococcyx californianus) | 1 (3)a | 0 (0)a |
| Other | ||
| Human (Homo sapiens) | 8 (21)a | 0 (0)b |
| Ornate tree lizard (Urosaurus ornatus) | 1 (3)a | 0 (0)a |
| Total | 39 (100) | 20 (100) |
Values are no. (%). Different letters after numbers indicate a significant difference in results from outbreak and control sites for that species (P < 0.05, by Fisher exact test, two-tailed).
Significantly more blood meals from mourning doves were found at outbreak sites (43%) than at control sites (0%) (P = 0.006, by Fisher exact test). The numbers of blood meals from house sparrows at outbreak sites versus control sites was not significantly different (P = 0.74, by Fisher exact test). Blood meals were taken more often at control sites than at outbreak sites from curve-billed thrashers (P = 0.012, by Fisher exact test), and domestic chickens (P = 0.031, by Fisher exact test), but these hosts only accounted for 3% each of blood hosts at outbreak sites. All 3 identified Cx. tarsalis blood feeds were from birds, and the single identified blood meal from Ps. columbiae was from a human.
The proportion of blood meals that could not be identified to host at control sites was higher (15 of 35, 43%) than at outbreak sites (14 of 53, 26%). This difference was worrisome because of concerns that the length of daily travel among sites may have inadvertently biased sample storage conditions, and reduced our ability to identify blood meal hosts at control sites. The null hypothesis was tested that there was no difference in the proportion of unidentified blood meals between the two locations. In this case the null hypothesis was confirmed (χ2 with Yates continuity correction = 1.89, degrees of freedom = 1, P = 0.16), indicating that no difference in the distribution of unidentified specimens between sites could be detected.
Discussion
The results of our investigation indicate that Cx. quinquefasciatus was the primary enzootic and epidemic vector of the 2010 WNV outbreak in Maricopa County. Two entomologic parameters that differed significantly between outbreak and control sites support this conclusion. First, Cx. quinquefasciatus was 2.6-fold more abundant at outbreak sites than at control sites. Second, Cx. quinquefasciatus blood meals taken from a human host were detected only at outbreak sites. Taken together, these data suggest that contact between humans and WNV-infected Cx. quinquefasciatus would occur more frequently in the outbreak area compared with the control area. This finding, combined with the detection of WNV in numerous pools of this species, incriminates Cx. quinquefasciatus as the primary enzootic and epidemic vector in this outbreak, and also may help explain why no human WNV disease was detected in the control area despite the Cx. quinquefasciatus IR being almost two-fold greater (21.8 versus 11.4) at control sites than at outbreak sites. These factors might have reduced human exposure to infection with WNV in the control area sufficiently so that the few persons who became ill were not tested for WNV, and thus, were not detected by the surveillance system.
Other factors may also be responsible for the lack of human cases in the control area. A number of demographic and environmental factors have been studied in other regions of the United States as risk factors for WNV disease, but some of these factors have not shown a consistent association with disease risk. In our outbreak area, most residences were newer than those in the control area, were more expensive, and a substantially smaller proportion of the population was ≥ 50 years old (17.8% versus 32.4% in the control area), the cohort at greatest risk of severe WNV disease. In contrast, in Chicago, IL, higher WNV case rates were observed in census tracts with a higher proportion of older and white residents, and in Chicago and Detroit, MI, higher WNV case rates were associated with middle class neighborhoods with older homes located in the inner suburbs, rather than with poorer inner city neighborhoods or more affluent outer suburban areas.8,9 In Suffolk County, NY, disease risk was also associated with middle class suburban neighborhoods, but not with population age,10 and in Harris County, TX (metropolitan Houston), and Orange County CA, south of Los Angeles, WNV disease risk was highest in low-income areas.11,12
Our control area most closely resembles an older, inner suburban residential area, and the outbreak area comprises newer, more affluent outer suburban neighborhoods. The precise behavioral and ecologic correlates that led to increased disease risk in the above studies are not well understood. However, a case–control study conducted in the East Valley concurrently with our study identified three environmental or behavioral risk factors for WNV infection: the presence of water-holding containers around the residence, not working or attending school outside the home, and residence in an area served by irrigation.2 This study also identified proximity to a neglected swimming pool as a risk factor by univariate analysis, but not by multivariate analysis. Neglected pools can serve as larval habitats capable of producing large numbers of both Cx. quinquefasciatus and Cx. tarsalis, and have been associated with increased risk of WNV disease.13
Culex quinquefasciatus was the second most abundant species collected, and was found at all sites in outbreak and control areas. Twenty-four of 28 WNV-positive mosquito pools were Cx. quinquefasciatus, and the single positive pool of Culex-containing specimens that were too damaged to identify to species was from a gravid trap, and likely also contained Cx. quinquefasciatus. Fourteen positive pools were detected in 1,308 Cx. quinquefasciatus collected at outbreak sites, and 10 positive pools were detected in 527 Cx. quinquefasciatus from control sites. Despite the control sites IR being approximately two-fold greater than that of the outbreak sites, this difference was not statistically significant. However, the relatively small number of mosquitoes tested, and the resulting wide CI, likely account for this lack of significance. Avian species accounted for 85% of identified Cx. quinquefasciatus blood meals, and humans (the only mammalian source detected) accounted for 14% of blood meals. These results are consistent with research indicating that Cx. quinquefasciatus is the main enzootic and epidemic vector of WNV to humans in southern California.14,15 Culex quinquefasciatus is a competent laboratory vector of WNV, although considerable variation exists among different geographic populations.16,17 Numerous field studies have implicated this species as an important vector of WNV in different parts of the southern United States.1,18–22
At outbreak and control sites combined, 75% of Cx. quinquefasciatus blood meals were taken from three species: mourning doves (33%), house sparrows (21%), and humans (21%) at outbreak sites, and house sparrows (20%), curve-billed thrashers (30%), and domestic chickens (25%) at control sites. This finding is consistent with those of a recent review of Cx. quinquefasciatus blood feeding literature, which emphasizes that this species takes a preponderance of its feeds on a small number of the available host species.23 Although an important blood source for mosquitoes at outbreak sites, mourning doves probably contribute little to WNV maintenance because of their lower viremia profiles and period of infectiousness to mosquitoes compared with house sparrows and house finches.17,24 The proportion of blood feeds taken from house sparrows was approximately the same at outbreak and control sites. This species likely is an important amplifier of WNV because of the magnitude and duration of the viremia when infected with WNV, and because it is ubiquitous in many parts of the United States.14,17,24
Domestic chickens constituted 25% of blood feeds at control sites, but prior studies indicate that adult chickens contribute little to WNV maintenance or amplification because of low and short duration viremias,25 although young chicks may develop sufficient viremias to infect vector species.26 The curve-billed thrasher, which constituted 30% of blood feeds at control sites, has not been evaluated for its susceptibility to WNV infection or ability to infect engorging mosquitoes. However, another member of the Family Mimidae, the northern mockingbird (Mimus polyglottos) is a weakly competent amplifier host.27 Our results are consistent with those of serologic studies of wild resident birds collected from mid-September through late October, after our (Komar N, unpublished data). This study, which was conducted in the East Valley north of our study sites, found the highest WNV antibody prevalence in house sparrows, house finches, great-tailed grackles (Quiscalus mexicanus), and mourning doves.
It is somewhat surprising that all of the mammalian blood feeds that we detected were from humans. A study conducted in residential areas of Tucson, AZ, approximately 195 km south of our study area, found that approximately 50% of Cx. quinquefasciatus blood meals were obtained from humans, 32% from birds, and the remainder from domestic mammals or mixed bird/mammal sources.28 Recent studies of Cx. quinquefasciatus blood-feeding behavior in other parts of the United States have found that 52.5% of feeds were mammalian (0.4% human) in Harris County, TX,20 11.6% mammalian (1.9% human) in southern California,14 and 40.1% mammalian (7% human) in East Baton Rouge, LA.29 However, our data on blood meal host preferences need to be interpreted cautiously because they represent a snapshot of host preferences taken during a brief period approximately mid-way through the outbreak. Also, we did not conduct bird or mammal censuses in our study areas to quantify availability of various host species. Thus, we cannot say whether the differences seen between outbreak and control sites reflect longer term trends in host use, or only reflect temporary fluctuations in host availability.
Culex tarsalis, the only other known WNV vector that we collected, accounted for only 2% of captures, and WNV was not detected in this species. This finding was likely caused by the low number that we collected, as on-going surveillance by the Maricopa County Department of Environmental Services, Vector Control surveillance and the Arizona Department of Health Services identified numerous WNV-positive pools of Cx. tarsalis in Maricopa County and statewide, indicating that this species is a vector in Arizona, but perhaps of lesser importance than Cx. quinquefasciatus (Smith K, Levy C, unpublished data). Research in southeastern California indicates a primary role for Cx. tarsalis as a WNV enzootic maintenance and early season amplification vector in agricultural and wetland areas around the Salton Sea, and Cx. quinquefasciatus was the primary vector in urbanized Palm Springs, CA, an area ecologically similar to Phoenix.15 Likewise, research in New Mexico also suggests that Cx. tarsalis is the most important vector in rural, agricultural areas, and Cx. quinquefasciatus is the predominant vector in urban areas.30,31
The detection of WNV in three pools of Ps. columbiae is of interest, in light of research identifying WNV in nine pools of this species in New Mexico,31 and in two pools captured at alligator farms in Louisiana.32 Although this species seems to feed preferentially on livestock,33 the degree to which it also feeds on small mammals, birds, or other vertebrates is not known. Our three positive pools came from a single outbreak site, a horse ranch, and those in New Mexico came primarily from semi-arid or agricultural sites.31 Experimental studies have shown that cottontail rabbits (Sylvilagus floridanus), chipmunks (Tamias striatus), and fox squirrels (Sciurus niger) can develop titers of WNV sufficient to infect some mosquito species,34–37 suggesting the possibility of secondary amplification cycles of WNV involving small rodents or Leporidae and mammal-feeding mosquitoes. The vector competence of Ps. columbiae for WNV should be assessed.
In summary, our study incriminated Cx. quinquefasciatus as the main vector of this outbreak, and adds to previous work suggesting that blood meal host preference, or availability, may affect the risk of infection with WNV. Additional multi-faceted studies such as ours, conducted during periods of increased transmission of WNV to humans, may be useful in the development of more effective outbreak intervention strategies.
ACKNOWLEDGMENTS
We thank K. Cox, D. Damian, and other staff of the Maricopa County Department of Environmental Services, Vector Control, and N. Panella and N. Komar (CDC, Fort Collins, CO) for technical assistance; J. Colborn (CDC Atlanta, GA) for suggesting Figure 2; and the property owners who allowed us to trap mosquitoes on their property.
Disclaimer: The opinions expressed in this article are those of the authors, and do not necessarily represent those of the Centers for Disease Control and Prevention.
Footnotes
Financial support: This study was supported by Centers for Disease Control and Prevention.
Authors' addresses: Marvin S. Godsey, Jr., Kristen Burkhalter, Ginger Young, Mark Delorey, and John-Paul Mutebi, Division of Vector-Borne Diseases, Arboviral Diseases Branch, Centers for Disease Control and Prevention, Fort Collins, CO, E-mails: mgodsey@cdc.gov, kburkhalter@cdc.gov, gyoung@inviragen.com, mdelorey@cdc.gov, and grv0@cdc.gov. Kirk Smith and John Townsend, Maricopa County Environmental Services, Vector Control, Phoenix, AZ, E-mails: jtownsen@mail.maricopa.gov and ksmith@mail.maricopa.gov. Craig Levy, Vector-Borne Diseases Program, Arizona Department of Health Services, Phoenix, AZ, E-mail: craiglevy@mail.maricopa.gov.
References
- 1.Reisen W, Lothrop H, Chiles R, Madon M, Cossen C, Woods L, Husted S, Kramer V, Edman J. West Nile virus in California. Emerg Infect Dis. 2004;10:1369–1378. doi: 10.3201/eid1008.040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibney KB, Colborn J, Baty S, Patterson AMB, Sylvester T, Briggs G, Stewart T, Levy C, Komatsu K, MacMillan K, Delorey MJ, Mutebi J-P, Fischer M, Staples JE. Modifiable risk factors for West Nile virus infection during an outbreak, Arizona, 2010. Am J Trop Med Hyg. 2012;86:895–901. doi: 10.4269/ajtmh.2012.11-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasci RS, Gottfried KL, Burkhalter KL, Kulasekera VL, Lambert AJ, Lanciotti RL, Hunt AR, Ryan JR. Comparison of Vero cell plaque assay, TaqMan® reverse transcriptase polymerase chain reaction RNA assay, and Vectest™ antigen assay for detection of West Nile virus in field-collected mosquitoes. J Am Mosq Control Assoc. 2002;18:294–300. [PubMed] [Google Scholar]
- 4.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. Rapid detection of West Nile virus from human clinical specimens, field collected mosquitoes and avian samples by a TaqMan RT-PCR assay. J Clin Microbiol. 2000;38:4066–4071. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent RJ, Juluisson L, Wessmann M, Evans S, Komar N. Seasonal blood feeding behavior of Culex tarsalis in Weld County, Colorado. J Med Entomol. 2009;46:380–390. doi: 10.1603/033.046.0226. [DOI] [PubMed] [Google Scholar]
- 6.Biggerstaff BJ. PooledInfRate: A Microsoft Excel Add-in to Compute Prevalence Estimates from Pooled Samples. 2006. http://www.cdc.gov/ncidod/dvbid/westnile/software.htm Available at. Accessed October 15, 2010.
- 7.Gujral IB, Zielinski-Gutierrez EC, LeBailly A, Nasci R. Behavioral risk factors for West Nile virus disease, northern Colorado, 2003. Emerg Infect Dis. 2007;13:419–425. doi: 10.3201/eid1303.060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz MO, Tedesco C, McTighe TJ, Austin C, Kitron U. Environmental and social determinants of human risk during a West Nile virus outbreak in the greater Chicago area, 2002. Int J Health Geogr. 2004;3:8. doi: 10.1186/1476-072X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz MO, Walker ED, Foster ES, Haramis LD, Kitron UD. Association of West Nile virus illness and urban landscapes in Chicago and Detroit. Int J Health Geogr. 2007;6:10. doi: 10.1186/1476-072X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rochlin I, Turbow D, Gomez F, Ninivaggi DV, Campbell SR. Predictive mapping of human risk for West Nile virus (WNV) based on environmental and socioeconomic factors. PLoS ONE. 2011;6:e23280. doi: 10.1371/journal.pone.0023280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rios J, Hacker CS, Hailey CA, Parsons RE. Demographic and spatial analysis of West Nile virus and St. Louis encephalitis in Houston, Texas. J Am Mosq Control Assoc. 2006;22:254–263. doi: 10.2987/8756-971X(2006)22[254:DASAOW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Harrigan R, Thomassen H, Buermann W, Cummings R, Kahn M, Smith T. Economic conditions predict prevalence of West Nile virus. PLoS ONE. 2010;5:e15437. doi: 10.1371/journal.pone.0015437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reisen W, Takahashi R, Carroll B, Quiring R. Delinquent mortgages, neglected swimming pools, and West Nile virus, California. Emerg Infect Dis. 2008;14:1747–1749. doi: 10.3201/eid1411.080719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molaei G, Cummings RF, Su T, Armstrong PM, Williams GA, Cheng M-L, Webb JP, Andreadis TG. Vector-host interactions governing epidemiology of West Nile virus in southern California. Am J Trop Med Hyg. 2010;83:1269–1282. doi: 10.4269/ajtmh.2010.10-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reisen WK, Lothrop HD, Wheeler SS, Kennsington M, Gutierrez A, Fang Y, Garcia S, Lothrop B. Persistent West Nile virus transmission and the apparent displacement of St. Louis encephalitis virus in southeastern California, 2003–2006. J Med Entomol. 2008;45:494–508. doi: 10.1603/0022-2585(2008)45[494:pwnvta]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goddard LB, Roth AE, Reisen WK, Scott TW. Vector competence of California mosquitoes for West Nile virus. Emerg Infect Dis. 2002;8:1385–1391. doi: 10.3201/eid0812.020536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reisen WK, Fang Y, Martinez VM. Avian host and mosquito (Diptera: Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. J Med Entomol. 2005;42:367–375. doi: 10.1093/jmedent/42.3.367. [DOI] [PubMed] [Google Scholar]
- 18.Godsey MS, Jr, Nasci R, Savage HM, Aspen S, King R, Powers AM, Burkhalter K, Colton L, Charnetzky D, Lasater S, Taylor V, Palmisano CT. West Nile virus-infected mosquitoes, Louisiana, 2002. Emerg Infect Dis. 2005;11:1401–1406. doi: 10.3201/eid1109.040443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmisano CT, Taylor V, Caillouet K, Byrd B, Wesson DM. Impact of West Nile virus outbreak upon St. Tammany Parish mosquito abatement district. J Am Mosq Control Assoc. 2005;21:33–38. doi: 10.2987/8756-971X(2005)21[33:IOWNVO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.Molaei G, Andreadis TG, Armstrong PM, Bueno R, Jr, Dennett JA, Real SV, Sargent C, Bala A, Radle Y, Guzman H, Travassos de Rosa A, Wuithiranyagool T, Tesh RB. Host feeding pattern of Culex quinquefasciatus (Diptera:Culicadae) and its role in transmission of West Nile virus in Harris County, Texas. Am J Trop Med Hyg. 2007;77:73–81. [PubMed] [Google Scholar]
- 21.Gleiser RM, Mackay AJ, Roy A, Yates MM, Vaeth RH, Faget GM, Folsom AE, Augustine WF, Jr, Wells RA, Perich MJ. West Nile virus surveillance in East Baton Rouge Parish, Louisiana. J Am Mosq Control Assoc. 2007;23:29–36. doi: 10.2987/8756-971X(2007)23[29:WNVSIE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 22.Mackay AJ, Roy A, Yates MM, Foil LD. West Nile virus detection in mosquitoes in East Baton Rouge Parish, Louisiana, from November 2002 to October 2004. J Am Mosq Control Assoc. 2008;24:28–35. doi: 10.2987/5681.1. [DOI] [PubMed] [Google Scholar]
- 23.Savage HM, Kothera L. The Culex pipiens complex in the Mississippi River basin: identification, distribution, and bloodmeal hosts. J Am Mosq Control Assoc. 2012 doi: 10.2987/8756-971X-28.4.93. In press. [DOI] [PubMed] [Google Scholar]
- 24.Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg Infect Dis. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langevin SA, Bunning M, Davis B, Komar N. Experimental infection of chickens as candidate sentinels for West Nile virus. Emerg Infect Dis. 2001;7:726–729. doi: 10.3201/eid0704.010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Styer LM, Bernard KA, Kramer LD. Enhanced early West Nile virus infection in young chickens infected by mosquito bite: effect of viral dose. Am J Trop Med Hyg. 2006;75:337–345. [PubMed] [Google Scholar]
- 27.Komar N, Panella NA, Langevin SA, Brault AC, Amador M, Edwards E, Owen JC. Avian hosts for West Nile virus in St. Tammany Parish, Louisiana, 2002. Am J Trop Med Hyg. 2005;73:1031–1037. [PubMed] [Google Scholar]
- 28.Zinser M, Ramberg F, Willott E. Culex quinquefasciatus (Diptera: Culicidae) as a potential West Nile virus vector in Tucson, Arizona: blood meal analysis indicates feeding on both humans and birds. J Insect Sci. 2004;4:20. doi: 10.1093/jis/4.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay AJ, Kramer WL, Meece JK, Brumfield RT, Foil LD. Host feeding patterns of Culex mosquitoes (Diptera:Culicidae) in East Baton Rouge Parish, Louisiana. J Med Entomol. 2010;47:238–248. doi: 10.1603/me09168. [DOI] [PubMed] [Google Scholar]
- 30.Dimenna MA, Bueno R, Jr, Parmenter RR, Norris DE, Sheyka JM, Molina JL, LaBeau EM, Hatton ES, Roberts CM, Glass GE. Emergence of West Nile virus in mosquito (Diptera:Culicidae) communities of the New Mexico Rio Grande Valley. J Med Entomol. 2006;43:594–599. doi: 10.1603/0022-2585(2006)43[594:eownvi]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitzer JB, Byford RL, Vuong HB, Steiner RL, Creamer RJ, Caccamise DF. Potential vectors of West Nile virus in a semiarid environment: Doña Ana County, New Mexico. J Med Entomol. 2009;46:1474–1482. doi: 10.1603/033.046.0634. [DOI] [PubMed] [Google Scholar]
- 32.Unlu I, Kramer WL, Roy AF, Foil LD. Detection of West Nile virus RNA in mosquitoes and identification of mosquito blood meals collected at alligator farms in Louisiana. J Med Entomol. 2010;47:625–633. doi: 10.1603/me09087. [DOI] [PubMed] [Google Scholar]
- 33.Kuntz KJ, Olson JK, Rade J. Role of domestic animals as hosts for blood-seeking females of Psorophora columbiae and other mosquito species in Texas rice lands. Mosq News. 1982;42:202–210. [Google Scholar]
- 34.Tiawsirisup S, Platt KB, Tucker BJ, Rowley WA. Eastern cottontail rabbits (Sylvilagus floridanus) develop West Nile virus viremias sufficient for infecting select mosquito species. Vector Borne Zoonotic Dis. 2005;5:342–350. doi: 10.1089/vbz.2005.5.342. [DOI] [PubMed] [Google Scholar]
- 35.Platt KB, Tucker BJ, Halbur PG, Tiawsirisup S, Blitvitch BJ, Fabiosa FG, Bartholomay LC, Rowley WA. West Nile virus viremia in eastern chipmunks (Tamias striatus) sufficient for infecting different mosquitoes. Emerg Infect Dis. 2007;13:831–837. doi: 10.3201/eid1306.061008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Root JJ, Oesterle PT, Nemeth NM, Klenk K, Gould DH, McLean RG, Clark L, Hall JS. Experimental infection of fox squirrels (Scirus niger) with West Nile virus. Am J Trop Med Hyg. 2006;75:697–701. [PubMed] [Google Scholar]
- 37.Platt KB, Tucker BJ, Halbur PG, Blitvitch BJ, Fabiosa FG, Mullin K, Parikh GR, Kitikoon P, Bartholomay LC, Rowley WA. Fox squirrels (Sciurus niger) develop West Nile virus viremias sufficient for infecting select mosquito species. Vector Borne Zoonotic Dis. 2008;8:225–233. doi: 10.1089/vbz.2007.0182. [DOI] [PubMed] [Google Scholar]