Abstract
Autosomal-dominant polycystic kidney disease (ADPKD) and von Hippel-Lindau (VHL) disease lead to large kidney cysts that share pathogenetic features. The polycystin-1 (PC1) and pVHL proteins may therefore participate in the same key signaling pathways. Jade-1 is a pro-apoptotic and growth suppressive ubiquitin ligase for beta-catenin and transcriptional coactivator associated with histone acetyltransferase activity that is stabilized by pVHL in a manner that correlates with risk of VHL renal disease. Thus, a relationship between Jade-1 and PC1 was sought. Full-length PC1 bound, stabilized and colocalized with Jade-1 and inhibited Jade-1 ubiquitination. In contrast, the cytoplasmic tail or the naturally occurring C-terminal fragment of PC1 (PC1-CTF) promoted Jade-1 ubiquitination and degradation, suggesting a dominant-negative mechanism. ADPKD-associated PC1 mutants failed to regulate Jade-1, indicating a potential disease link. Jade-1 ubiquitination was mediated by Siah-1, an E3 ligase that binds PC1. By controlling Jade-1 abundance, PC1 and the PC1-CTF differentially regulate Jade-1-mediated transcriptional activity. A key target of PC1, the cyclin-dependent kinase inhibitor p21, is also up-regulated by Jade-1. Through Jade-1, PC1 and PC1 cleaved forms may exert fine control of beta-catenin and canonical Wnt signaling, a critical pathway in cystic renal disease. Thus, Jade-1 is a transcription factor and ubiquitin ligase whose activity is regulated by PC1 in a manner that is physiologic and may correlate with disease. Jade-1 may be an important therapeutic target in renal cystogenesis.
INTRODUCTION
Autosomal-dominant polycystic kidney disease (ADPKD) is one of the most common monogenic disorders (1–3), affecting an estimated 12.5 million people worldwide. ADPKD is marked by numerous fluid-filled cysts in ductal organs, primarily the kidneys and the liver. The enlarging cysts cause end-stage renal disease (ESRD) in 50–75% of ADPKD patients (4,5). In addition, ADPKD is responsible for 4% of ESRD cases in the USA and 8–10% of cases in Europe and Australia (6). ADPKD is caused in 85% of instances by mutation in the PKD1 gene, which maps to chromosome 16p13.3 (7–9). The PKD1 gene encodes polycystin-1 (PC1), an integral membrane glycoprotein comprising 4302 amino acids. PC1 is predicted to span the plasma membrane 11 times and contains a large, extracellular amino-terminus of ∼3109 amino acids and an intracellular carboxyl-terminus of ∼200 amino acids (10). PC1 undergoes cleavage at a G-protein coupled receptor proteolytic site (GPS) between residues 3048 and 3049 in a process that requires the receptor for egg jelly motif (11,12). Proteolysis results in the generation of an N-terminal and C-terminal fragment (PC1-NTF and PC1-CTF, respectively). PC1 plays a role in cell–cell and cell–matrix adhesion, cell signaling cascades and renal tubulogenesis (13). GPS cleavage of PC1 may be required for full biological activity (11,12,14), and the PC1-NTF and PC1-CTF may have functions different from the full-length PC1 protein.
von Hippel-Lindau (VHL) disease is characterized by retinal angiomas, central nervous system hemangioblastomas, pheochromocytomas, renal, pancreatic and epididymal cysts, and clear-cell renal cancers (15). VHL renal disease shares a number of features with ADPKD, which raises the possibility of a common molecular mediator in these disorders. Cyst formation in ADPKD and VHL disease occurs in a similar manner. Both diseases are transmitted in an autosomal-dominant fashion, and the initiating cystogenic event is inactivation of the second allele of the respective gene (16,17). Both PC1 and pVHL proteins slow the G1-S transition of the cell cycle by promoting the expression of cyclin-dependent kinase inhibitors, such as p21 and p27 (18–20), and protect the cell against apoptosis (19,21,22). Increased proliferation and apoptosis are general features of renal cystogenesis. Thus, PC1 and pVHL may control common signaling pathways involving proliferation and apoptosis.
Jade-1 is a transcription factor (23,24) and ubiquitin ligase (25) that was cloned based on its interaction with pVHL. Jade-1 is the first member of a small family of Jade proteins containing plant homeodomains and PEST protein degradation motifs (23,26,27). As a protein stabilized by pVHL (23), Jade-1 itself has properties of a tumor suppressor (28). Jade-1 inhibits proliferation and promotes apoptosis. Jade-1 is also a ubiquitin ligase for beta-catenin (25), an important molecule in renal cancer and renal cystic disease pathogenesis. Moreover, pVHL stabilizes Jade-1 in a manner that correlates with risk of VHL renal disease (27), which suggests that Jade-1 may participate in the pathogenesis of renal cystic disease more generally. Consequently, a relationship between PC1 and Jade-1 was examined. Jade-1 may represent a key target shared widely in renal cystic diseases.
RESULTS
PC1 binds Jade-1
Jade-1 binds and partially colocalizes with the tumor suppressor protein pVHL (23). This interaction increases Jade-1 protein half-life. Given the similarities in the mechanism of renal cyst formation in ADPKD and VHL disease and the potential role for Jade-1 in VHL renal disease (27), an interaction between Jade-1 and PC1 proteins was sought. Schematics of the Jade-1 truncations and PC1 constructs appear in Figure 1. In cotransfection experiments in 293T17 cells, Jade-1 bound the cytoplasmic domain of PC1 linked to the CD16.7 membrane-targeting domain (cPC1) (Fig. 2A) (29–31). The cPC1 construct contains an extracellular domain from CD16 fused to a transmembrane domain from CD7 linked to the cytoplasmic domain of PC1 (Fig. 1B), and the empty vector control for this construct contains the CD16-CD7 fusion. Thus, Jade-1 and cPC1 may interact primarily in the plasma membrane or in the endoplasmic reticulum.
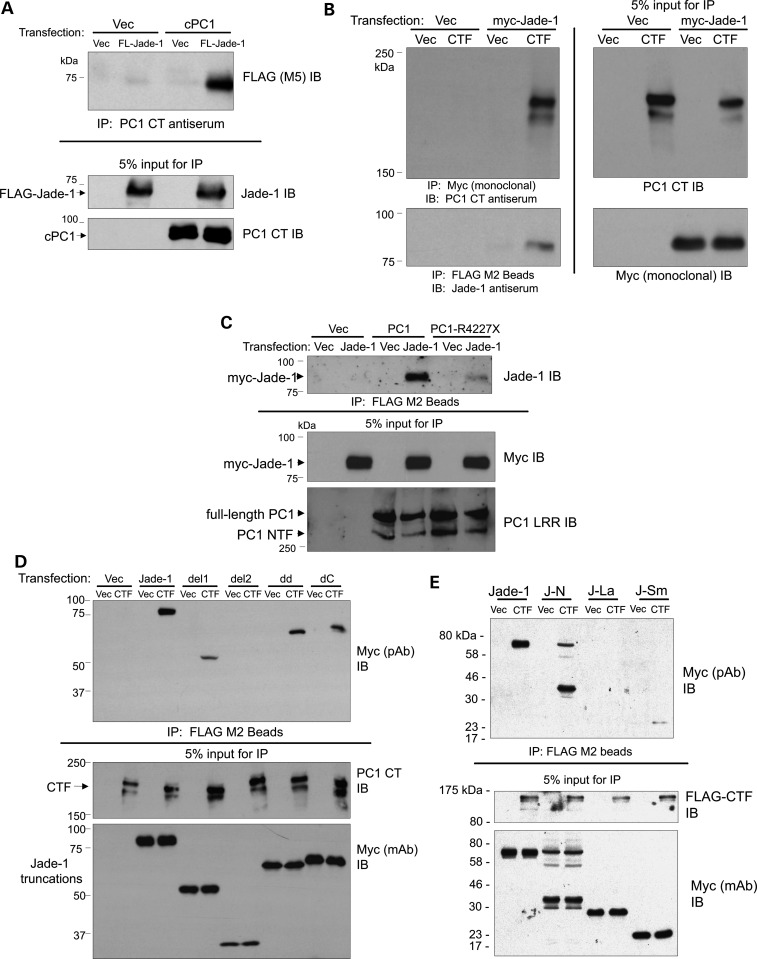
Figure 1.
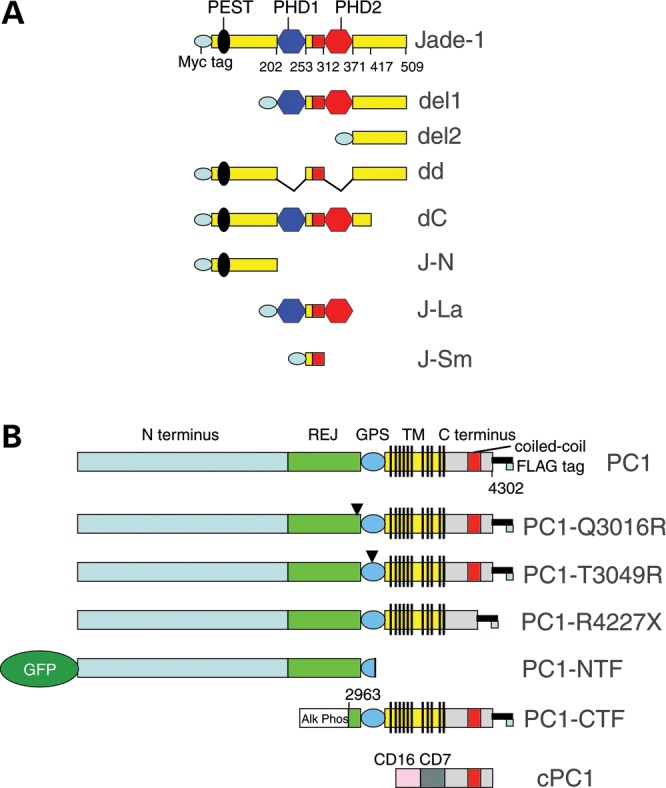
Schematics of Jade-1 truncations (A) and PC1 constructs (B).
Figure 2.
Jade-1 and PC1 proteins interact. (A) Jade-1 and cPC1 strongly interact. FLAG (FL)-tagged Jade-1 was cotransfected with empty vector (Vec) and cPC1 in 293T17 cells. PC1 CT antiserum immunoprecipitations were followed by immunoblotting with FLAG (M5) antibody. (B) Jade-1 and PC1-CTF (CTF) are binding partners. Myc-tagged Jade-1 was cotransfected with either vector or FLAG-tagged PC1-CTF. Immunoprecipitations were performed with myc monoclonal antibody and FLAG M2 beads, and western blotting was performed with PC1 CT or Jade-1 antiserum, respectively. (C) Jade-1 binds to full-length PC1 through the PC1 coiled-coil motif. Myc-tagged Jade-1 was cotransfected with vector, FLAG-tagged PC1 or PC1-R4227X. Immunoprecipitations employed FLAG M2 beads, and western blotting was performed using Jade-1 antiserum. PC1 input was assessed using rabbit antiserum to the PC1 N-terminal leucine-rich region (PC1 LRR). (D and E) Identification of the major Jade-1 PC1-binding domains as the Jade-1 amino terminal region and, to a lesser extent, the inter-PHD motif. FLAG-tagged PC1-CTF was cotransfected with the full series of myc-tagged Jade-1 truncations (Fig. 1A). Immunoprecipitations with FLAG M2 beads were followed by immunoblot with myc polyclonal antibody (pAb). Myc monoclonal antibody (mAb) reveals Jade-1 truncation input.
Binding of Jade-1 to different forms of PC1 was also analyzed in 293T17 cells. In cotransfection experiments, Jade-1 bound the PC1-CTF (Fig. 2B). In its C-terminal cytoplasmic tail, PC1 contains a coiled-coil motif that is required for interaction with PC2 (29,32). PC1-R4227X is a naturally occurring PC1 mutation that truncates the protein after residue 4226 within the coiled-coil domain, thus interfering with the PC1–PC2 protein interaction (33). Jade-1 bound full-length PC1, but not PC1-R4227X in cotransfection experiments (Fig. 2C). The Jade-1–PC1 interaction therefore requires the PC1 coiled-coil domain.
Protein truncations of Jade-1 were also examined for binding to PC1-CTF (Fig. 2D and E). Jade-1 truncations containing the Jade-1 amino terminus [Jade-1, Jade-1 dd, Jade-1 dC and Jade-1 N terminus (J-N)] bound most strongly to PC1-CTF. Persistence of binding in the absence of the Jade-1 amino terminus (Jade-1 del1 and J-Sm) suggested that the Jade-1 inter-PHD region also contributes, but to a lesser degree. The Jade-1 C terminus or PHD zinc fingers did not contribute substantially to PC1-CTF binding. The complexity of this protein–protein interaction may reflect the nature of Jade-1 protein tertiary structure, which includes a central Enhancer of Polycomb-N-terminus domain that folds together part of the J-N, the PHD-extended PHD module and part of the C terminus (34).
Colocalization of Jade-1 and PC1 was then sought. In 293T17 cells, transfected GFP-Jade-1 colocalized with PC1 mostly at the plasma membrane in a punctate pattern (Fig. 3A). Percent colocalization by NIH ImageJ64 was 2.61 ± 0.60 (SE), whereas PC1-R4227X exhibited 1.22 ± 0.32 (SE) percent colocalization with GFP-Jade-1, also at the plasma membrane (Fig. 3B), P= 0.0328 by the two-tailed t-test (35). The PC1-CTF exhibited extensive colocalization with GFP-Jade-1 in a different pattern throughout the cytoplasm and perhaps in the plasma membrane (Fig. 3C). Unlike GFP-Jade-1, GFP alone was expressed either equally in the nucleus and the cytoplasm or predominantly in the nucleus and was unaffected by contransfection of PC1 (data not shown). Thus, transfected Jade-1 binds and colocalizes with PC1 at the plasma membrane. The interaction and co-distribution of Jade-1 with PC1 is reduced by the elimination of the PC1 coiled-coil motif.
Figure 3.
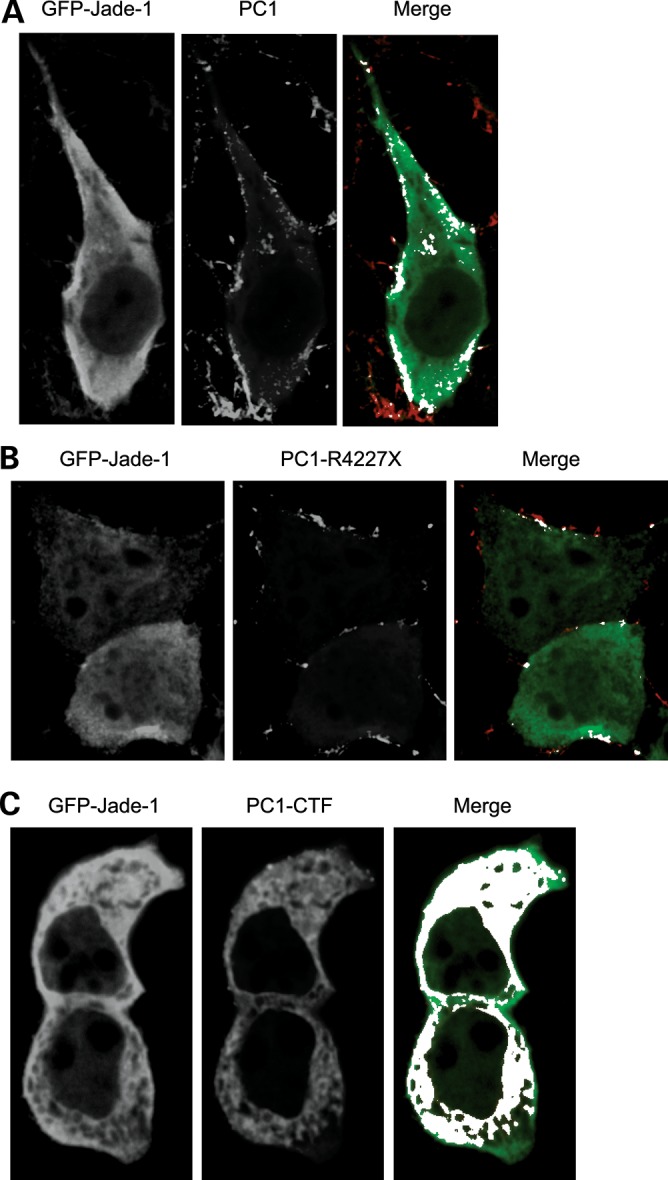
Jade-1 colocalizes with PC1 by confocal microscopy. GFP-Jade-1 was cotransfected with (A) FLAG-PC1, (B) FLAG-PC1-R4227X and (C) FLAG-PC1-CTF in 293T17 cells. Anti-GFP polyclonal antibody and Alexa Fluor 647-labeled anti-rabbit secondary antibody were used to localize transfected Jade-1. Anti-FLAG monoclonal antibody and rhodamine-red labeled anti-mouse secondary antibody were used for transfected PC1, PC1-CTF and PC1-R4227X. (A) Colocalization of Jade-1 and full-length PC1 is observed at the plasma membrane. The merged panel shows GFP-Jade-1 (green), PC1 (red) and white areas of signal overlap as determined by NIH ImageJ64. (B) Colocalization of Jade-1 and PC1 is reduced by truncation of the PC1 coiled-coil domain. (C) Jade-1 colocalizes extensively with PC1-CTF in the cytosol and perhaps at the plasma membrane.
PC1 regulates Jade-1 protein abundance
To determine whether PC1, like pVHL (23), regulates Jade-1 abundance, cotransfection experiments were performed. As expected, pVHL increased Jade-1 abundance (Fig. 4A). Surprisingly, cPC1 sharply decreased the levels of transfected Jade-1 protein, whereas PC2 had no substantial effect. The effect of full-length PC1 on Jade-1 was then determined. Interestingly, full-length PC1 and cPC1 differentially regulate Jade-1 abundance in a dose-dependent manner (Fig. 4B). cPC1 decreased Jade-1 protein levels, whereas full-length PC1 increased Jade-1 protein levels. Thus, full-length PC1 acts like pVHL on Jade-1, whereas cPC1 exerts an apparent dominant-negative effect on Jade-1.
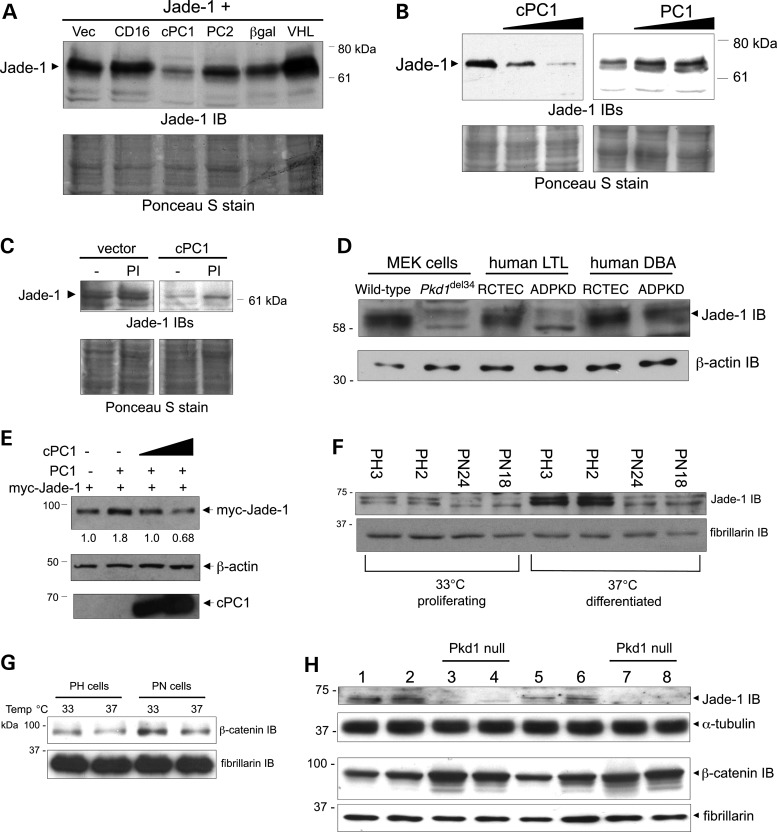
Figure 4.
Full-length PC1 and cPC1 differentially regulate Jade-1 abundance. (A) The cytoplasmic tail of PC1 decreases transfected Jade-1 protein levels. HA-tagged Jade-1 was cotransfected with vector controls (Vec and CD16), cPC1, PC2, β-galactosidase (βgal) and pVHL in 293T17 cells. Jade-1 abundance was measured by western blotting. (B) cPC1 and full-length PC1 differentially regulate transfected Jade-1 protein in a dose-dependent manner. HA-tagged Jade-1 was cotransfected with increasing amounts of either cPC1 or full-length PC1. (C and D) cPC1 and full-length PC1 differentially regulate endogenous Jade-1 protein levels. (C) Pooled IMCD cells stably expressing vector or cPC1 were assessed for the expression of endogenous Jade-1. Cells were examined at comparable early confluence or after 12-h incubation with 10 µm MG132 (PI). (D) Loss of both copies of a PKD gene results in reduction in endogenous Jade-1 protein abundance. Jade-1 immunoblotting was performed on mouse embryonic kidney (MEK) cells of distal origin [dolichos biflorus agglutinin (DBA) positive] from a Pkd1-null mouse in comparison with littermate control cells (wild-type); and human ADPKD cell lines of proximal [lotus tetragonolobos lectin (LTL) positive] or distal origin (DBA positive) in comparison with renal cortical tubule epithelial cells (RCTEC) of the corresponding nephron segment. (E) Increasing doses of cPC1 interfere with the increase in Jade-1 protein abundance mediated by full-length PC1 in a dominant-negative manner. (F) Endogenous Jade-1 protein is reduced in Pkd1 Null (PN) mouse proximal tubule cells transiently transfected with Cre recombinase and clonally derived from clonal, parental Pkd1 Heterozygous (PH) mouse proximal tubule cells harboring the temperature-sensitive T antigen, particularly under differentiating conditions. (G) Endogenous beta-catenin levels are increased in nuclear extracts from PN cells in a manner that is reciprocal to Jade-1 levels in (F). (H) Endogenous Jade-1 protein levels are reduced and endogenous beta-catenin protein levels are increased in cystic livers from Pkd1 null mice in comparison with control littermates. Each numbered lane represents one animal.
Effects of PC1 on endogenous Jade-1 protein abundance were then assessed in murine inner medullary collecting duct (IMCD) cells retrovirally transfected with either cPC1 or vector control (31) and MDCK cells stably transfected with either full-length PC1 or vector control (21). To confirm the identity of Jade-1, these cell lines were also treated with MG132, a proteasome inhibitor, which increases Jade-1 abundance (28). In concordance with the cotransfection data, endogenous Jade-1 levels were decreased in IMCD cells stably expressing cPC1 (Fig. 4C). The Jade-1 immunoblot panels in Figure 4C were from the same blot processed identically. Moreover, endogenous Jade-1 levels were increased in MDCK cells stably expressing full-length PC1 (data not shown). In further support of the effect of full-length PC1 on endogenous Jade-1, Jade-1 levels were reduced in dolichos biflorus agglutinin (DBA) positive mouse embryonic kidney (MEK) cells derived from the Pkd1del34/del34mouse in comparison with wild-type littermate control DBA positive MEK cells (Fig. 4D) (36). Furthermore, endogenous Jade-1 levels were reduced in immortalized human ADPKD cyst lines of proximal [lotus tetragonolobus lectin (LTL) positive] or a distal (DBA positive) origin, in comparison with immortalized renal cortical tubular epithelial cells (RCTEC) of proximal or distal origin, respectively (Fig. 4D) (37). Thus, both transfected and endogenous Jade-1 protein levels are decreased by cPC1 and increased by full-length PC1. This differential regulation suggested that the truncated form of PC1 might be capable of interfering with the function of full-length PC1 through a dominant-negative mechanism, which was tested and confirmed (Fig. 4E).
As perhaps the most genetically similar cell culture model of Pkd1 gene deletion, clonal Pkd1 null mouse proximal tubule cell lines (PN cells) derived from parental heterozygous Pkd1fl/−; temperature-sensitive large T antigen clones (PH cells) by transient transfection of Cre recombinase (38–40) were examined for levels of endogenous Jade-1 protein. Whether proliferating at 33°C or particularly when differentiated, the PN lines had substantially less Jade-1 than the PH cells (Fig. 4F). In the PN cells, the lower Jade-1 band runs faster than in the PH cells. Intriguingly, we have observed this same shift in mobility of the Jade-1 lower band in Figure 4D and in Vhlh-null mouse tissues (data not shown). Differential Jade-1 protein abundance in these lines was confirmed by Jade-1 immunoprecipitation (data not shown). As a marker of the biological activity of Jade-1 (25), endogenous nuclear beta-catenin was reciprocally increased in the PN cells (Fig. 4G). Nuclear extracts were used in this experiment to show the major pool of transcriptionally active beta-catenin. Results with Jade-1 and nuclear beta-catenin in the cell lines were confirmed in cystic liver tissues from conditional Pkd1cond/del2−4; tamoxifen-Cre mice (Fig. 4H) (41,42). The frequency of Pkd1 loop-out is highest in the liver in these mice. Thus, endogenous Pkd1 is capable of regulating endogenous Jade-1 protein.
To lend physiological relevance to the dominant-negative pathway of cPC1, the naturally occurring PC1-CTF was tested for regulation of Jade-1 protein abundance. The 293T17 cells were cotransfected with Jade-1 and either full-length PC1, PC1-CTF, mutants of PC1 that do not undergo cleavage at the GPS site, or PC1-R4227X. PC1-T3049R and PC1-Q3016R are engineered and naturally occurring mutations, respectively, that disrupt PC1 cleavage (11). While full-length PC1 stabilized Jade-1, PC1-CTF sharply decreased Jade-1 abundance (Fig. 5). Thus, PC1-CTF functions in the same dominant-negative manner as cPC1 with respect to Jade-1. In contrast, the missense mutation constructs did not significantly affect Jade-1 protein levels. The PC1-NTF, which does not contain the coiled-coil domain, also had no effect on Jade-1 (data not shown). Therefore, intact, full-length PC1 may be required to up-regulate Jade-1, whereas cleavage of PC1 at the GPS site may be necessary to down-regulate Jade-1. The inability of a naturally occurring, disease-associated PC1 mutant, PC1-Q3016R, to regulate Jade-1 supports a correlation between PC1-mediated Jade-1 regulation and ADPKD.
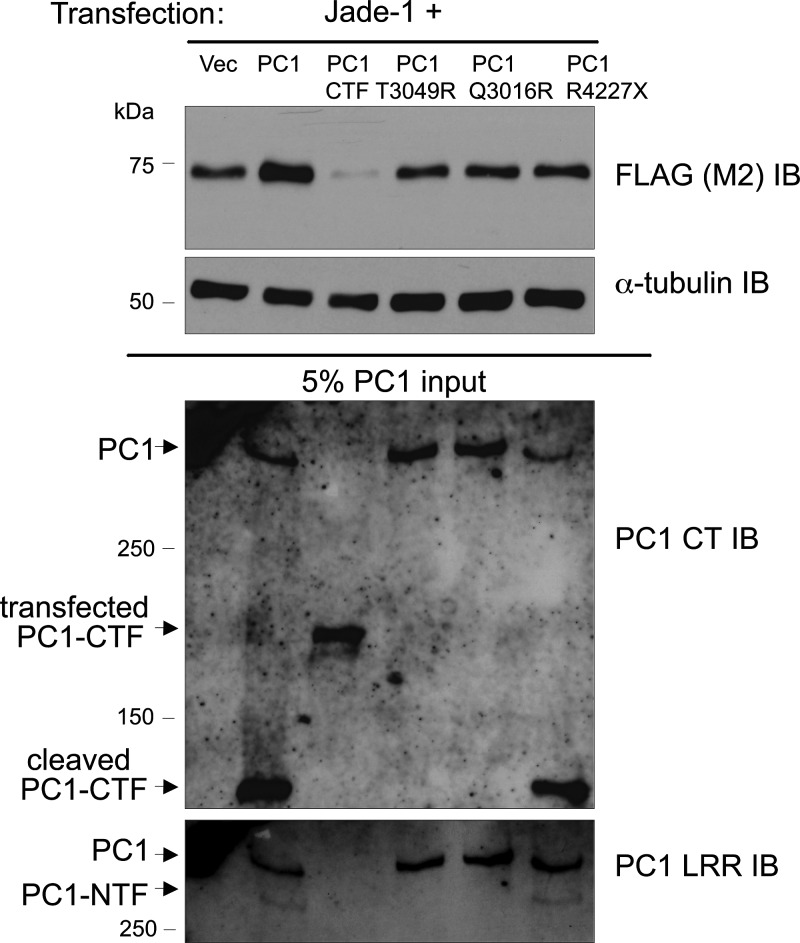
Figure 5.
Jade-1 is regulated by PC1-CTF but not by mutated PC1. Jade-1 protein levels are decreased by PC1-CTF but unaffected by mutated PC1 that does not undergo cleavage at the GPS site. Furthermore, the PC1 coiled-coil domain is required for regulation of Jade-1 abundance. In 293T17 cells, FLAG-tagged Jade-1 was cotransfected with vector, full-length PC1, PC1-CTF, PC1 containing a mutation that prevents cleavage (PC1-T3049R or PC1-Q3016R) or PC1-R4227X. To allow for equal protein expression of the PC1 proteins, the quantity of input plasmid was altered, and additional empty vector plasmid was included in the transfection when necessary. Transfected PC1-CTF contains alkaline phosphatase and its leader sequence, which increase the size of the protein by roughly 63 kDa to ∼200 kDa total in comparison with naturally cleaved PC1-CTF, which is ∼135 kDa.
In contrast to full-length PC1, PC1-R4227X does not alter Jade-1 protein levels relative to the vector control (Fig. 5). Thus, the coiled-coil motif of PC1 is required not only for binding to Jade-1, but also for regulating Jade-1 abundance. A coiled-coil domain is found in both PCs and mediates the PC1–PC2 interaction. The inability of PC1-R4227X to regulate Jade-1 abundance may indicate a requirement for PC2 as a cofactor for PC1 in the regulation of Jade-1 expression. Furthermore, PC1-R4227X is another disease-associated mutation of PC1, and its inability to regulate Jade-1 further strengthens the correlation between Jade-1 and ADPKD.
PC1 and PC1-CTF differentially affect Jade-1 protein half-life
Jade-1 is a short-lived protein with a half-life of 39 min, as determined by pulse-chase metabolic labeling (23). Jade-1 half-life was increased by pVHL to 106 min. To assess the mechanism by which full-length PC1 increases and PC1-CTF decreases Jade-1 abundance, protein half-life studies were performed in 293T17 cells using emetine, a protein translation inhibitor. Jade-1 was cotransfected with either vector, full-length PC1 or PC1-CTF, and cells were treated with emetine over a 120 min time course. Jade-1 and β-actin were detected by immunoblotting (Fig. 6A), and Jade-1 half-life was determined by plotting the 50% remaining intercepts, based on densitometry followed by normalization of Jade-1 to β-actin values (Fig. 6B). Transfected Jade-1 protein half-life was estimated at 20.3 min. This half-life was increased by full-length PC1 to 40.4 min and decreased by PC1-CTF to 9.2 min. PC1 therefore regulates Jade-1 protein half-life over a 4.4-fold range. PC2, PC1-NTF and PC1-R4227X, which did not affect Jade-1 abundance, did not affect Jade-1 protein half-life (data not shown). In addition, transfected or endogenous Jade-1 message levels were not altered by any form of transfected PC1, including those shown to regulate Jade-1 protein abundance by western blot (data not shown). Effects of Pkd1 gene deletion on endogenous Jade-1 protein half-life were confirmed in the PH and PN cells (Fig. 6C and D). Proteasome inhibitor treatment of the PH and PN cells normalized their levels of Jade-1 protein (data not shown), supporting the role of protein stability in their differential regulation of Jade-1. Thus, Jade-1 protein undergoes differential protein processing in the presence of full-length PC1 versus PC1-CTF, perhaps related to differences in post-translational modification.
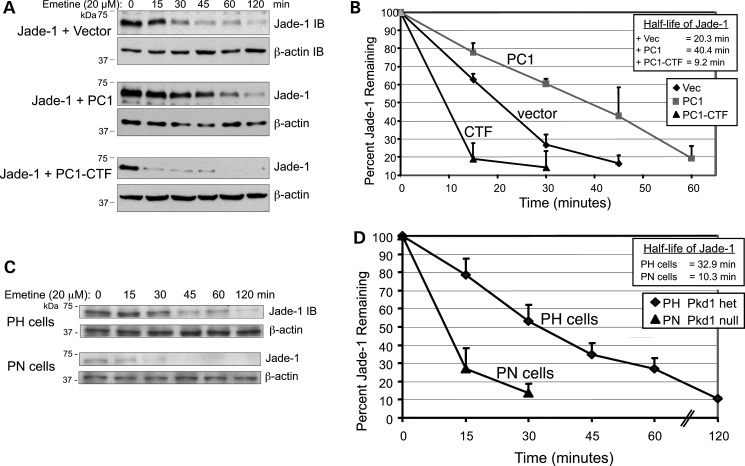
Figure 6.
Full-length PC1 increases and PC1-CTF decreases Jade-1 protein half-life. (A and B) HA-tagged Jade-1 was cotransfected with vector, full-length PC1 or PC1-CTF in 293T17 cells. Cells were treated with 20 µm emetine for the times shown. (A) Representative western blots for transfected Jade-1 and endogenous β-actin are shown. (B) Linear plots of the percent transfected Jade-1 protein remaining after emetine treatment. Densitometry was performed on Jade-1 and β-actin, and Jade-1 values were normalized to β-actin. Data plotted are an average of n = 6 for vector control, n = 4 for PC1 and n = 3 for PC1-CTF ± SE. Half-life values were determined on the linear plot by calculating the intercept at which 50% of Jade-1 protein remained. (C and D) Loss of Pkd1 reduces the half-life of endogenous Jade-1 protein in Pkd1 Null PN cells versus Pkd1 Heterozygous PH mouse proximal tubule cells. (C) Representative immunoblots for endogenous Jade-1 and endogenous β-actin. (D) As in (B), linear plots of the percent endogenous Jade-1 remaining after emetine treatment in PH and PN cells ±SE, n = 3.
PC1-CTF promotes degradation of Jade-1 via the proteasome pathway
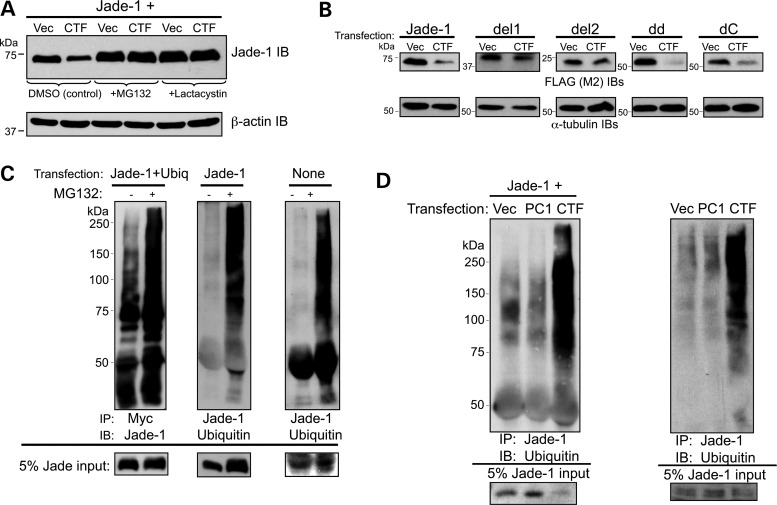
The ubiquitin-proteasome pathway was a strong candidate for the route of Jade-1 protein degradation by PC1-CTF (28). To test this possibility, Jade-1 was cotransfected with either PC1-CTF or vector, and cells were treated with proteasome inhibitors MG132 or lactacystin. The effect of PC1-CTF on Jade-1 degradation was completely abrogated by proteasome inhibition (Fig. 7A). Thus, PC1-CTF most likely promotes Jade-1 degradation through the proteasome pathway.
Figure 7.
The ubiquitination of Jade-1 is regulated by PC1. (A) PC1-CTF reduces Jade-1 abundance by the proteasome pathway. FLAG-tagged Jade-1 was cotransfected with vector or PC1-CTF in 293T17 cells. Cells were treated with 10 µm MG132, 10 µm lactacystin or DMSO for 12 h. (B) The N terminus of Jade-1 is required for degradation by PC1-CTF. FLAG-tagged Jade-1 and Jade-1 truncations were cotransfected with vector or PC1-CTF. (C) Transfected and endogenous Jade-1 undergoes ubiquitination. The 293T17 cells were transfected with FLAG-Jade-1 and myc-tagged ubiquitin (ubiq) or FLAG-Jade-1 alone, or nothing. Cells were treated with or without 10 µm MG132 for 12 h. (D) PC1-CTF augments the ubiquitination of transfected and endogenous Jade-1. (left panel) FLAG-Jade-1 was cotransfected with vector, PC1 or PC1-CTF, and cells were treated with 10 µm MG132 for 1 h. (right panel) The 293T17 cells were transfected with vector, PC1 or PC1-CTF, and cells were treated with 10 µm MG132 for 6 h.
The domain of Jade-1 required for degradation by PC1-CTF was then determined. The Jade-1 truncations were cotransfected with either vector or PC1-CTF. Like wild-type Jade-1, abundance of both Jade-1 dd and Jade-1 dC protein was decreased in response to PC1-CTF (Fig. 7B). In contrast, levels of Jade-1 del1 and Jade-1 del2 were not substantially reduced by PC1-CTF. Both Jade-1 del1 and Jade-1 del2 lack the N-terminus containing the PEST domain, which suggests the Jade-1 N-terminus is responsible for PC1-CTF-mediated degradation.
Ubiquitination of transfected and endogenous Jade-1 protein was assessed. Cotransfection of Jade-1 with myc-tagged-ubiquitin results in Jade-1 ubiquitination, which is enhanced with MG132 treatment (Fig. 7C, left panel). Transfected Jade-1 alone also undergoes ubiquitination, which is increased with MG132 (Fig. 7C, middle panel). Furthermore, endogenous Jade-1 protein sustains ubiquitination that is augmented by proteasome inhibition (Fig. 7C, right panel).
The effect of PC1 and PC1-CTF on Jade-1 ubiquitination was determined in transient transfections. As expected, transfected Jade-1 undergoes ubiquitination (Fig. 7D, left panel). Jade-1 ubiquitination was slightly decreased in the presence of full-length PC1, despite an increase in the input Jade-1 protein. Conversely, the ubiquitination of Jade-1 was dramatically increased in the presence of PC1-CTF, in spite of a substantial decrease in the input Jade-1 protein. Furthermore, similar effects were observed on endogenous Jade-1 protein (Fig. 7D, right panel). These data correlate well with the observed effects of full-length PC1 and PC1-CTF on Jade-1 protein abundance and half-life.
Siah-1 decreases Jade-1 abundance and enhances Jade-1 ubiquitination
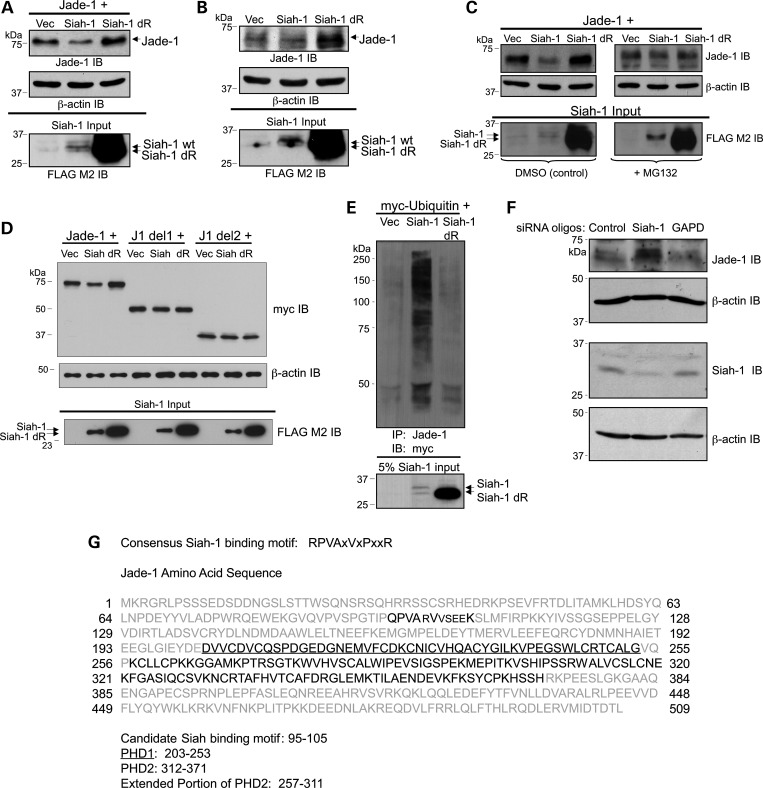
Siah-1 is a RING zinc-finger protein that functions as an E3 ubiquitin ligase (43,44). A previous report demonstrated an in vivo interaction between the cytoplasmic tail of PC1, either as a soluble or membrane-targeted form, and Siah-1, which induced ubiquitination and degradation of PC1 (45). Because Jade-1 interacts with cPC1, Siah-1 was a strong candidate to ubiquitinate Jade-1. Jade-1 was cotransfected with vector, Siah-1 or a dominant-negative form of Siah-1 (Siah-1 dR) that lacks the catalytic RING finger domain. As predicted, Siah-1 decreased protein levels of transfected Jade-1 in a manner similar to PC1-CTF and cPC1, whereas Siah-1 dR increased Jade-1 abundance (Fig. 8A). Siah-1 regulated endogenous Jade-1 similarly (Fig. 8B). Siah-1 dR is expressed at a much higher level than the wild-type protein (Fig. 8A and B), as mutations in the Siah-1 RING domain are stabilizing (46). In addition, Jade-1 degradation by Siah-1 was partially abrogated by MG132, thus implicating the proteasome pathway as the mechanism of Siah-1-mediated Jade-1 degradation (Fig. 8C). Degradation of Jade-1 by Siah-1 was dependent on the presence of the Jade-1 amino terminus (Fig. 8D), as it was for PC1-CTF-mediated Jade-1 degradation (Fig. 7B). The effect of Siah-1 on Jade-1 ubiquitination was then tested by cotransfection of 293T17 cells with myc-tagged ubiquitin and empty vector, Siah-1, or Siah-1 dR, followed by a 6-h incubation with MG132. Siah-1 dramatically increased the ubiquitination of endogenous Jade-1 protein, whereas Siah-1 dR did not alter Jade-1 ubiquitination, which indicates that the RING domain of Siah-1 is necessary for regulating Jade-1 ubiquitination (Fig. 8E). Moreover, silencing of endogenous Siah-1 increased endogenous Jade-1 protein abundance (Fig. 8F). Siah-1 is therefore a likely E3 ligase for Jade-1. Siah-1, but not Siah-1 dR, binds GST-Jade-1 (data not shown). Furthermore, Jade-1 protein contains a sequence (QPVArVvseeK) that bears substantial homology to the consensus Siah-binding motif (RPVAxVxPxxR) (44) (Fig. 8G). As anticipated, this Jade-1 candidate Siah-1-binding motif resides in the N-terminal portion of the protein (Figs 7B and 8D).
Figure 8.
E3 ligase Siah-1 regulates Jade-1 ubiquitination. (A) Siah-1 decreases the abundance of transfected Jade-1, but a RING-domain-deleted version of Siah-1 increases transfected Jade-1. FLAG-tagged Jade-1 was cotransfected with vector, FLAG-tagged Siah-1 or Siah-1 dR in 293T17 cells. (B) Full-length Siah-1 decreases and Siah-1 dR increases endogenous Jade-1 abundance. The 293T17 cells were transfected with vector, Siah-1 and Siah-1 dR. (C) The Siah-1-mediated decrease in Jade-1 is dependent on the proteasome pathway. Untagged-Jade-1 was cotransfected with vector, Siah-1 or Siah-1 dR. Cells were treated with either 10 µm MG132 or DMSO (vehicle control) for 12 h. (D) The N terminus of Jade-1 is required for degradation by Siah-1, as by the PC1-CTF. Jade-1, Jade-1 del1 or Jade del2 were cotransfected with vector, full-length Siah-1 (Siah) or Siah-1 dR (dR). (E) Siah-1 strongly increases the ubiquitination of endogenous Jade-1, which is dependent on the RING domain of Siah-1. Myc-tagged ubiquitin was cotransfected with vector, Siah-1 or Siah-1 dR in 293T17 cells. Immunoprecipitation with Jade-1 antiserum was followed by western blotting using a myc monoclonal antibody to detect ubiquitinated endogenous Jade-1. (F) Silencing of endogenous Siah-1 with siRNA oligonucleotides increases endogenous Jade-1 protein in 293T17 cells. (G) The Jade-1 N terminus contains a strong candidate Siah binding motif, shown in bold font.
Jade-1 is a transcriptional effector of PC1
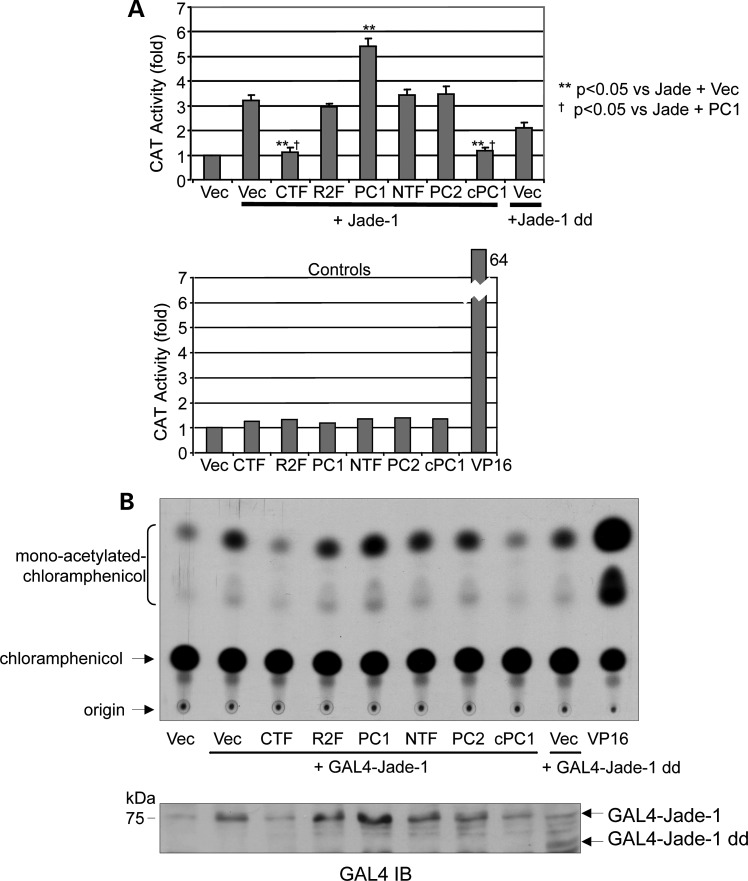
Jade-1 is a transcriptional coactivator associated with histone acetyltransferase (HAT) activity (24). Therefore, the effect of full-length PC1 and PC1-CTF on transcriptional activity of GAL4-Jade-1 was examined by the CAT assay in 293T17 cells. The promoter–reporter construct contained five GAL4 DNA binding sites and the adenoviral major late (AdML) promoter upstream of the CAT reporter gene (24). Jade-1 transcriptional activity was quantitated by liquid scintillation analysis of spots excised from TLC plates and normalized to β-galactosidase activity in order to control for transfection efficiency. Jade-1 increased AdML promoter activity by 3.2-fold (Fig. 9A and B). As expected, full-length PC1 further increased Jade-1-mediated transcription by 1.7-fold, whereas both PC1-CTF and cPC1 completely abrogated Jade-1-mediated transactivation and reduced transcription to baseline levels. PC1-mediated regulation of Jade-1 transcriptional activity is likely a dosage effect of Jade-1 protein, rather than altered Jade-1 transcriptional activity itself, as shown by the expression of GAL4-Jade-1 by western blot (Fig. 9B). Thus, PC1 and PC1-CTF differentially regulate Jade-1 transcriptional activity over a 5.4-fold range. As expected, PC1-R4227X, PC1-NTF and PC2 did not alter Jade-1-mediated transcription.
Figure 9.
Jade-1 is a transcriptional effector of PC1. (A) By controlling Jade-1 abundance, PC1 and PC1-CTF regulate Jade-1 transcriptional activity over a 5.4-fold range. GAL4-tagged Jade-1 was cotransfected with polycystin constructs in 293T17 cells, and CAT assays were performed. Values are relative to the empty GAL4 vector alone ±SE. R2F = PC1-R4227X. (B) Representative autoradiograph of Jade-1 transcriptional activity by the CAT assay. GAL4-Jade-1 input was detected by western blot with a GAL4 monoclonal antibody. GAL4-Jade-1 dd served as a negative control, and GAL4-VP16 was a positive control (74).
Jade-1 up-regulates cell cycle inhibitor p21
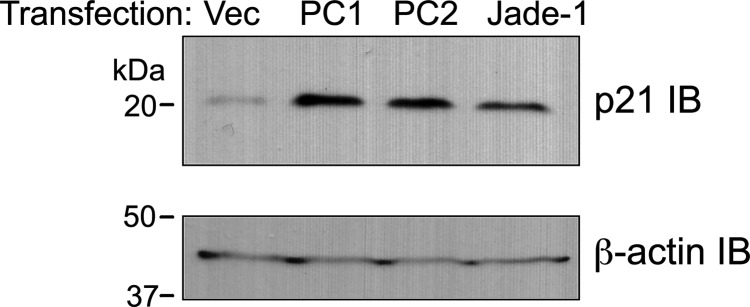
PC1 activates multiple signaling pathways, and an important transcriptional target is cyclin-dependent kinase inhibitor p21. PC1 activates the JAK2-STAT1 pathway, which results in increased p21 protein levels and slowing of the cell cycle at the G1/S transition (20). PC2 also binds and sequesters Id2, a transcriptional inhibitor of p21 (47). The effect of Jade-1 on p21 was assessed. Like PC1 and PC2, Jade-1 increased endogenous p21 protein levels (Fig. 10A). The series of Jade-1 deletion constructs was tested for effects on endogenous p21 protein abundance. Intact Jade-1 was required to up-regulate p21 (data not shown). Regulation of p21 by both PC1 and Jade-1 may place these proteins on a common signaling pathway. In addition, up-regulation of p21 likely plays a role in the ability of Jade-1 to inhibit cellular proliferation (28) and may have important implications for cyst formation in ADPKD. A schematic summarizing the effects of wild-type, cleaved and mutated PC1 on Jade-1 and downstream consequences is shown in Figure 11.
Figure 10.
Jade-1 increases p21 protein levels. (A) PC1, PC2 and Jade-1 increase p21 protein levels. Vector, FLAG-PC1, HA-PC2 and FLAG-Jade-1 were transfected into 293T17 cells, and endogenous p21 levels were assessed by western blot.
Figure 11.
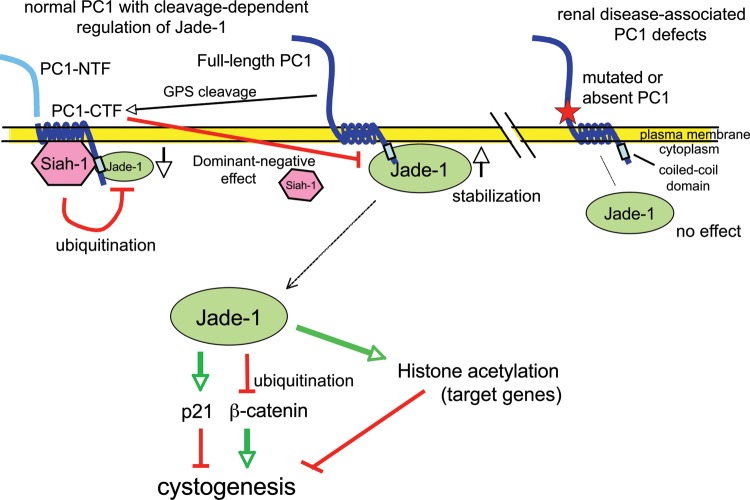
Model of the differential regulation of Jade-1 protein by full-length PC1 and the PC1-CTF, lack of regulation by mutated PC1, and downstream effects on Jade-1 targets. Wild-type full-length PC1 binds and stabilizes Jade-1 protein (center). Jade-1 binds the coiled-coil domain of PC1. Accumulated Jade-1 can exert its full range of biological activities, affecting downstream targets through histone acetylation (24) or by controlling levels of oncoprotein beta-catenin (25) and cyclin-dependent kinase inhibitor p21, which have all been implicated in the pathogenesis of renal cyst formation. By regulating beta-catenin (39,49) and p21 (20,47) Jade-1 can be placed along the same signaling pathways as PC1, suggesting it is a key PC1 effector that inhibits proliferation. PC1 also undergoes GPS cleavage (left), generating a PC1-CTF and PC1-NTF, which remain associated (11). PC1-CTF binds and promotes the ubiquitination and degradation of Jade-1, likely through the E3 ubiquitin ligase Siah-1, which also binds the PC1 coiled-coil motif (45). PC1-CTF can act in a dominant-negative fashion to block the stabilization of Jade-1 by full-length PC1, perhaps through differential control of Siah-1. Thus, PC1 cleavage may be physiologically regulated, with important downstream consequences for Jade-1 and its targets. ADPKD-associated mutations of PC1 that either truncate the coiled-coil motif or prevent GPS cleavage (right) prevent regulation of Jade-1, further supporting a connection between Jade-1 and disease pathogenesis.
DISCUSSION
The mechanisms of renal cyst formation in VHL disease and ADPKD share several striking similarities, suggesting that pVHL and PC1 proteins might participate in the same key signaling pathways. pVHL binds and stabilizes transcription factor and ubiquitin ligase Jade-1 and, importantly, does so in a manner that correlates with risk of VHL renal disease (23–25,27,28). Thus, the relationship between the PC proteins and Jade-1 was examined to explore a wider role for Jade-1 in renal cystogenesis. The major findings of this study are the following: (i) Jade-1 is a binding partner of PC1; (ii) Jade-1 abundance, protein half-life and ubiquitination are differentially regulated by PC1 and PC1-CTF; (iii) regulation of Jade-1 by PC1 is abrogated by ADPKD disease-causing mutations; (iv) Jade-1 is targeted for degradation by Siah-1, an E3 ligase associated with PC1; (v) Jade-1 transcriptional activity is modulated by PC1 and (vi) in addition to decreasing beta-catenin levels, Jade-1 increases p21 protein levels, thus placing Jade-1 and PC1 on a common signaling pathway (Fig. 11).
Full-length PC1 functions in a manner similar to pVHL, as both PC1 and pVHL stabilize Jade-1 protein (Figs 4B, D and 5). In contrast, the membrane-targeted cytoplasmic tail of PC1 alone reduces Jade-1 protein levels through an apparent dominant-negative effect (Fig. 4A, C and E). The C-terminal fragment of PC1, resulting from proteolysis at the GPS site, also sharply decreases Jade-1 protein abundance (Fig. 5). This effect of the PC1-CTF, a naturally occurring form of PC1, lends physiological relevance to the dominant-negative activity of cPC1. Full-length PC1 and the PC1-CTF also differentially regulate Jade-1 protein half-life and ubiquitination (Figs 6 and 7D). Stabilization of Jade-1 thus appears dependent on the status of the extracellular domain of PC1, which may affect interaction of the intracellular domain with Siah-1. Roughly half of cellular PC1 exists as full-length protein, while the remainder consists of the GPS-cleaved transmembrane PC1-CTF and tethered PC1-NTF (11,12), although these amounts may be subject to physiological regulation. The balance between these forms of PC1 protein may be essential for maintaining proper Jade-1 protein levels in the cell to balance Jade-1 growth-suppressive and apoptotic effects.
Specific ADPKD-associated PC1 mutations prevent Jade-1 regulation, which strongly suggests that a correlation exists between ADPKD and control of Jade-1 by the PCs (Fig. 11). The R4227X nonsense mutation in the coiled-coil domain of PC1 is naturally occurring and ADPKD associated (48). Unlike full-length PC1 and PC1-CTF, PC1-R4227X does not regulate Jade-1 (Fig. 5). The PC1 coiled-coil motif is thus required not only for binding to Jade-1, but also for regulation of Jade-1 abundance and, most likely, protein half-life. Both full-length PC1 and PC1-CTF likely require the coiled-coil motif to exert their distinct regulation of Jade-1. A non-conservative amino acid substitution, Q3016R, is a naturally occurring, disease-associated mutation of PC1 (48). This mutation prevents both cleavage of PC1 at the GPS motif (11) and regulation of Jade-1 levels (Fig. 5). In addition, an engineered mutation that specifically alters the GPS cleavage site, T3049R, also does not affect Jade-1 abundance (Fig. 5). These mutations highlight the importance of PC1 GPS cleavage in the regulation of Jade-1. Furthermore, stabilization of Jade-1 as assessed by western blotting may prove to be a useful functional assay to distinguish PC1 mutations from polymorphisms.
Jade-1 is the first example of a protein whose ubiquitination is promoted by the PC1-CTF (Fig. 7D). Moreover, the full-length PC1 protein inhibits the ubiquitination of Jade-1 (Fig. 7D), which is another novel PC1 function. In contrast, cPC1 has also been shown to stabilize β-catenin and c-Jun by inhibiting the activity of GSK-3β and thereby inhibiting β-catenin and c-Jun ubiquitination (49). The Pkd1 status has also been shown to affect the ubiquitination of the HGF receptor c-Met (50). However, there was no direct link identified between PC1 and c-Met; the c-Met ubiquitin ligase c-Cbl is mislocalized in Pkd1-null cells. Most likely, PC1 and PC1-CTF affect ubiquitination of Jade-1 by binding and differentially controlling the E3 ubiquitin ligase Siah-1. The PC1-CTF may adopt a different conformation following cleavage at the GPS site, which may permit interaction of the cleaved PC1 with Siah-1 and components of the ubiquitin-proteasome pathway, which full-length PC1 may be unable to access due to sterical constraints.
Siah-1 was a good candidate E3 ligase for Jade-1, as Siah-1 binds and promotes the ubiquitination of the cytoplasmic domain of PC1 (45) and also regulates beta-catenin (51–53). The evidence supporting a role for Siah-1 in Jade-1 ubiquitination is strong, as a mutated form of Siah-1 that lacks the RING domain increases Jade-1 abundance in a dominant-negative manner. This notion is further supported by the candidate Siah-1 binding motif in Jade-1 and the increase in endogenous Jade-1 protein abundance with Siah-1 siRNA. Siah-1 has been shown to function both in E3 ubiquitin ligase complexes (43) or as a single subunit E3 ligase (54), even for a single substrate such as beta-catenin (51–53). Siah-1 binds the PC1 coiled-coil domain (45), as does Jade-1, which may permit interaction between Jade-1 and Siah-1. Siah-1 may therefore represent another important molecule in ADPKD pathogenesis, promoting the degradation of PC1 and PC1-interacting proteins, such as Jade-1.
Although not the major focus of the current work, PC1, Jade-1 and Siah-1 appear to be part of a regulatory circuit in controlling beta-catenin protein abundance. Siah-1 can ubiquitinate PC1 (45), beta-catenin (51–53) and Jade-1, whereas Jade-1 ubiquitinates beta-catenin as well (25). Beta-catenin abundance may depend on the relative amounts of full-length and cleaved PC1 and their effect on Jade-1, and on the particular subcellular compartment involved. Because Siah-1 preferentially ubiquitinates unphosphorylated beta-catenin (51,52), whereas Jade-1 preferentially ubiquitinates phospho-beta-catenin (25), increases in Siah-1 might favor relative increases in phospho-beta-catenin, which could have functional implications. Beta-catenin is emerging as a key effector of the PCs (39,49,55,56) that is important in polycystic kidney disease pathogenesis (57–63). Control of beta-catenin by PC1 through Jade-1 is likely to be critical to this process (25,64). In general, wild-type PCs are associated with decreased, whereas cystic disease is associated with increased canonical Wnt signaling, which is consistent with the relationship observed here between PC1 and Jade-1. The interplay between PC1 and beta-catenin ubiquitin ligases Siah-1 and Jade-1 may help explain some of the complexities of PC1 regulation of canonical Wnt signaling in ADPKD and, potentially, in other forms of cystic renal disease (65,66).
Jade-1 is also a transcriptional coactivator that associates with HAT activity (24,67). Increasing Jade-1 expression promotes global histone H4 acetylation (24), suggesting Jade-1 may regulate PC transcriptional targets. As expected, full-length PC1 increased and PC1-CTF decreased Jade-1 transcriptional activity over a 5.4-fold range (Fig. 9). The altered transcription is apparently a consequence of a protein dosage effect of Jade-1. PC1 may therefore regulate Jade-1 activity by controlling its ubiquitination in the cytosol or membrane. Interestingly, the C-terminal tail (CTT) of PC1 can be cleaved and released from the membrane-bound portion of the protein (14,39,68). The PC1-CTT enters the nucleus and can activate the AP-1 (14) and STAT6 pathways (68) or inhibit nuclear beta-catenin signaling (39). Since Jade-1 binds to PC1 and also possesses nuclear functions, it may participate in PC1-CTT-mediated nuclear signaling events. The potential importance of Jade-1, as part of a HAT complex, in PC nuclear signaling is supported by the observation that histone deacetylase inhibitors, which increase histone acetylation, can ameliorate renal cyst formation in animal models of ADPKD (69,70).
Jade-1 up-regulates the cell cycle inhibitor p21, a key target of PC signaling (Fig. 10). PC1 expression activates the JAK2-STAT1 pathway, which in turn induces p21 (20). PC1 induction of p21 requires PC2 as an essential cofactor and causes cell cycle arrest in G0/G1. Furthermore, PC1 and PC2 modulate the cell cycle through regulation of transcription factor Id2 (47). PC2 sequesters Id2 in the cytosol, which allows the induction of p21 in the absence of nuclear Id2. As an interactor of the PC1 coiled-coil domain, the site of the PC1–Jade-1 interaction, PC2 may play an important role in PC1 regulation of Jade-1. PC2 may even be a required cofactor for the PC1–Jade-1 relationship. However, as observed here, PC2 overexpression may not have a substantial effect on Jade-1 protein levels. In most cells types, endogenous PC2 is at abundance in comparison with endogenous PC1, which is often difficult to detect. Thus, PC2 overexpression may not enhance occupancy of a PC1–PC2–Jade-1 complex to further augment Jade-1 protein levels. Nevertheless, Jade-1 may be yet another protein that mediates the effect of the PCs on p21. Jade-1, PC1 and PC2 may therefore participate in a common signaling cascade. The induction of p21 may in part explain how Jade-1 inhibits cell proliferation (28). Dysregulation of this potential PC1/Jade-1/p21 pathway may result in loss of the G1/S cell cycle checkpoint and, consequently, increased cell proliferation, and renal cystogenesis.
In summary, we have shown that the fate of transcriptional coactivator and ubiquitin ligase Jade-1 is intricately regulated by PC1. The differential ubiquitination of Jade-1 by PC1 and PC1-CTF is a novel mechanism that may also be applicable to other PC1 targets. In addition, our data indicate that there may be a link between Jade-1 regulation by PC1 and the pathogenesis of ADPKD. As a growth suppressive factor that is inducible with proteasome inhibition (28), Jade-1 may ultimately prove to be a useful therapeutic target for both ADPKD and VHL disease, and perhaps other forms of renal cystic disease.
MATERIALS AND METHODS
Plasmid DNA constructs
pFLAG-CMV2 Jade-1, pCR3.1 uni-HA-Jade-1 and Jade-1 deletion constructs in pFLAG-CMV2 were previously described (23,24). Jade-1 was cloned into pEGFP-N3 (Clontech) in a C-terminal position to EGFP. Full-length Jade-1 and Jade-1 deletion constructs were cloned into pCS2+ myc tag expression vector, which was a generous gift from D. Seldin (Boston University School of Medicine). For myc-tagged J-N, Jade-1 PHD-extended PHD module or large fragment (J-La) and Jade-1 inter-PHD region or small fragment (J-Sm) constructs, Jade-1 nucleotide sequences encoding amino acids 1–202, 203–271 and 254–311, respectively, were PCR-cloned into pCS2+MT vector using XhoI and SnaBI sites (Fig. 1A). cPC1 encodes the extracellular domain of CD16 followed by the CD7 transmembrane domain fused to the cytoplasmic tail of PC1 (29,30) (Fig. 1B). pCI-CMV-PKD1-FLAG (PC1) encodes the full-length cDNA for PC1 fused to a C-terminal FLAG epitope, and pCI-CMV-PKD1-R4227X-FLAG (PC1-R4227X) contains a FLAG epitope inserted after codon 4226, yielding a truncation of PC1 in the coiled-coil domain (33). pCI-CMV-AP-GPS-CTF-FLAG (PC1-CTF) encodes the PC1-CTF that results from cleavage at the GPS site (11). All domains N terminal to the GPS cleavage site were replaced with alkaline phosphatase. pCI-CMV-PKD1-T3049R-FLAG (PC1-T3049R) is an artificial PC1 mutation construct (11). pCI-CMV-PKD1-Q3016R-FLAG (PC1-Q3016R) contains a naturally occurring mutation in PC1. Neither PC1-Q3016R nor PC1-T3049R undergo cleavage at the GPS site (11). pCI-CMV-GFP-PKD1-T3049X (PC1-NTF) contains an early stop codon at residue 3049 and represents the PC1-NTF (11). HA-tagged PKD2 in pcDNA3.0 (HA-PC2) (71) and pFLAG-CMV2-pVHL were previously described (23,72). pcDNA 3.0-FLAG-Siah-1 and pcDNA 3.0-FLAG-Siah-1-dR were generously provided by E.R. Fearon (University of Michigan) (46). pCMV2-myc-ubiquitin was a generous gift from J. Xiao (Boston University School of Medicine). The CAT reporter construct contains five GAL4-binding sites cloned upstream of the adenovirus major late (AdML) promoter and was a generous gift from T. Kouzarides (Wellcome/CRC Institute) (73). GAL4-VP16, GAL4-Jade-1 and GAL4-Jade-1 dd fusions in pSG424 were described previously (24,74). β-Galactosidase was expressed under the control of a CMV-driven promoter in pCB.β-gal (75).
Compounds
Z-leu-leu-leu-CHO (MG132) and clasto-lactacystin β-lactone (lactacystin) were from Boston Biochem, Inc. Dimethylsulfoxide (DMSO) was from Fisher Scientific.
Antibodies
Jade-1 rabbit antiserum has been previously described (23), as have rabbit antisera to the C-terminus (PC1 CT) (21) and the leucine-rich region of human PC1 (PC1 LRR) (11). FLAG M5, FLAG M2 and α-tubulin monoclonal antibodies, and an anti-FLAG M2 affinity gel (M2 beads) were from Sigma. β-Actin monoclonal and myc polyclonal antibodies were from Abcam. Myc-tag monoclonal antibody was from Cell Signaling Technology. HA, GAL4 (DBD) and ubiquitin monoclonal antibodies were from Santa Cruz Biotechnology. p21 monoclonal antibody was from BD Pharmingen. Antibodies for immunofluorescence, including GFP rabbit polyclonal, Alexa Fluor® 647 goat anti-rabbit immunoglobulin (IgG) and Rhodamine RedTM-X goat anti-mouse IgG antibodies were from Molecular Probes.
Cell lines and transfection
Cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and penicillin and streptomycin (Invitrogen). Human embryonic kidney 293T17 cells were generously provided by Z. Luo (Boston University School of Medicine). The 293T17 cells were transfected using Lipofectamine 2000 (Invitrogen). Murine IMCD cells retrovirally infected to express empty vector (containing the CD16.7 cassette) or cPC1 were generously provided by L.G. Cantley (Yale University) (31). For immunoprecipitations, cells were harvested 24 h post-transfection (Fig. 2). For Jade-1 abundance and ubiquitination assays, cells were harvested 72 h post-transfection (Figs 4, 5, 7 and 8). Immortalized mouse DBA positive MEK cells from the Pkd1del34/del34mouse and littermate controls (36), as well as immortalized human ADPKD lines 9–12 and human RCTECs of proximal and distal origin were generously provided by J. Zhou and W. El-Jouni (Harvard University) (37). Cells were cultured in DMEM with 2% FBS, interferon-γ (3 U/ml), ITS (15 mg/ml), hydrocortisone (36 ng/ml) and T3 (10–7 m) at 33°C until 90% confluent, then transferred to 37°C for differentiation for 3 days without interferon. Mouse proximal tubule cell lines null (PN18 and PN24) and heterozygous (PH2 and PH3) for Pkd1 were generously provided by Z. Yu and S. Somlo (Yale University) (38–40). The lines null and heterozygous for Pkd1 were made from a single mouse carrying one null and one floxed Pkd1 allele and the conditionally immortalizing ImmortoMouse (H-2Kb-tsA58) transgene. The null lines were clonally derived from parental Pkd1fl/− clones following transient transfection with Cre recombinase and were maintained in DMEM/F12 supplemented with 10% FBS and interferon-γ (5 U/ml, Sigma-Aldrich, St Louis, MO, USA) at 33°C and 5% CO2. Cells were changed to interferon-γ-free and 37°C growth conditions 5 days before experiments to suppress large T antigen. Two independent cell lines for each genotype were used to avoid clonal artifacts.
Immunoprecipitation
Cells were lysed in lysis buffer [50 mm Tris–HCl, pH 7.6, 150 mm NaCl, 3 mm EDTA, pH 8.0, and 0.5% Triton X-100 with complete protease inhibitor (Roche)] on ice for 30 min. For immunoprecipitation, 0.5–2 mg of lysate protein was incubated with 1 µg primary antibody or with anti-FLAG M2 affinity gel (M2 beads). SDS–PAGE analysis and immunoblotting were performed as previously described (23). For 5% input of PC1, PC1-CTF, PC1-R4227X, PC1-T3049R or PC1-Q3016R in immunoprecipitations or Jade-1 abundance assays, FLAG M2 beads were used to immunoprecipitate PC1 protein from 5% of the lysate, followed by immunoblotting with PC1 CT or PC1 LRR antisera.
Mouse tissues
Livers from Pkd1cond/del2–4; tamoxifen-Cre mice and littermate controls (41,42) were used to assess endogenous Jade-1 and beta-catenin protein abundance. Mouse liver whole tissue lysates (for Jade-1) and nuclear fraction (for beta-catenin) were analyzed by SDS–PAGE and western blot. Nuclear fractions were prepared as reported (25).
Immunofluorescence
The 293T17 cells were plated on coverslips (Fisher Scientific) and cotransfected with GFP-Jade-1 and either pCI-FLAG-PC1, PC1-CTF or PC1-R4227X. Two days post-transfection, coverslips were washed in PBS, fixed for 15 min at 4°C in fixative solution (20 mm HEPES, pH 7.4, 140 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 2% formaldehyde and 0.2% glutaraldehyde), and washed in PBS. Coverslips were blocked at room temperature for 90 min in blocking buffer (1% BSA and 0.2% Triton X-100 in PBS), and then incubated at room temperature for 90 min with primary antibodies in blocking buffer. Coverslips were washed three times in PBS, blocked for 30 min at room temperature and incubated at room temperature for 60 min with secondary antibodies in blocking buffer. Coverslips were washed in PBS and mounted onto slides with gelvatol. Cells were visualized by confocal microscopy using an UltraView Live Cell Imager (Perkin Elmer). Images were analyzed for percent colocalization of GFP-Jade-1 with full-length PC1 or PC1-R4227X using NIH Image J64 colocalization finder. Statistical significance was determined by the two-tailed t-test (35).
Jade-1 protein half-life studies
The 293T17 cells were transfected with HA-Jade-1 and either empty vector, pCI-PC1 or PC1-CTF. Forty-eight hours post-transfection, cells were treated with 20 µm emetine dihydrochloride hydrate (Sigma) for 0, 15, 30, 45, 60 or 120 min. Samples were analyzed by SDS–PAGE and western blot for both Jade-1 and β-actin. Densitometry was performed on both Jade-1 and β-actin signals using the Quantity One® Alias and Multi-Analyst software (Bio-Rad). Jade-1 values were normalized to β-actin, and Jade-1 protein half-life was determined by linear regression. Data plotted are an average of n = 6 for vector, n = 4 for PC1 and n = 3 for PC1-CTF ± SE. For endogenous Jade-1 protein half-life, PN and PH cell lines were cultured at 37°C for 5 days before testing. The cells were treated with 20 μM emetine for 0, 15, 30, 45, 60 or 120 min. Nuclear fraction samples were analyzed by SDS–PAGE and western blot for both Jade-1 and β-actin. Densitometry was performed on both Jade-1 and β-actin signals using the NIH ImageJ software. Data plotted are an average of n = 3 for each sample ± SE.
CAT reporter assay
The 293T17 cells were seeded in 60-mm dishes and transfected with a total of 7.6 µg plasmid DNA, including 0.1 µg pCMV-β-gal, 2.5 µg AdML CAT-reporter plasmid, 2.5 µg either SV40-promoter-driven GAL4 DNA binding domain, or SV40-promoter-driven GAL4-Jade-1, GAL4-Jade-1 dd or GAL4-VP16 (as a positive control) and 2.5 µg effector plasmid, either pCI empty vector or PC expression vector. CAT assays were performed as described (24). Data presented in bar graphs are means of three experiments ±SE. Jade-1 transcriptional activity was analyzed using one-way non-parametric ANOVA (Kruskal–Wallis) followed by the Bonferroni multiple-comparison test. P<0.05 was considered statistically significant.
FUNDING
This work was supported by National Institutes of Health grants to H.T.C. (R01 DK067569 and R01 CA079830), National Institutes of Health training grant for R.L.F. (T32 DK07053), and fellowship grants from the National Kidney Foundation and Polycystic Kidney Disease Foundation to V.C.C. PH and PN cells were provided by the Mouse Genetics and Cell Line Core of the George M O'Brien Kidney Center at Yale (grant P30 DK079310).
ACKNOWLEDGMENTS
We thank Drs S. Alper, L. Cantley, W. El-Jouni, E. Fearon, T. Kouzarides, Z. Luo, D. Seldin, S. Somlo, J. Xiao, Z. Yu and J. Zhou for generously providing reagents. We are also grateful to Drs S. Singh and S. Krishnan for technical assistance with confocal microscopy.
Conflict of Interest statement. None declared.
References
- 1.Igarashi P., Somlo S. Genetics and pathogenesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2002;13:2384–2398. doi: 10.1097/01.asn.0000028643.17901.42. [DOI] [PubMed] [Google Scholar]
- 2.Wilson P.D. Polycystic kidney disease. N. Engl. J. Med. 2004;350:151–164. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- 3.Torres V.E., Harris P.C., Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 4.Parfrey P.S., Bear J.C., Morgan J., Cramer B.C., McManamon P.J., Gault M.H., Churchill D.N., Singh M., Hewitt R., Somlo S., et al. The diagnosis and prognosis of autosomal dominant polycystic kidney disease. N. Engl. J. Med. 1990;323:1085–1090. doi: 10.1056/NEJM199010183231601. [DOI] [PubMed] [Google Scholar]
- 5.Gabow P.A., Johnson A.M., Kaehny W.D., Kimberling W.J., Lezotte D.C., Duley I.T., Jones R.H. Factors affecting the progression of renal disease in autosomal- dominant polycystic kidney disease. Kidney Int. 1992;41:1311–1319. doi: 10.1038/ki.1992.195. [DOI] [PubMed] [Google Scholar]
- 6.Freedman B.I., Soucie J.M., Chapman A., Krisher J., McClellan W.M. Racial variation in autosomal dominant polycystic kidney disease. Am. J. Kidney Dis. 2000;35:35–39. doi: 10.1016/S0272-6386(00)70298-8. [DOI] [PubMed] [Google Scholar]
- 7.Kimberling W.J., Fain P.R., Kenyon J.B., Goldgar D., Sujansky E., Gabow P.A. Linkage heterogeneity of autosomal dominant polycystic kidney disease. N. Engl. J. Med. 1988;319:913–918. doi: 10.1056/NEJM198810063191405. [DOI] [PubMed] [Google Scholar]
- 8.The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. The European Polycystic Kidney Disease Consortium. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 9.Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. The International Polycystic Kidney Disease Consortium. Cell. 1995;81:289–298. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 10.Hughes J., Ward C.J., Peral B., Aspinwall R., Clark K., San Millan J.L., Gamble V., Harris P.C. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 1995;10:151–160. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 11.Qian F., Boletta A., Bhunia A.K., Xu H., Liu L., Ahrabi A.K., Watnick T.J., Zhou F., Germino G.G. Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc. Natl Acad. Sci. USA. 2002;99:16981–16986. doi: 10.1073/pnas.252484899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S., Hackmann K., Gao J., He X., Piontek K., Garcia-Gonzalez M.A., Menezes L.F., Xu H., Germino G.G., Zuo J., et al. Essential role of cleavage of Polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc. Natl Acad. Sci. USA. 2007;104:18688–18693. doi: 10.1073/pnas.0708217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvet J.P. Ciliary signaling goes down the tubes. Nat. Genet. 2003;33:113–114. doi: 10.1038/ng1078. [DOI] [PubMed] [Google Scholar]
- 14.Chauvet V., Tian X., Husson H., Grimm D.H., Wang T., Hiesberger T., Igarashi P., Bennett A.M., Ibraghimov-Beskrovnaya O., Somlo S., et al. Mechanical stimuli induce cleavage and nuclear translocation of the polycystin-1 C terminus. J. Clin. Invest. 2004;114:1433–1443. doi: 10.1172/JCI21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen H.T., McGovern F.J. Renal-cell carcinoma. N. Engl. J. Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 16.Lubensky I.A., Gnarra J.R., Bertheau P., Walther M.M., Linehan W.M., Zhuang Z. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am. J. Pathol. 1996;149:2089–2094. [PMC free article] [PubMed] [Google Scholar]
- 17.Qian F., Watnick T.J., Onuchic L.F., Germino G.G. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell. 1996;87:979–987. doi: 10.1016/s0092-8674(00)81793-6. [DOI] [PubMed] [Google Scholar]
- 18.Pause A., Lee S., Lonergan K.M., Klausner R.D. The von Hippel-Lindau tumor suppressor gene is required for cell cycle exit upon serum withdrawal. Proc. Natl Acad. Sci. USA. 1998;95:993–998. doi: 10.1073/pnas.95.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenfeld A.R., Parris T., Eisenberger A., Davidowitz E.J., De Leon M., Talasazan F., Devarajan P., Burk R.D. The von Hippel-Lindau tumor suppressor gene protects cells from UV- mediated apoptosis. Oncogene. 2000;19:5851–5857. doi: 10.1038/sj.onc.1203985. [DOI] [PubMed] [Google Scholar]
- 20.Bhunia A.K., Piontek K., Boletta A., Liu L., Qian F., Xu P.N., Germino F.J., Germino G.G. PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell. 2002;109:157–168. doi: 10.1016/s0092-8674(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 21.Boletta A., Qian F., Onuchic L.F., Bhunia A.K., Phakdeekitcharoen B., Hanaoka K., Guggino W., Monaco L., Germino G.G. Polycystin-1, the gene product of PKD1, induces resistance to apoptosis and spontaneous tubulogenesis in MDCK cells. Mol. Cell. 2000;6:1267–1273. doi: 10.1016/s1097-2765(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 22.Devarajan P., De Leon M., Talasazan F., Schoenfeld A.R., Davidowitz E.J., Burk R.D. The von Hippel-Lindau gene product inhibits renal cell apoptosis via Bcl-2-dependent pathways. J. Biol. Chem. 2001;276:40599–40605. doi: 10.1074/jbc.M103424200. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M.I., Wang H., Ross J.J., Kuzmin I., Xu C., Cohen H.T. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J. Biol. Chem. 2002;277:39887–39898. doi: 10.1074/jbc.M205040200. [DOI] [PubMed] [Google Scholar]
- 24.Panchenko M.V., Zhou M.I., Cohen H.T. von Hippel-Lindau partner Jade-1 is a transcriptional co-activator associated with histone acetyltransferase activity. J. Biol. Chem. 2004;279:56032–56041. doi: 10.1074/jbc.M410487200. [DOI] [PubMed] [Google Scholar]
- 25.Chitalia V.C., Foy R.L., Bachschmid M.M., Zeng L., Panchenko M.V., Zhou M.I., Bharti A., Seldin D.C., Lecker S.H., Dominguez I., et al. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat. Cell Biol. 2008;10:1208–1216. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzouanacou E., Tweedie S., Wilson V. Identification of Jade1, a gene encoding a PHD zinc finger protein, in a gene trap mutagenesis screen for genes involved in anteroposterior axis development. Mol. Cell. Biol. 2003;23:8553–8562. doi: 10.1128/MCB.23.23.8553-8562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M.I., Wang H., Foy R.L., Ross J.J., Cohen H.T. Tumor suppressor von Hippel-Lindau (VHL) stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res. 2004;64:1278–1286. doi: 10.1158/0008-5472.can-03-0884. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M.I., Foy R.L., Chitalia V.C., Zhao J., Panchenko M.V., Wang H., Cohen H.T. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc. Natl Acad. Sci. USA. 2005;102:11035–11040. doi: 10.1073/pnas.0500757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsiokas L., Kim E., Arnould T., Sukhatme V.P., Walz G. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc. Natl Acad. Sci. USA. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnould T., Kim E., Tsiokas L., Jochimsen F., Gruning W., Chang J.D., Walz G. The polycystic kidney disease 1 gene product mediates protein kinase C alpha-dependent and c-Jun N-terminal kinase-dependent activation of the transcription factor AP-1. J. Biol. Chem. 1998;273:6013–6018. doi: 10.1074/jbc.273.11.6013. [DOI] [PubMed] [Google Scholar]
- 31.Nickel C., Benzing T., Sellin L., Gerke P., Karihaloo A., Liu Z.X., Cantley L.G., Walz G. The polycystin-1 C-terminal fragment triggers branching morphogenesis and migration of tubular kidney epithelial cells. J. Clin. Invest. 2002;109:481–489. doi: 10.1172/JCI12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian F., Germino F.J., Cai Y., Zhang X., Somlo S., Germino G.G. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat. Genet. 1997;16:179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- 33.Hanaoka K., Qian F., Boletta A., Bhunia A.K., Piontek K., Tsiokas L., Sukhatme V.P., Guggino W.B., Germino G.G. Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature. 2000;408:990–994. doi: 10.1038/35050128. [DOI] [PubMed] [Google Scholar]
- 34.Perry J. The Epc-N domain: a predicted protein-protein interaction domain found in select chromatin associated proteins. BMC Genomics. 2006;7:6. doi: 10.1186/1471-2164-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrie R.J., Deans J.P. Colocalization of the B cell receptor and CD20 followed by activation-dependent dissociation in distinct lipid rafts. J. Immunol. 2002;169:2886–2891. doi: 10.4049/jimmunol.169.6.2886. [DOI] [PubMed] [Google Scholar]
- 36.Nauli S.M., Alenghat F.J., Luo Y., Williams E., Vassilev P., Li X., Elia A.E., Lu W., Brown E.M., Quinn S.J., et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 37.Loghman-Adham M., Nauli S.M., Soto C.E., Kariuki B., Zhou J. Immortalized epithelial cells from human autosomal dominant polycystic kidney cysts. Am. J. Physiol. Renal Physiol. 2003;285:F397–F412. doi: 10.1152/ajprenal.00310.2002. [DOI] [PubMed] [Google Scholar]
- 38.Joly D., Ishibe S., Nickel C., Yu Z., Somlo S., Cantley L.G. The polycystin 1-C-terminal fragment stimulates ERK-dependent spreading of renal epithelial cells. J. Biol. Chem. 2006;281:26329–26339. doi: 10.1074/jbc.M601373200. [DOI] [PubMed] [Google Scholar]
- 39.Lal M., Song X., Pluznick J.L., Di Giovanni V., Merrick D.M., Rosenblum N.D., Chauvet V., Gottardi C.J., Pei Y., Caplan M.J. Polycystin-1 C-terminal tail associates with beta-catenin and inhibits canonical Wnt signaling. Hum. Mol. Genet. 2008;17:3105–3117. doi: 10.1093/hmg/ddn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei F., Karihaloo A., Yu Z., Marlier A., Seth P., Shibazaki S., Wang T., Sukhatme V.P., Somlo S., Cantley L.G. Neutrophil gelatinase-associated lipocalin suppresses cyst growth by Pkd1 null cells in vitro and in vivo. Kidney Int. 2008;74:1310–1318. doi: 10.1038/ki.2008.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piontek K.B., Huso D.L., Grinberg A., Liu L., Bedja D., Zhao H., Gabrielson K., Qian F., Mei C., Westphal H., et al. A functional floxed allele of Pkd1 that can be conditionally inactivated in vivo. J. Am. Soc. Nephrol. 2004;15:3035–3043. doi: 10.1097/01.ASN.0000144204.01352.86. [DOI] [PubMed] [Google Scholar]
- 42.Piontek K., Menezes L.F., Garcia-Gonzalez M.A., Huso D.L., Germino G.G. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat. Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorick K.L., Jensen J.P., Fang S., Ong A.M., Hatakeyama S., Weissman A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.House C.M., Frew I.J., Huang H.L., Wiche G., Traficante N., Nice E., Catimel B., Bowtell D.D. A binding motif for Siah ubiquitin ligase. Proc. Natl Acad. Sci. USA. 2003;100:3101–3106. doi: 10.1073/pnas.0534783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H., Jeong W., Ahn K., Ahn C., Kang S. Siah-1 interacts with the intracellular region of polycystin-1 and affects its stability via the ubiquitin-proteasome pathway. J. Am. Soc. Nephrol. 2004;15:2042–2049. doi: 10.1097/01.ASN.0000133490.00348.59. [DOI] [PubMed] [Google Scholar]
- 46.Hu G., Fearon E.R. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X., Luo Y., Starremans P.G., McNamara C.A., Pei Y., Zhou J. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat. Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 48.Rossetti S., Strmecki L., Gamble V., Burton S., Sneddon V., Peral B., Roy S., Bakkaloglu A., Komel R., Winearls C.G., et al. Mutation analysis of the entire PKD1 gene: genetic and diagnostic implications. Am. J. Hum. Genet. 2001;68:46–63. doi: 10.1086/316939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim E., Arnould T., Sellin L.K., Benzing T., Fan M.J., Gruning W., Sokol S.Y., Drummond I., Walz G. The polycystic kidney disease 1 gene product modulates Wnt signaling. J. Biol. Chem. 1999;274:4947–4953. doi: 10.1074/jbc.274.8.4947. [DOI] [PubMed] [Google Scholar]
- 50.Qin S., Taglienti M., Nauli S.M., Contrino L., Takakura A., Zhou J., Kreidberg J.A. Failure to ubiquitinate c-Met leads to hyperactivation of mTOR signaling in a mouse model of autosomal dominant polycystic kidney disease. J. Clin. Invest. 2010;120:3617–3628. doi: 10.1172/JCI41531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J., Stevens J., Rote C.A., Yost H.J., Hu Y., Neufeld K.L., White R.L., Matsunami N. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 52.Matsuzawa S.I., Reed J.C. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- 53.Dimitrova Y.N., Li J., Lee Y.T., Rios-Esteves J., Friedman D.B., Choi H.J., Weis W.I., Wang C.Y., Chazin W.J. Direct ubiquitination of beta-catenin by Siah-1 and regulation by the exchange factor TBL1. J. Biol. Chem. 2010;285:13507–13516. doi: 10.1074/jbc.M109.049411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu G., Zhang S., Vidal M., Baer J.L., Xu T., Fearon E.R. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes. Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boca M., D'Amato L., Distefano G., Polishchuk R.S., Germino G.G., Boletta A. Polycystin-1 induces cell migration by regulating phosphatidylinositol 3-kinase-dependent cytoskeletal rearrangements and GSK3beta-dependent cell cell mechanical adhesion. Mol. Biol. Cell. 2007;18:4050–4061. doi: 10.1091/mbc.E07-02-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim I., Ding T., Fu Y., Li C., Cui L., Li A., Lian P., Liang D., Wang D.W., Guo C., et al. Conditional mutation of Pkd2 causes cystogenesis and upregulates beta-catenin. J. Am. Soc. Nephrol. 2009;20:2556–2569. doi: 10.1681/ASN.2009030271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huan Y., van Adelsberg J. Polycystin-1, the PKD1 gene product, is in a complex containing E- cadherin and the catenins. J. Clin. Invest. 1999;104:1459–1468. doi: 10.1172/JCI5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saadi-Kheddouci S., Berrebi D., Romagnolo B., Cluzeaud F., Peuchmaur M., Kahn A., Vandewalle A., Perret C. Early development of polycystic kidney disease in transgenic mice expressing an activated mutant of the beta-catenin gene. Oncogene. 2001;20:5972–5981. doi: 10.1038/sj.onc.1204825. [DOI] [PubMed] [Google Scholar]
- 59.Rodova M., Islam M.R., Maser R.L., Calvet J.P. The polycystic kidney disease-1 promoter is a target of the beta - catenin/T-cell factor pathway. J. Biol. Chem. 2002;277:29577–29583. doi: 10.1074/jbc.M203570200. [DOI] [PubMed] [Google Scholar]
- 60.Lin F., Hiesberger T., Cordes K., Sinclair A.M., Goldstein L.S., Somlo S., Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl Acad. Sci. USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roitbak T., Ward C.J., Harris P.C., Bacallao R., Ness S.A., Wandinger-Ness A. A polycystin-1 multiprotein complex is disrupted in polycystic kidney disease cells. Mol. Biol. Cell. 2004;15:1334–1346. doi: 10.1091/mbc.E03-05-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qian C.N., Knol J., Igarashi P., Lin F., Zylstra U., Teh B.T., Williams B.O. Cystic renal neoplasia following conditional inactivation of apc in mouse renal tubular epithelium. J. Biol. Chem. 2005;280:3938–3945. doi: 10.1074/jbc.M410697200. [DOI] [PubMed] [Google Scholar]
- 63.Simons M., Gloy J., Ganner A., Bullerkotte A., Bashkurov M., Kronig C., Schermer B., Benzing T., Cabello O.A., Jenny A., et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat. Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berndt J.D., Moon R.T., Major M.B. beta-catenin gets jaded and von Hippel-Lindau is to blame. Trends Biochem. Sci. 2009;34:101–104. doi: 10.1016/j.tibs.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 65.Benzing T., Simons M., Walz G. Wnt signaling in polycystic kidney disease. J. Am. Soc. Nephrol. 2007;18:1389–1398. doi: 10.1681/ASN.2006121355. [DOI] [PubMed] [Google Scholar]
- 66.Lancaster M.A., Gleeson J.G. Cystic kidney disease: the role of Wnt signaling. Trends Mol. Med. 2010;16:349–360. doi: 10.1016/j.molmed.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doyon Y., Cayrou C., Ullah M., Landry A.J., Cote V., Selleck W., Lane W.S., Tan S., Yang X.J., Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 68.Low S.H., Vasanth S., Larson C.H., Mukherjee S., Sharma N., Kinter M.T., Kane M.E., Obara T., Weimbs T. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev. Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Cao Y., Semanchik N., Lee S.H., Somlo S., Barbano P.E., Coifman R., Sun Z. Chemical modifier screen identifies HDAC inhibitors as suppressors of PKD models. Proc. Natl Acad. Sci. USA. 2009;106:21819–21824. doi: 10.1073/pnas.0911987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia S., Li X., Johnson T., Seidel C., Wallace D.P., Li R. Polycystin-dependent fluid flow sensing targets histone deacetylase 5 to prevent the development of renal cysts. Development. 2010;137:1075–1084. doi: 10.1242/dev.049437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsiokas L., Arnould T., Zhu C., Kim E., Walz G., Sukhatme V.P. Specific association of the gene product of PKD2 with the TRPC1 channel. Proc. Natl Acad. Sci. USA. 1999;96:3934–3939. doi: 10.1073/pnas.96.7.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukhopadhyay D., Knebelmann B., Cohen H.T., Ananth S., Sukhatme V.P. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol. Cell. Biol. 1997;17:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Balbas M.A., Bannister A.J., Martin K., Haus-Seuffert P., Meisterernst M., Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 75.Gashler A.L., Swaminathan S., Sukhatme V.P. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol. Cell. Biol. 1993;13:4556–4571. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]