Abstract
The role of non-avian vertebrates in the ecology of eastern equine encephalomyelitis virus (EEEV) is unresolved, but mounting evidence supports a potential role for snakes in the EEEV transmission cycle, especially as over-wintering hosts. To determine rates of exposure and infection, we examined serum samples from wild snakes at a focus of EEEV in Alabama for viral RNA using quantitative reverse transcription polymerase chain reaction. Two species of vipers, the copperhead (Agkistrodon contortrix) and the cottonmouth (Agkistrodon piscivorus), were found to be positive for EEEV RNA using this assay. Prevalence of EEEV RNA was more frequent in seropositive snakes than seronegative snakes. Positivity for the quantitative reverse transcription polymerase chain reaction in cottonmouths peaked in April and September. Body size and sex ratios were not significantly different between infected and uninfected snakes. These results support the hypothesis that snakes are involved in the ecology of EEEV in North America, possibly as over-wintering hosts for the virus.
Introduction
Eastern equine encephalomyelitis virus (EEEV; family Togaviridae, genus Alphavirus) is an extremely pathogenic arbovirus endemic to New England south to Florida, extending west as far as Michigan.1 This virus circulates year round in Florida, but outside of Florida its transmission is seasonal.
Recent studies have suggested that the virus is periodically introduced from Florida to the northeastern United States, where it establishes itself in defined foci and is capable of maintaining itself for several seasons.2–6 How EEEV over-winters in these foci remains unresolved. However, recent studies have implicated ectothermic animals, and snakes in particular, as potential over-wintering reservoir hosts for EEEV. For example, studies on the ecology of EEEV conducted in the Tuskegee National Forest in Alabama documented the presence of EEEV in pools of Culex peccator, Culex territans, and Uranotaenia sapphirina mosquitoes, with some of the EEEV positive pools in these species detected early in the transmission season.7,8 These mosquito species feed primarily upon ectothermic hosts, with Cx. territans primarily feeding upon amphibians and Cx. peccator and Ur. sapphirina primarily feeding upon reptiles.7,8
Laboratory studies have also supported the hypothesis that ectotherms might play a role in over-wintering of EEEV. Recently, it was reported that snakes experimentally infected with EEEV developed circulating levels of viremia that were sufficient to infect mosquitoes and maintained these potentially infectious viral titers for 7–10 days.9 This period was longer than the period that infectious titers persist in passerine birds, the accepted enzootic hosts for EEEV. Furthermore, viremic snakes, when induced to hibernate, maintained a circulating viremia upon exiting hibernation.9
Snakes also appear to be commonly exposed to EEEV. A recent serosurvey of ectothermic species from a focus of EEEV transmission in Tuskegee National Forest in Alabama showed that more than 35% of the cottonmouths (Agkistrodon piscivorus), the ectothermic species most frequently fed upon by mosquitoes in Tuskegee National Forest,7 contained antibodies to EEEV.10 However, these data must be interpreted with caution because the presence of antibodies recognizing EEEV in these animals might merely reflect exposure to the virus, but not the development of a patent infection.
Snakes and other ectothermic animals mount relatively inefficient antibody responses to pathogens,11 suggesting that antibodies produced by these animals against EEEV might not be efficient in clearing the infection with the virus. In support of this hypothesis, plasma from cottonmouths experimentally infected with EEEV, while containing antibodies to EEEV that were detectable by the luminex assay, were found to lack detectible antiviral activity in plaque reduction neutralization assays.10 These findings suggested the hypothesis that snakes exposed to EEEV in the wild might maintain a low level of circulating virus. To test this hypothesis, plasma samples collected from snakes at Tuskegee National Forest were tested for the presence of EEEV by using quantitative reverse transcription polymerase chain reaction (qRT-PCR).
Materials and Methods
Collection of plasma from snakes at the Tuskegee National Forest has been described in detail in a previous publication.10 In brief, samples were collected from an EEEV-endemic area in Tuskegee National Forest, located in east-central Alabama. Samples were collected during April–September 2007–2009 during surveys of the herpetofauna present at the site, as described.10 Procedures used for collection of blood samples were approved by the Institutional Review Board for Animal Use and Care of Auburn University. Blood samples (1 mL) were collected from the caudal sinus with a 26-gauge heparinized syringe. At the time of blood draw, body size and sex of each snake were recorded. Animals were marked to prevent re-sampling and released at the point of capture. Blood samples were transferred to 1.5-mL microcentrifuge tubes, placed on wet ice, and transported to the laboratory. Samples were centrifuged briefly and the plasma was decanted from the cell pellet. The luminex assay (recognizing antibodies to EEEV) was then used to determine snake exposure to EEEV only, as described.10 Samples determined to be antibody positive by luminex were then tested by qRT-PCR for EEEV RNA. In addition, samples from 66 randomly selected seronegative snakes (11 per month, April–September) were tested by qRT-PCR for EEEV RNA.
Total RNA was prepared from 140 μL of plasma by using the QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. The process was automated with the Qiacube system (QIAGEN), and isolated RNA (60 μL) was stored at –80°C. Each batch of 12 samples processed in the Qiacube consisted of 11 serum samples and one sham extraction as a negative control.
The qRT-PCR was performed using the iScript one step RT-PCR kit for probes (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Primers and reaction conditions used to detect EEEV RNA were those recommended by Lambert and others,12 with the exception that reactions were performed in a final volume of 25 μL and used 5 μL of the RNA template. These primers produced an amplicon spanning positions 9298–9456 in the EEEV genome sequence (GenBank accession no. X67111).13 Samples (and associated sham extractions) were run in a 96-well plate format, with each plate containing two qRT-PCR negative control wells. Amplicons were detected by using a 5′ 6-FAM, 3′ BHQ1a-Q probe spanning positions 9411–9431 in the EEEV genome. Samples producing a signal at a cycle threshold (Ct) value ≤ 37 were considered putatively positive.
RNA samples found to be putatively positive in the screening assay were subjected to a confirmatory qRT-PCR assay provided by the Centers for Disease Control and Prevention (Atlanta, GA) to state Department of Health Laboratories conducting arboviral surveillance activities.14 The confirmatory assay used two primers (5′-ACCTTGCTGACGACCAGGTC-3′ and 5′-GTTGTTGGTCGCTCAATCCA-3′), which produced an amplicon spanning positions 9428–9497 in the EEEV genome. The sequence of the probe used in this assay was 5′-CTTGGAAGTGATGCAAATCCAACTCGACA-3′, which spanned positions 9449–9477 in the genome. The probe contained the same fluorescent and quencher molecules as the first qRT-PCR. Putatively positive samples that produced a detectable amplicon in the confirmatory assay (Ct < 40) were considered confirmed as positive for EEEV RNA. The biochemical limit of detection of both assays was determined to be ≤ 1 plaque-forming unit when assayed against cultured viral stocks of known titers.
Virus isolation was attempted from all confirmed qRT-PCR–positive samples by inoculating individual T-25 flasks of confluent Vero cell cultures with 1 mL of plasma. Flasks were incubated for two hours at 37°C, with gentle rocking every 15 minutes. After the incubation, 9 mL of maintenance media (1× Earle's minimal essential medium, 2% fetal bovine serum, 200 U/mL penicillin, 200 μg/mL streptomycin, and 2.5 μg/mL amphotericin B) were added to each flask. Cells were then monitored daily for a cytopathic effect.
Fisher's exact test was used to test for the significance of the proportion of qRT-PCR–positive samples in seropositive and seronegative snakes. The Pearson's chi-square test was used to test differences in sex ratio in EEEV qRT-PCR–positive and EEEV qRT-PCR–negative snakes. Statistical differences in body sizes of infected and uninfected snakes (separated by sex) were determined using a t-test. All analyses were performed by using SAS version 9.1 statistical software (SAS Institute, Cary, NC).
Results
Cottonmouths (Agkistrodon piscivorus) were the most common and commonly sampled snakes at our site, representing 41% of the ectotherm biomass,8 and were previously shown to exhibit high rates of EEEV exposure, with 35.4% of cottonmouth serum samples tested containing antibodies to EEEV.10 For this reason, initial studies concentrated upon determining if EEEV RNA could be detected in seropositive and seronegative cottonmouths. Although EEEV RNA was detected in seropositive and seronegative snakes, cottonmouths with detectable antibodies against EEEV in their serum were significantly more likely to be qRT-PCR positive than seronegative cottonmouths (P < 0.001, by Fisher's exact test). Of the 66 seronegative cottonmouths tested, only one (1.5%) was qRT-PCR positive, and 12 (22.2%) of the 54 seropositive snakes were positive for EEEV RNA. The Ct values for the snakes ranged from 33.4 to 37 for the screening assay and from 30.9 to 40 for the confirmatory assay. Attempts to culture EEEV from all the qRT-PCR–positive samples were not successful.
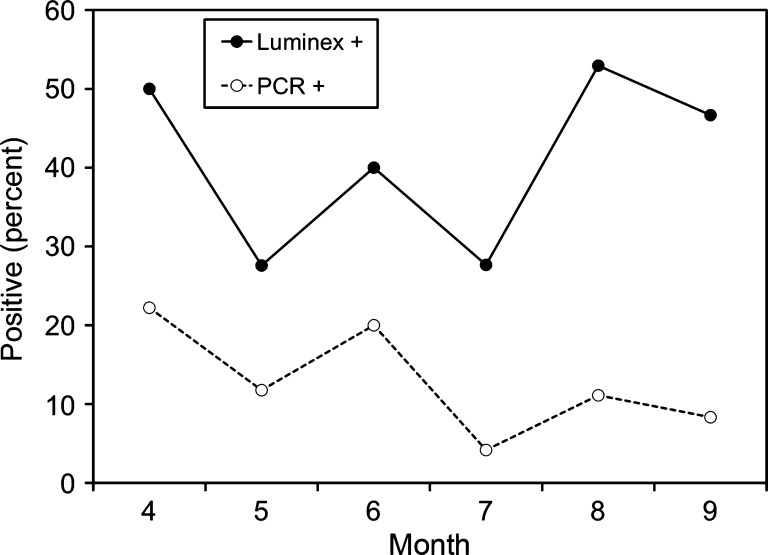
Previous studies had shown that temporal distribution of EEEV exposure in cottonmouths (as measured by antibody positivity) was relatively constant throughout the transmission season, with some suggestion of an increased prevalence of seropositivity in the spring and fall.10 A similar biphasic distribution of qRT-PCR positivity in seropositive cottonmouths was seen (Figure 1). In April, 22.2% of the total cottonmouths tested (seropositive and seronegative) were qRT-PCR positive. The proportion of qRT-PCR positive cottonmouths then decreased through the May–July period, reaching a nadir of 3.2% in July, and began to increase again in August. The single seropositive cottonmouth sample collected in September was found to be qRT-PCR-positive for EEEV (Figure 1).
Figure 1.
Exposure to (Antibody +) and infection with (polymerase chain reaction [PCR] +) eastern equine encephalomyelitis virus in cottonmouth snakes (Agkistrodon piscivorus) from Tuskegee National Forest, Alabama, USA. Data on seropositivity rates were taken from previously published sources.10
Seropositive serum samples collected from other species of snakes exposed to EEEV at Tuskegee National Forest were then tested for EEEV RNA by qRT-PCR. Two of the eight snake species tested (the cottonmouth and the copperhead [Agkistrodon contortrix]) were found to contain qRT-PCR-positive animals. Of the three copperheads sampled, one was positive for EEEV RNA. Serum samples from two other seropositive snake species (Plain-bellied watersnake [Nerodia erythrogaster] and black racer [Coluber constrictor]) were not positive for EEEV RNA (Table 1).
Table 1.
Proportion of Luminex-positive snakes that were positive by RT-PCR for eastern equine encephalomyelitis virus from Tuskegee National Forest, Alabama, USA*
| Species | Common name | No. tested | % RT-PCR positive | 95% CI |
|---|---|---|---|---|
| Coluber constrictor | Racer | 4 | 0 | ND* |
| Agkistrodon contortrix | Copperhead | 3 | 33.3 | 0–87 |
| Agkistrogon piscivorus | Cottonmouth | 54 | 22.2 | 13–35 |
| Storeria dekayi | Dekay's brownsnake | 1 | 0 | ND |
| Nerodia sipedon pleuralis | Midland water snake | 2 | 0 | ND |
| Nerodia erythrogaster | Plain-bellied water snake | 7 | 0 | ND |
| Diadophis punctatus | Ringneck snake | 1 | 0 | ND |
| Crotalus horridus | Timber rattlesnake | 1 | 0 | ND |
RT-PCR = quantitative reverse transcription polymerase chain reaction; CI = confidence interval; ND = not determined.
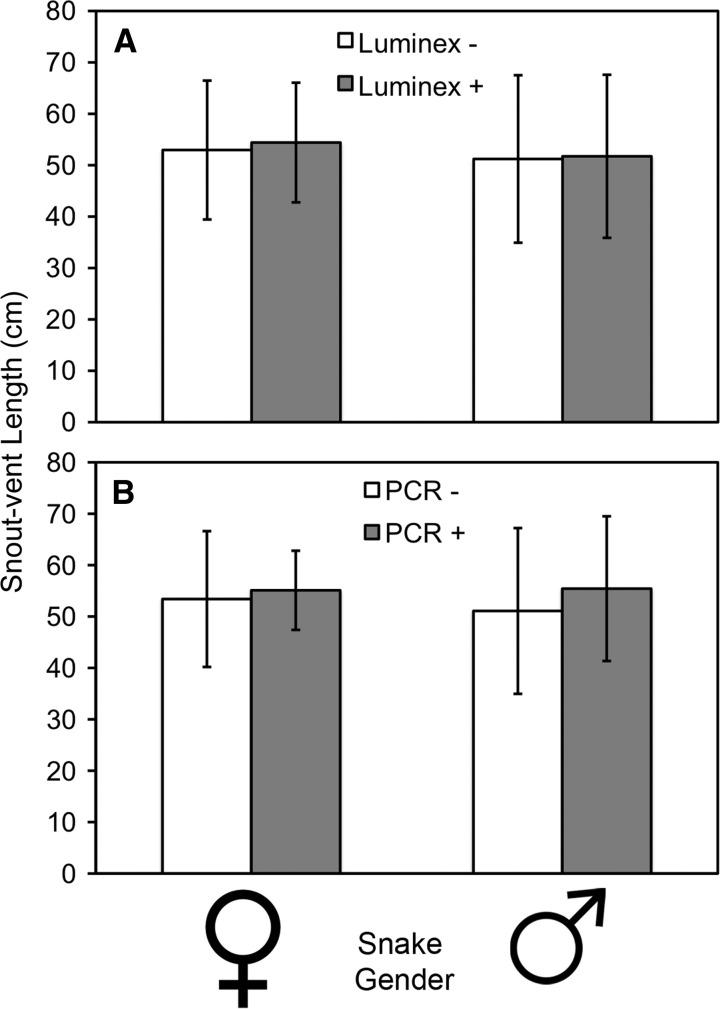
Because host body size8 and behavior15 can have strong effects on selection of ectothermic hosts by mosquitoes, and thus vector-host contact rates (a major driver in pathogen exposure), we investigated potential relationships between EEEV prevalence and snake body size and sex. No difference was observed between mean body size (measured by snout-vent length of male or female cottonmouths exposed to EEEV and those not exposed to EEEV (Figure 2A) (females: P = 0.5779, t = 0.5595, degrees of freedom [df] = 61; males: P = 0.2898, t = 1.0718, df = 43). A slightly greater body size (mean snout-vent length) was observed for cottonmouths that were qRT-PCR positive than those that were qRT-PCR negative (Figure 2B), although the difference was not statistically significant (females: P = 0.6962, t = 0.3923, df = 61; males: P = 0.8798, t = 0.1521, df = 43). Female cottonmouths constituted a greater proportion of the EEEV qRT-PCR–positive snakes than males (Figure 3), although this ratio was not significantly different from the sex ratio of qRT-PCR–negative snakes (χ2 = 0.142, df = 1, P = 0.707).
Figure 2.
Body size (snout-vent length) of male and female cottonmouth snakes (Agkistrodon piscivorus) exposed to (A) and infected with (B) eastern equine encephalomyelitis virus from Tuskegee National Forest, Alabama, USA. Error bars show SD.
Figure 3.
Sex ratio of eastern equine encephalomyelitis virus (EEEV) quantitative reverse transcription polymerase chain reaction (qRT-PCR)–positive (A) and EEEV qRT-PCR–negative (B) cottonmouth snakes (Agkistrodon piscivorus) from Tuskegee National Forest, Alabama, USA.
Discussion
The data presented demonstrate that snakes at Tuskegee National Forest were not only exposed to EEEV (antibody positive), but that a proportion of snakes have detectable infections (qRT-PCR positive for EEEV RNA). Two snake species (cottonmouth and copperhead), both of the genus Agkistrodon, were found to have detectable levels of EEEV in serum samples collected at our study site in Tuskegee National Forest. Cottonmouths, the greatest source of reptilian biomass at our site,8 were frequently exposed to and infected with EEEV.
To our knowledge, this is the first report of the detection of virus (as opposed to antibodies) detected in field-collected serum samples from ectothermic vertebrates. Karstad16 detected neutralizing antibodies to EEEV from wild ectotherms, but could not isolate virus from the samples. Dalrymple and others17 also detected EEEV antibodies in several ectothermic species, but made no mention of whether virus isolation was attempted for serum samples from these same hosts. However, other snakes have been shown to be competent hosts for western equine encephalomyelitis virus,18,19 an Alphavirus related to EEEV, with one study showing viremia in western equine encephalomyelitis virus–infected snakes lasting 70 days post hibernation.18 Attempts to culture EEEV were unsuccessful in this study, which might reflect sample degradation because the serum samples had been subjected to multiple freeze–thaw cycles. Another possibility is that the relatively inefficient adaptive immune response of snakes20 was insufficient to completely clear the infection, permitting the maintenance of a low-titer circulating viremia.
The data suggest that the proportion of qRT-PCR–positive cottonmouths was highest in the spring. These data are in concordance with those of previous laboratory studies, which demonstrated that garter snakes (Thamnophis sirtalis) experimentally infected with EEEV held at low temperatures (18°C) were found to maintain circulating viremias for longer periods than did animals held at higher temperatures (25°C or 30°C).9 It is possible this might be a consequence of the temperature dependence of the ectothermic adaptive immune system. During cooler months, snakes may not be able to raise body temperatures to levels that would enable them to clear infections because the adaptive immune response of ectotherms is more efficient at higher temperatures.20–23
The relationship between host body size and infection is complicated, and positive and negative associations have been found in different host/vector-borne pathogen systems. Body size can be affected by infection, when physiological cost of infection is high.24 Larger body size can contribute to greater infection through increased exposure because vectors are believed to feed more frequently upon larger hosts.25 In our own study, we found no significant difference in the body size of exposed and unexposed snakes. However, body size was slightly (but not significantly) larger in qRT-PCR–positive snakes than in qRT-PCR–negative snakes.
For many vertebrate pathogens, males have higher prevalence and intensity of infection because of their larger home ranges, mate attracting/guarding activities, and hormonal influences.26,27 In contrast, in this study, we found that compared with the uninfected population, females made up a larger proportion of the population of infected cottonmouths, although the difference was not statistically significant from that of the uninfected population.
Previous laboratory studies demonstrated that snakes experimentally infected with EEEV can remain viremic through hibernation, and that viremia in these animals is affected by the ambient temperature, with infected animals held at lower temperatures having lower viral titers of circulating virus, but maintaining viremia for longer periods than animals held at higher temperatures.9 The demonstration that wild-caught snakes contain EEEV virus in circulating blood and that the proportion of animals with circulating viremia is highest in the spring months provides further support to the hypothesis that snakes play an important role in over-wintering and early season enzootic amplification of EEEV.
Footnotes
Financial support: This study was supported by a grant from the National Institute of Allergy and Infectious Diseases (Project no. R01AI049724) to Thomas R. Unnasch.
Authors' addresses: Andrea M. Bingham, Nathan D. Burkett-Cadena, Hassan K. Hassan, and Thomas R. Unnasch, Global Health Infectious Disease Research Program, Department of Global Health, College of Public Health, University of South Florida, Tampa, FL, E-mails: abingha1@health.usf.edu, nburkett@health.usf.edu, and tunnasch@health.usf.edu. Sean P. Graham, Department of Biology, The Pennsylvania State University, University Park, PA, E-mail: szg170@psu.edu. Gregory S. White, Coachella Valley Mosquito and Vector Control District, Indio, CA, E-mail: gwhite@cvmvcd.org.
References
- 1.Bigler WJ, Lassing EB, Buff EE, Prather EC, Beck EC, Hoff GL. Endemic eastern equine encephalomyelitis in Florida: a twenty-year analysis, 1955–1974. Am J Trop Med Hyg. 1976;25:884–890. doi: 10.4269/ajtmh.1976.25.884. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong PM, Andreadis TG, Anderson JF, Stull JW, Mores CN. Tracking eastern equine encephalitis virus perpetuation in the northeastern United States by phylogenetic analysis. Am J Trop Med Hyg. 2008;79:291–296. [PubMed] [Google Scholar]
- 3.Weaver SC, Hagenbaugh A, Bellew LA, Gousset L, Mallampalli V, Holland JJ, Scott TW. Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol. 1994;68:158–169. doi: 10.1128/jvi.68.1.158-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, Scott TW, Rico-Hesse R. Molecular evolution of eastern equine encephalomyelitis virus in North America. Virology. 1991;182:774–784. doi: 10.1016/0042-6822(91)90618-l. [DOI] [PubMed] [Google Scholar]
- 5.Young DS, Kramer LD, Maffei JG, Dusek RJ, Backenson PB, Mores CN, Bernard KA, Ebel GD. Molecular epidemiology of eastern equine encephalitis virus, New York. Emerg Infect Dis. 2008;14:454–460. doi: 10.3201/eid1403.070816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White GS, Pickett BE, Lefkowitz EJ, Ottendorfer CL, Stark LM, Unnasch TR. Phylogenetic analysis of eastern equine encephalitis virus isolates from Florida. Am J Trop Med Hyg. 2011;84:709–717. doi: 10.4269/ajtmh.2011.10-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cupp EW, Zhang D, Yue X, Cupp MS, Guyer C, Korves T, Unnasch TR. Identification of reptilian and amphibian bloodmeals from mosquitoes in an eastern equine encephalomyelitis virus focus in central Alabama. Am J Trop Med Hyg. 2004;71:272–276. [PMC free article] [PubMed] [Google Scholar]
- 8.Burkett-Cadena ND, Graham SP, Hassan HK, Guyer C, Eubanks MD, Katholi CR, Unnasch TR. Blood feeding patterns of potential arbovirus vectors of the genus Culex targeting ectothermic hosts. Am J Trop Med Hyg. 2008;79:809–815. [PMC free article] [PubMed] [Google Scholar]
- 9.White G, Ottendorfer C, Graham S, Unnasch TR. Competency of reptiles and amphibians for eastern equine encephalitis virus. Am J Trop Med Hyg. 2011;85:421–425. doi: 10.4269/ajtmh.2011.11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham SP, Chapman T, Hassan HK, White G, Guyer C, Unnasch TR. Serosurveillance of eastern equine encephalitis virus in amphibians and reptiles from Alabama, USA. Am J Trop Med Hyg. 2012;86:540–544. doi: 10.4269/ajtmh.2012.11-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu E. Mutation, selection, and memory in B lymphocytes of exothermic vertebrates. Immunol Rev. 1998;162:25–36. doi: 10.1111/j.1600-065x.1998.tb01426.x. [DOI] [PubMed] [Google Scholar]
- 12.Lambert AJ, Martin DA, Lanciotti RS. Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays. J Clin Microbiol. 2003;41:379–385. doi: 10.1128/JCM.41.1.379-385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volchkov VE, Volchkova VA, Netesov SV. Complete nucleotide sequence of the Eastern equine encephalomyelitis virus genome [in Russian] Mol Gen Mikrobiol Virusol May. 1991:8–15. [PubMed] [Google Scholar]
- 14.Florida Department of Health . Real-time RT-PCR Arbovirology Protocol, 2011. Tampa, FL: Florida Department of Health Bureau of Laboratories; 2011. [Google Scholar]
- 15.Burkett-Cadena ND, McClure CJ, Ligon RA, Graham SP, Guyer CG, Hill GE, Ditchkoff SS, Eubanks MD, Hassan HK, Unnasch TR. Host reproductive phenology drives seasonal patterns of host use in mosquitoes. PLoS ONE. 2011;7:e17681. doi: 10.1371/journal.pone.0017681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karstad L. Reptiles as Possible Reservoir Hosts for Eastern Encephalitis Virus. 1961. pp. 186–202. Transactions of the 26th North American Wildlife Conference. [Google Scholar]
- 17.Dalrymple JM, Young OP, Eldridge BF, Russell PK. Ecology of arboviruses in a Maryland freshwater swamp. 3. Vertebrate hosts. Am J Epidemiol. 1972;96:129–140. doi: 10.1093/oxfordjournals.aje.a121439. [DOI] [PubMed] [Google Scholar]
- 18.Gebhardt LP, Hill DW. Overwintering of western equine encephalitis virus. Proc Soc Exp Biol Med. 1960;104:695–698. doi: 10.3181/00379727-104-25955. [DOI] [PubMed] [Google Scholar]
- 19.Thomas LA, Eklund CM. Overwintering of western equine encephalomyelitis virus in garter snakes experimentally infected by Culex tarsalis. Proc Soc Exp Biol Med. 1962;109:421–424. doi: 10.3181/00379727-109-27225. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerman LM, Vogel LA, Bowden RM. Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol. 2010;213:661–671. doi: 10.1242/jeb.038315. [DOI] [PubMed] [Google Scholar]
- 21.Allen FW, McDaniel EC. A study of the relation of temperature to antibody formation in cold-blooded animals. J Immunol. 1937;32:143–152. [Google Scholar]
- 22.Tait NN. The effect of temperature on the immune response in cold-blooded vertebrates. Physiol Zool. 1969;42:29–35. [Google Scholar]
- 23.LeMorvan C, Troutaud D, Deschaux P. Differential effects of temperature on specific and nonspecific immune defenses in fish. J Exp Biol. 1998;201:165–168. doi: 10.1242/jeb.201.2.165. [DOI] [PubMed] [Google Scholar]
- 24.Rätti O, Dufva R, Rauno V, Alatalo RV. Blood parasites and male fitness in the Pied Flycatcher. Oecologia. 1993;96:410–414. doi: 10.1007/BF00317512. [DOI] [PubMed] [Google Scholar]
- 25.Daviews CR, Ayres JM, Dye C, Deane LM. Malaria infection rate of Amazonian primates increases with body weight and group size. Funct Ecol. 1991;5:655–662. [Google Scholar]
- 26.Poulin R. Sexual inequalities in helminth infections: a cost of being a male? Am Nat. 1996;147:287–295. [Google Scholar]
- 27.Zuk M, McKean KA. Sex differences in parasite infections: patterns and processes. Int J Parasitol. 1996;26:1009–1023. [PubMed] [Google Scholar]