Abstract
5-Hydroxymethylcytosine (5-hmC) is a newly discovered modified form of cytosine that has been suspected to be an important epigenetic modification in neurodevelopment. While DNA methylation dynamics have already been implicated during neurodevelopment, little is known about hydroxymethylation in this process. Here, we report DNA hydroxymethylation dynamics during cerebellum development in the human brain. Overall, we find a positive correlation between 5-hmC levels and cerebellum development. Genome-wide profiling reveals that 5-hmC is highly enriched on specific gene regions including exons and especially the untranslated regions (UTRs), but it is depleted on introns and intergenic regions. Furthermore, we have identified fetus-specific and adult-specific differentially hydroxymethylated regions (DhMRs), most of which overlap with genes and CpG island shores. Surprisingly, during development, DhMRs are highly enriched in genes encoding mRNAs that can be regulated by fragile X mental retardation protein (FMRP), some of which are disrupted in autism, as well as in many known autism genes. Our results suggest that 5-hmC-mediated epigenetic regulation may broadly impact the development of the human brain, and its dysregulation could contribute to the molecular pathogenesis of neurodevelopmental disorders.
Accession number: Sequencing data have been deposited to GEO with accession number GSE40539.
INTRODUCTION
Methylation at the 5-positon of cytosine (5-mC), which is catalyzed by DNA methyltransferases (DNMTs), plays important roles in mammalian neuronal systems (1). The proper establishment of DNA methylation is critical for embryonic and postnatal development (2,3). DNMT3a is required for maintaining neural stem cell self-renewal, and loss of the protein significantly impairs postnatal neurogenesis (4), suggesting a regulatory role of DNA methylation in neurodevelopmental process (5). Dnmt1 and Dnmt3a depletion induces abnormal neuronal phenotypes, including learning and memory deficits and abnormal synaptic plasticity (6). Furthermore, pharmacological inhibition of Dnmt activity can block hippocampus-dependent memory formation (7). Mutation of methyl-CpG binding protein 2 (MECP2), which binds to methylated DNA and acts as a transcriptional repressor or activator, causes Rett syndrome, and related neurodevelopmental disorders (8). These observations indicate that the proper establishment and maintenance of DNA methylation are essential for normal development and function of the mammalian brain.
5-Hydroxymethylcytosine (5-hmC), which is converted from 5-mC by ten-eleven translocation (TET) proteins, is present in the mammalian genome (9–11). All TET family proteins can catalyze the conversion of 5-mC to 5-hmC. TET1 was first reported to have a role in maintaining the pluripotent state of embryonic stem cells (ESCs) (12), and together with TET2, it also regulates the cell lineage commitment of ESCs (13). TET2 modulates the balance between self-renewal and differentiation in hematopoietic stem cells, making them critical for normal myelopoiesis; TET2 mutations are seen in multiple types of leukemia (14–18). TET3 contributes to the global DNA methylation erasure during the zygote stage of embryonic development (19,20). Taken together, these studies point to a critical role of TET-mediated 5-hmC modification in developmental processes and the possibility that dysregulation of 5-hmC may be associated with disease.
Although DNA methylation has generally been regarded as a highly stable epigenetic mark, recent studies have uncovered DNA methylation changes during brain development and aging, suggesting that epigenetic changes like 5-mC could function as an intermediate step for the internal or external environmental regulation of the brain genome. Studies from our own and other groups have identified strong enrichment of 5-hmC in mammalian brains (9,10,21). However, compared with 5-mC, little is known about the roles of 5-hmC in the mammalian brain. Our previous study revealed the dynamics of DNA hydroxymethylation during postnatal development and ageing in mouse brain (21,22). Another study found that 5-hmC is enriched at promoters and gene bodies, and its enrichment on gene bodies is positively correlated with gene expression in the frontal lobe tissue of human brain (23). Nevertheless, the features of 5-hmC during human brain development remain a mystery.
Here, we extend our previous work to profile the genome-wide distribution of 5-hmC during cerebellum development in human fetal and adult brains. We find that the overall 5-hmC level increases during cerebellum development. Most differentially hydroxymethylated regions (DhMRs) between the fetus and the adult overlap with genes, and are strongly associated with CpG island shores. Strikingly, these 5-hmC changes are highly enriched in genes whose transcripts can be regulated by fragile X mental retardation protein (FMRP), as well as in many genes linked with autism.
RESULTS
Dynamics of DNA hydroxymethylation and its genomic features in the human cerebellum
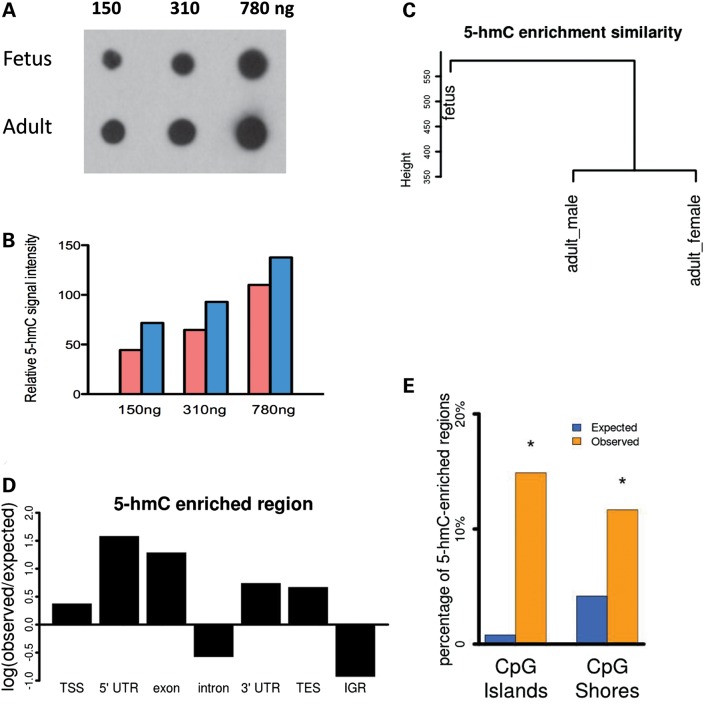
Previous studies indicated that 5-hmC levels in the mouse cerebellum are high and can change during developmental processes (10,21). To extend this work, here we focused on the human cerebellum to study the features of 5-hmC in the developing human brain. We first performed 5-hmC-specific dot blot with fetus and adult cerebellum DNA samples, and found that the total levels of 5-hmC increased significantly (42.1% increase) from the fetus to the adult stage (Fig. 1A and B), consistent with the observation in mouse cerebellums.
Figure 1.
Global DNA hydroxymethylation dynamics in the developing human cerebellum. (A) 5-hmC dot blot analysis shows a significant increase of total 5-hmC levels in the adult cerebellum compared with the fetal cerebellum. Each column indicates the total amount of DNA used. (B) Quantitative measurement of total intensity of 5-hmC levels shown in (A). (C) Cluster dendrogram analysis using the 5-hmC-enriched regions either present in fetus or two adult samples across all samples. (D) The relative enrichment of 5-hmC distribution across distinct genomic regions. (E) 5-hmC is preferentially enriched in CpG islands and CpG island shores. Asterisk indicates statistical significance of observed distribution compared with expected distribution (P < 0.001, Pearson's chi-squared test).
To profile the genome-wide distribution of 5-hmC in the human cerebellum, we isolated genomic DNA containing 5-hmC using a chemical capture technique developed previously (22) and then sequenced those DNA fragments. A genome-wide correlation analysis showed that the 5-hmC profiles from two adult cerebellums are more similar to each other than either is to the fetal cerebellum (Fig. 1C). We then determined the genomic features associated with 5-hmC-enriched regions and found that 5-hmC is highly enriched at genes (Supplementary Material, Fig. S1), particularly 5′-UTR and exons, but it is depleted at introns and intergenic regions in both the fetal and adult samples (Fig. 1D). Furthermore, 5-hmC is strongly associated with CpG islands and CpG island shores (P < 0.001, comparing observed and expected frequencies) (Fig. 1E). Together, these data suggest that 5-hmC is significantly increased during the development of the human cerebellum and is associated with specific genomic regions.
5-hmC genomic features during cerebellum development
To examine the genomic features of 5-hmC during the development of the human cerebellum, we used a Poisson-based model calling method to determine the fetus-specific DhMRs (fetus has higher 5-hmC levels than adult) and adult-specific DhMRs (adult has higher 5-hmC levels than fetus). Across the genome, we identified 28 015 DhMRs between the fetus and adult samples, of which 15 829 are adult-specific and 12 186 are fetus-specific DhMRs (Fig. 2A). Hierarchical clustering of the top 500 most significant DhMRs indicated a greater similarity between adult samples, and a significant difference between the fetus and the adult, most showing an increase of 5-hmC levels in adult (Fig. 2B). We noted that the fold-change of adult-specific DhMRs is much greater than fetus-specific DhMRs (Fig. 2C). The majority of DhMRs we identified also showed a strong bias towards CpG islands and CpG island shores (P < 0.001) (Fig. 2D and E). Fetus-specific DhMRs had a stronger tendency to be associated with CpG islands (10.4 versus 5.2%) and CpG shores (22 versus 14%) than adult-specific DhMRs. Furthermore, both fetus- and adult-specific DhMRs overlapped more with CpG shores than with CpG islands (Fig. 2D and E).
Figure 2.
Unique genomic features of dynamic 5-hmC in fetal and adult cerebellums. (A) Total number of fetus-specific DhMRs and adult-specific DhMRs. (B) Heatmap of the top 500 DhMRs that show the most significant differences between the fetus and the adult. Green color represents more 5-hmC counts and red color represents less 5-hmC counts. (C) Average fold change of adult-specific DhMRs and fetus-specific DhMRs. (D) Fetus- and adult-specific DhMRs distribution in CpG islands. (E) Fetus- and adult-specific DhMRs distribution in CpG island shores. In (D) and (E), yellow bar indicates numbers overlapping with CpG islands or shores, and blue bar indicates numbers not overlapping with CpG islands or shores.
To examine the potential relationship between DhMRs and gene function, we aligned the identified DhMRs to the annotated human genes and found that 69% (8,407) of fetus-specific DhMRs overlapped with genes (Fig. 3A) and 72% (11,396) of adult-specific DhMRs overlapped with genes (Fig. 3B). Both the fetus-specific and adult-specific DhMRs are also largely localized to UTR and exons, but are depleted at introns and intergenic regions (Fig. 3C and D). A similar pattern has been seen in DhMRs identified in mouse cerebellum (21). Intriguingly, the fetus-specific DhMRs are more strongly associated with transcription start sites (TSS) than the adult-specific DhMRs (Fig. 3C and D). Furthermore, around half of the genes associated with DhMRs also show differential hydroxymethylation during cerebellum development in mouse (observed-to-expected ratio: 1.45, 3.46 and 6.83 for genes that are associated with at least 1, 2 and 4 DhMRs) (Fig. 3E). Gene ontology (GO) analysis indicated that the adult-specific DhMRs are enriched at genes involved in ion channel binding and cell–cell adhesion, and the fetus-specific DhMRs preferentially localize at genes associated with the ion channel and neuronal development (Supplementary Material, Fig. S2A and B). Figure 3E shows DhMRs in two genes, MYOD1 and MAP1B, both of which encode proteins involved in neuronal function.
Figure 3.
Genomic features of DhMRs during cerebellum development. (A) Number of fetus-specific DhMRs overlapping with genes. (B) The relative enrichment of fetus-specific DhMRs distribution across distinct genomic features. (C) Number of adult-specific DhMRs overlapping with genes. (D) The relative enrichment of adult-specific DhMRs distribution across distinct genomic features. (E) Genes associated with DhMRs are highly conserved between human and mouse. Shown are observed and expected percentages of human genes associated with at least 1, 2 and 4 DhMRs that are conserved in the mouse. (F) IGV Genome Browser track showing 5-hmC levels of MYOD1 and MAP1b in the fetus and two adult cerebellums.
The fetus-specific DhMRs displays pluripotent epigenetic memories
Epigenetic properties of human ESCs dictate proper fetal development, and presumably, hESCs share more epigenetic features with the fetus than with the adult (24,25). Previous studies have also found 5-hmC enriched at genes in hESCs (26,27). We compared 5-hmC profiling between H1 hESCs and human cerebellum, and found that 5-hmC is dramatically depleted at TSS in human brain relative to ESCs, but still, retain a bimodal distribution (Fig. 4A). Moreover, 5-hmC exhibits less enrichment at CpG islands and shores in human brain than in hESCs (Fig. 4B–D). These results indicate that the genomic distribution of 5-hmC has a different pattern in human brain, consisting mainly of post-mitotic cells, proliferating hESCs.
Figure 4.
Fetus-specific DhMRs show more epigenetic memories that are present in embryonic stem cells (ESCs). (A) Enrichment of 5-hmC at RefSeq gene TSS boundary regions (± 3 kb) in H1 hESCs, fetal and adult cerebellums. (B) Enrichment of 5-hmC at CpG island shores upstream boundary regions (−5 kb) in H1 hESCs, fetal and adult cerebellums. (C) Enrichment of 5-hmC at CpG island boundary regions (± 3 kb) in H1 hESCs, fetal and adult cerebellums. (D) Enrichment of 5-hmC at CpG island shores downstream boundary regions (−5 kb) in H1 hESCs, fetal and adult cerebellums. (E) 5-hmC in fetus-specific DhMRs compared with H1 hESCs. Regions also showing 5-hmC enrichment in H1 hESCs are highlighted in gray. Regions showing the absence of 5-hmC in H1 hESCs are highlighted in black. (F) Top 500 most significant DhMRs by fold-change compared with H1 hESCs; gray and black colors are the same as (E).
We next compared DhMRs identified in human brain with H1 ESCs 5-hmC enrichment peaks that we previously identified (28). We found that 57% of fetus-specific DhMRs were shared with hESCs, but only 19% of adult-specific DhMRs were found in hESCs (Fig. 4E). Furthermore, 338 of the 500 most highly enriched fetus-specific DhMRs were shared with ESCs while only 146 of the 500 most highly enriched adult DhMRs occurred in ESCs (Fig. 4F). These data indicate that the fetus shares more epigenetic memory with ESCs than does the adult; this could play a role in development-specific gene expression, as suggested by the GO analysis of fetus-specific DhMRs described above.
DhMRs are associated with genes involved in neurodevelopmental disorders
DNA methylation-mediated epigenetic modulation plays important roles in neurodevelopment. Dysregulation of DNA methylation can cause disorders such as Rett syndrome and fragile X syndrome (FXS) (1,29,30). Recent studies have revealed 5-hmC-mediated epigenetic dynamics during embryonic (19,20) and postnatal development (21), which indicates a potential function for 5-hmC in development and disease (31). More interestingly, both loss- and over-expression of MeCP2 not only affects the overall level of 5-hmC, but also modulates its distribution in the genome during mouse cerebellum development (21). These studies pointed out to a potential role of 5-hmC in neurodevelopmental disorders.
To better understand the roles of 5-hmC in neurodevelopmental disorders, here we asked whether 5-hmC-enriched regions we identified were associated with neurodevelopmental disorders such as FXS, and more broadly, autism spectrum disorder (ASD). We first compared 5-hmC enrichment on all UC Santa Cruz (UCSC) RefSeq genes and FMRP target genes that were identified previously (32); FMRP targets displayed more 5-hmC enrichment than the RefSeq genes (Fig. 5A). We then stratified the RefSeq genes into three groups: highly, moderately and weakly expressed genes based on the RNA expression level in the human cerebellum. FMRP target genes displayed more 5-hmC enrichment than any of the three groups (P < 0.001, Wilcoxon rank test). To rule out the possibility that enrichment of hydroxymethylation in FMRP target genes is a general characteristic of RNA binding proteins in cerebellum, we also looked at TAR DNA-binding protein 43 (TDP-43), which is involved in pre-mRNA splicing and translational regulation of its RNA ligand. Compared with FMRP target genes, we did not see strong 5-hmC enrichment in TDP-43 target genes (Fig. 5A). We then analyzed 5-hmC mapping reads on all RefSeq genes, highly expressed genes, TDP-43 target genes and FMRP target genes and found 5-hmC showed higher mapping reads on FMRP target genes in both fetus and adult brain versus all other groups (P < 0.001, Wilcoxon rank test) (Fig. 5B and C). Moreover, DhMRs also had a stronger tendency to localize on FMRP target genes than any other group (Fig. 5D and E).
Figure 5.
FMRP target genes have strong 5-hmC enrichment and are significantly associated with DhMRs. (A) Number of 5-hmC peaks per 10 kb length in RefSeq genes weakly, moderately, highly expressed genes TDP-43 target genes and FMRP target genes. (B) Box plots of hydroxymethylation levels among all RefSeq genes, highly expressed genes, TDP-43 target genes and FMRP target genes in fetus (left panel) and two adults (middle and right panels). Asterisk indicates significantly more 5-hmC levels compared with all others (P < 0.001, Wilcoxon rank test). (C) Histograms of gene number summary by number of 5-hmC peaks per 10 kb in each gene of all RefSeq genes, highly expressed genes, TDP-43 target genes and FMRP target genes. (D) The composition of RefSeq genes weakly, moderately and highly expressed genes top 2000 most expressed genes TDP-43 target genes FMRP target genes that are associated with DhMRs between the fetus and adult cerebellums. (E) Box plots of normalized DhMRs among all groups mentioned in (D).
FXS is one of the leading monogenic causes of ASD, with up to 30% of FXS patients showing autistic symptoms. Two recent studies found an unusual coincidence between autism-related genes and FMRP target genes, which had more than expected de novo mutations in children with ASD (32,33). Since we saw that 5-hmC significantly overlaps with FMRP target genes, we suspect that 5-hmC could also overlap more broadly with ASD genes. To examine this possibility, we investigated 190 autism candidate genes from the Simons Foundation Autism Research Initiative (SFARI) database (as of October 2011). Even though most of the FMRP target genes and autism candidate genes are associated with synaptic functions, in contrast to FMRP target genes, we did not see 5-hmC preferentially enriched in these autism candidate genes. However, consistent with FMRP target genes, we observed that the identified DhMRs between the fetus and the adult were enriched more (P < 0.001, Wilcoxon rank test; and observed-to-expected ratio, 1.65) in autism candidate genes relative to all RefSeq genes (Fig. 6A and B). The enrichment is also true when the 190 candidate genes were grouped into highly expressed genes, moderately expressed genes and weakly expressed genes (Fig. 6C). To further examine this correlation, we focused on the 22 syndromic genes and 21 strong candidate and suggestive genes linked to autism in the SFARI database. We found that both the total counts and DhMRs of 5-hmC are enriched in some autism syndromic genes including CACN1C, SHANK3 and TSC2 (Supplementary Material, Fig. S3A and C) as well as strong candidate and suggestive genes including CACNA1H, ATP10A and OXTR (Supplementary Material, Fig. S3B and D). Importantly, DhMRs significantly overlap with strong candidate and suggestive genes (observed-to-expected ratio, 1.52).
Figure 6.
SFARI autism candidate genes are preferentially associated with developmentally dynamic hydroxymethylation regions. (A) DhMRs distribution in all RefSeq genes and SFARI autism candidate genes. Genes are normalized by 10 kb length when counting DhMRs. Asterisk indicates a significant difference in 5-hmC levels (P < 0.001, Wilcoxon rank test). (B) Percentage of all RefSeq genes and SFARI autism genes that are associated with DhMRs. (C) Percentage of highly expressed genes and 61 highly expressed SFARI autism genes (left), moderately expressed genes and 55 related SFARI autism genes (middle), weakly expressed genes and 84 related SFARI autism genes (right) that are overlapped with DhMRs.
Taken together, these results indicate that both stable and dynamic 5-hmC strongly associated with FMRP target genes, and 5-hmC changes are associated with autism genes. Our results suggest that dysregulation of 5-hmC could be a potential contributor to the pathogenesis of neurodevelopmental disorders.
DISCUSSION
Previous studies have shown that 5-hmC is enriched in the mammalian brain (10,21,23), and its levels vary in different tissues (34–37). However, the genomic features of 5-hmC modification during the development of human brain are still unknown. To explore this, we profiled 5-hmC at a genome-wide level in fetal and adult human brain and found that 5-hmC is increased during the development of human cerebellum, suggesting strong DNA hydroxymethylation dynamics in this brain region. In our previous study, we have discovered that 5-hmC displays dynamics during the postnatal development of mouse brain (21). Together with this finding, our current results suggest that 5-hmC-mediated epigenetic pathways might play evolutionarily conserved roles in the mammalian brain. Compared with other brain regions, the cerebellum displays a distinct pattern of both DNA methylation and gene expression (38–40). The differences between the cerebellum and other brain regions may be partially attributable to cerebellum Purkinje neurons, which are considered a primary organizer in the development of the cerebellum. Purkinje neurons exhibit a greater proportion of 5-hmC versus other types of neurons. Therefore, the cerebellum may be a specific brain region susceptible to 5-hmC changes partially through Purkinje neurons.
DNA methylation is considered a relatively stable epigenetic mark compared with histone modifications. During the last several years, DNA methylation signature was found to show certain dynamics associated with brain development and aging. Since most approaches assessing DNA methylation levels cannot distinguish hydroxymethylation from methylation, the extent to which hydroxymethylation contributes to previously discovered methylation dynamics remains unknown. Our results suggest that, at least in the cerebellum, 5-hmC levels also change dramatically. In particular, some of the 5-hmC dynamically modified genes, such as MYOD1, have been reported to be associated with methylation dynamics during brain development and the aging process (39,40). It is likely that 5-hmC, together with 5-mC, acts as an intermediate step for upstream regulators to regulate these gene expression changes.
5-hmC is abundant in hESCs and brain regions, and the genomic features of 5-hmC in hESCs are well characterized (28,41). Similar to hESCs, in cerebellum, 5-hmC displayed a bimodal distribution around TSS and was enriched in gene body regions. Interestingly, compared with hESCs, we found less 5-hmC at CpG islands and CpG shores, suggesting that CpG islands and shores may have a specific protective mechanism against 5-hmC modification in cerebellum. Furthermore, we find that 5-hmC changes tend to occur more often outside of CpG islands (more likely to occur in CpG island shores than islands). This observation is also consistent with DNA methylation dynamics being seen most often occurring outside of CpG island regions during early brain development and aging, as well as tissue and cancer differences (42,43). For example, Numta et al. showed that DNA methylation changes associated with developmental and aging processes in the prefrontal cortex are more likely to occur outside of CpG islands (43).
Neurodevelopmental disorders, such as ASD, usually present years after birth. However, the molecular pathogenesis is thought to occur early during pregnancy or around birth. Consistent with the finding that DNA methylation changes in the brain during early life stages, we found that 5-hmC also shows a positive correlation with cerebellum development both in the mouse and in the human. The cerebellum of the human brain plays an important role in motor control as well as motor learning. In additional to this well-established role, there has been a growing recognition that the cerebellum is also involved in cognitive functions such as attention and language, and emotional control, such as fear and pleasure responses (44,45). Our findings in human cerebellum could have further implications for 5-hmC in the pathogenesis of neurodevelopmental disorders, which are associated with cognitive impairment, stereotypic movement, etc. Our earlier study revealed that the global level of 5-hmC is negatively correlated with the Rett syndrome protein MeCP2 dosage, implying that loss of MeCP2 could influence DNA methylation at DhMRs during brain development (21). In the present study, we have gained several new insights. We find that FMRP target genes are highly enriched with 5-hmC modifications and subject to changes during cerebellum development. This observation may be unique to FMRP target genes, since we did not see 5-hmC enrichment in the targets of other RNA binding proteins such as TDP-43, whose dysfunction can cause Amyotrophic lateral sclerosis (ALS), a neurodegenerative disease. FMRP can inhibit protein translation of target mRNAs that are involved in neuron plasticity, the balance between sensitization and desensitization responding to neuron activity. Therefore, FMRP-modulated protein concentration may be critical for normal neuronal function and sensitive to gene dosage. The statistically significant overlap between 5-hmC and FMRP target genes suggests that 5-hmC may contribute to proper brain function and development via epigenetic regulation of these gene transcription activities. For example, we see a dramatic decrease of 5-hmC levels in the FMRP target gene MAP1B, which encodes a protein that belongs to the microtubule-associated protein family. MAP1B protein is involved in microtubule assembly, which is an essential step for neurogenesis. Still, the molecular mechanism remains to be determined.
We found that DhMRs between the fetus and the adult are associated more with genes. Moreover, we also found that 5-hmC changes are statistically significantly associated with autism candidate genes, including several well-characterized autism candidate genes such as SHANK3, NLGN3 and TSC2. FMRP targets and ASD genes can be grouped into several functional categories, such as synaptic cell adhesion molecules, the NMDA (N-methyl-D-aspartate) receptor complex, and the mTOR pathway, most of which are associated with synaptic functions (32,46). Notably, synaptic dysfunction is critical to the development of autistic features and intellectual disabilities. Our observations suggest that aberrant DNA hydroxymethylation of these genes may potentially contribute to the etiology of autism and related neurodevelopmental disorders. TET1 is known to be involved in neuronal activity-induced DNA demethylation and subsequent gene expression in mouse brain (9), suggesting that 5-hmC dynamics may be regulated by TET1 during cerebellum development.
In summary, we profiled the genome-wide distribution of 5-hmC during the development of human brain. Our results indicate that 5-hmC levels increase during cerebellum development, which could contribute to proper brain function and development. Most importantly, we found that DhMRs preferentially locate in FMRP target genes and ASD genes, implying that abnormal alteration of 5-hmC may contribute to neurodevelopmental disorders.
MATERIALS AND METHODS
Cerebellum DNA sample preparation
The fetal cerebellum DNA sample was from The State Key Laboratory of Medical Genetics, Hunan, China. The female and male adult cerebellum samples were from The Emory Alzheimer's Disease Research Center. Genomic DNA was purified by Proteinase K digestion (0.667 g/l Proteinase K in 10 mM Tris–HCl, pH 8.5, 5 mM EDTA, 0.2% SDS and 200 mM NaCl, overnight at 55°C) followed by extraction with an equal volume of (25:24:1) phenol:chloroform:isoamyl alcohol saturated with 10 mM Tris–HCl, pH 8.0 and 1 mm EDTA. Purified genomic DNA was precipitated with an equal volume of isopropanol and resuspended in 10 mM Tris–HCl, pH 8.0.
5-hmC dot blot and capture
5-hmC dot-blot was performed as described previously (22). The primary antibody used is anti-5-hmC antibody (1:10 000, #39769, Active Motif). For 5-hmC capture, genomic DNA was sonicated into ∼200 bp size by Misonix 3000 (microtip, 4 pulses, 27 s each, 1 min rest, power output 2, on ice). The fragment size was verified by agarose gel electrophoresis. The following 5-hmC capture steps were performed as described previously (21).
Next generation sequencing of 5-hmC-enriched DNA
5-hmC captured libraries were generated by the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina according to the manufacturer's protocol. Briefly, 25 ng of input genomic DNA or 5-hmC-captured DNA were used. DNA fragments between 150 and 300 bp were gel-purified after the adapter ligation step. PCR-amplified DNA libraries were quantified on an Agilent 2100 Bioanalyzer and diluted to 6–8 pm for subsequent cluster generation and sequencing. We performed 38-cycle single-end sequencing using version 4 Cluster Generation and sequencing kits and version 7.0 recipes. Image processing and sequence extraction were done using the standard Illumina Genome Analyzer software and pipelines developed in house at the Department of Human Genetics, Emory University.
Sequence alignment and peak identification
Human FASTQ sequence files were aligned to human (NCBI36, hg18) references using Bowtie 0.12.6 with no more than two mismatches within the first 25 bp (47). After alignment, a custom computational pipeline was used to retain only non-duplicate unique genomic matches. 5-hmC enrichment peaks were determined using a model-based analysis of ChIP-Seq (MACS) against genomic DNA input. Parameters used for analysis were: effective genome size = 2.7e + 09; tag size = 38; bandwidth = 200; P-value cutoff = 1.00e–05.
DhMR identification and annotation
To identify fetus- and adult-specific DhMRs in human brain, we employed a Poisson-based peak identification algorithm (MACS) using aligned 5-hmC-enriched tags. DhMRs were determined among all pairs of 5-hmC-enriched samples by directly comparing one sample with another in each direction. Parameters used were: effective genome size = 2.7e + 09; tag size = 38; bandwidth = 200; P-value cutoff = 1.00e–08.
The association of DhMRs with genomic features was achieved by overlapping defined sets of DhMRs with known genomic features obtained from UCSC tables for NCBI36/hg18: RefSeq Whole Gene, 5′UTR, Exon, Intron, 3′UTR, ±500 bp of TSS, RefSeq Intergenic, CpG islands, and CpG island shores (±2 kb of CpG islands). DhMRs were assigned to a given genomic feature if overlapping ≥1 bp. In order to determine the fold change from expected values, the percentage of total DhMRs within a defined set was divided by the percentage expected to overlap each genomic feature by chance, based on the percentage of genomic space occupied by that genomic feature. All the statistical analysis and data processing were performed using R (http://www.r-project.org/).
FMRP target genes were obtained from Darnell et al. by HITS-CLIP experiment (32). TDP-43 target genes were obtained from Polymenidou et al. by HITS-CLIP experiment (48). Autism-related genes were obtained from the SFARI database of autism candidate genes (http://gene.sfari.org). Lists of highly, moderately or weakly expressed genes were generated according to human cerebellum RNA-Seq data in BrainSpan: Atlas of the Developing Human Brain database (http://www.brainspan.org/).
Gene ontology (GO) analysis
GO analyses were performed as previously described using the DAVID Bioinformatics Resources 6.7 Functional Annotation Tool (49). Gene sets were identified by joining subsets of DhMRs with RefSeq tables obtained from the UCSC genome browser tables.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported in part by the National Institutes of Health (NS051630 and NS079625 to P.J.; MH089606 and HD24064 to S.T.W.), the Emory Genetics Discovery Fund, International Rett Syndrome Foundation (P.J.), National Natural Science Foundation of China (81071028 and 81172513 to R.D.), the Autism Speaks grant (#7660 to X.L.) and the Simons Foundation Autism Research Initiative (P.J.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of Jin lab and Viren Patel at the Department of Human Genetics for their assistance. We thank Michael Santoro, Joshua Suhl and C. Strauss for critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 3.Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 4.Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirabayashi Y., Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 6.Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt J.D., Silva A.J., Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller C.A., Gavin C.F., White J.A., Parrish R.R., Honasoge A., Yancey C.R., Rivera I.M., Rubio M.D., Rumbaugh G., Sweatt J.D. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 9.Guo J.U., Su Y., Zhong C., Ming G.L., Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quivoron C., Couronne L., Della Valle V., Lopez C.K., Plo I., Wagner-Ballon O., Do Cruzeiro M., Delhommeau F., Arnulf B., Stern M.H., et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M.E., Vasanthakumar A., Patel J., Zhao X., et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Cai X., Cai C.L., Wang J., Zhang W., Petersen B.E., Yang F.C., Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S., An J., Lamperti E.D., Koh K.P., Ganetzky R., et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko M., Bandukwala H.S., An J., Lamperti E.D., Thompson E.C., Hastie R., Tsangaratou A., Rajewsky K., Koralov S.B., Rao A. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl Acad. Sci. USA. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G., et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 20.Inoue A., Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I., et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X., et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin S.G., Wu X., Li A.X., Pfeifer G.P. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S., Jacobsen S.E., Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 26.Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M., McLoughlin E.M., Brudno Y., Mahapatra S., Kapranov P., et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szulwach K.E., Li X., Li Y., Song C.X., Han J.W., Kim S., Namburi S., Hermetz K., Kim J.J., Rudd M.K., et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang T., Bray S.M., Warren S.T. New perspectives on the biology of fragile X syndrome. Curr. Opin. Genet. Dev. 2012;22:256–263. doi: 10.1016/j.gde.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoro M.R., Bray S.M., Warren S.T. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 31.Tan L., Shi Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W., et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A., et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruzov A., Tsenkina Y., Serio A., Dudnakova T., Fletcher J., Bai Y., Chebotareva T., Pells S., Hannoun Z., Sullivan G., et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinney S.M., Chin H.G., Vaisvila R., Bitinaite J., Zheng Y., Esteve P.O., Feng S., Stroud H., Jacobsen S.E., Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J. Biol. Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nestor C.E., Ottaviano R., Reddington J., Sproul D., Reinhardt D., Dunican D., Katz E., Dixon J.M., Harrison D.J., Meehan R.R. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ladd-Acosta C., Pevsner J., Sabunciyan S., Yolken R.H., Webster M.J., Dinkins T., Callinan P.A., Fan J.B., Potash J.B., Feinberg A.P. DNA methylation signatures within the human brain. Am. J. Hum. Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez D.G., Nalls M.A., Gibbs J.R., Arepalli S., van der Brug M., Chong S., Moore M., Longo D.L., Cookson M.R., Traynor B.J., et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christensen B.C., Houseman E.A., Marsit C.J., Zheng S., Wrensch M.R., Wiemels J.L., Nelson H.H., Karagas M.R., Padbury J.F., Bueno R., et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stroud H., Feng S., Morey Kinney S., Pradhan S., Jacobsen S.E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Numata S., Ye T., Hyde T.M., Guitart-Navarro X., Tao R., Wininger M., Colantuoni C., Weinberger D.R., Kleinman J.E., Lipska B.K. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmahmann J.D., Caplan D. Cognition, emotion and the cerebellum. Brain. 2006;129:290–292. doi: 10.1093/brain/awh729. [DOI] [PubMed] [Google Scholar]
- 45.Schmahmann J.D. The role of the cerebellum in cognition and emotion: personal reflections since 1982 on the dysmetria of thought hypothesis, and its historical evolution from theory to therapy. Neuropsychol. Rev. 2010;20:236–260. doi: 10.1007/s11065-010-9142-x. [DOI] [PubMed] [Google Scholar]
- 46.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C., et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.