Abstract
Genetic lesions such as BCR-ABL1, E2A-PBX1 and MLL rearrangements (MLLr) are associated with unfavorable outcomes in adult B-acute lymphoblastic leukemia (B-ALL). Leukemia oncoproteins may directly or indirectly disrupt cytosine methylation patterning to mediate the malignant phenotype. We postulated that DNA methylation signatures in these aggressive B-ALLs would point towards disease mechanisms and useful biomarkers and therapeutic targets. We therefore performed DNA methylation and gene expression profiling on a cohort of 215 adult B-ALL patients enrolled in a single phase III clinical trial (ECOG E2993) and normal control B-cells. In BCR-ABL1-positive B-ALL, aberrant cytosine methylation patterning centered around a cytokine network defined by hypomethylation and overexpression of IL2RA(CD25). The E2993 trial clinical data showed that CD25 expression was strongly associated with a poor outcome in ALL patients regardless of BCR-ABL1 status, suggesting CD25 as a novel prognostic biomarker for risk stratification in B-ALL. In E2A-PBX1-positive B-ALL, aberrant DNA methylation patterning was strongly associated with direct fusion protein binding as shown by the E2A-PBX1 ChIP sequencing (ChIP-seq), suggesting that E2A-PBX1 fusion protein directly remodels the epigenome to impose an aggressive B-ALL phenotype. MLLr B-ALL featured prominent cytosine hypomethylation, which was linked with MLL fusion protein binding, H3K79 dimethylation and transcriptional upregulation, affecting a set of known and newly identified MLL fusion direct targets with oncogenic activity such as FLT3 and BCL6. Notably, BCL6 blockade or loss of function suppressed proliferation and survival of MLLr leukemia cells, suggesting BCL6 targeted therapy as a new therapeutic strategy for MLLr B-ALL.
Keywords: Acute lymphoblastic leukemia (ALL), DNA methylation profiling, gene expression profiling, biomarker, therapeutic target
Introduction
Adult B-cell precursor acute lymphoblastic leukemia (B-ALL) is an aggressive disease with <40% long-term survival(1). This relatively poor outcome compared to childhood B-ALL is partly explained by an increased frequency of high-risk molecular lesions such as BCR-ABL1 (20%-40% in adults vs. 2%-5% in children), MLL rearrangements (MLLr, 10-20%) and E2A-PBX1 fusions (5%)(1, 2). The molecular mechanisms underlying poor outcome in these adult B-ALLs are only partially understood. However, each of these B-ALL subtypes feature distinct and perturbed gene expression profiles as compared to each other and to normal pre-B cells(3-6). A deeper understanding of the mechanisms driving aberrant gene expression as well as improved biomarkers and therapeutic targets are needed to improve risk stratification and therapy.
Transcriptional regulation and hence cellular phenotypes are increasingly understood to be programmed by epigenetic modifications of chromatin(7, 8). Epigenetic information is encoded in large part by patterns of cytosine methylation and histone modifications. An increased abundance of cytosine methylation at gene promoters, especially those containing CpG islands, is generally associated with transcriptional silencing, while decreased cytosine methylation may facilitate transcriptional activation. Importantly, perturbations in cytosine methylation patterning are epigenetically retained during cell division, enabling dividing cells to transmit transcriptional programming to their progeny(7, 8). This process allows cells within tissues including tumors to retain their specific phenotypes. Along these lines DNA methylation patterning has been shown to shift during normal hematopoiesis and is believed to play an essential role in lineage specification(9). Accordingly disruption of the function of DNA methyltransferases perturbs normal hematopoiesis(10, 11). By the same token DNA methylation patterning of terminally differentiated cells must be erased for them to return to a pluripotent and self-renewal state(12, 13). Similar to normal tissues, tumors may be dependent on specific DNA methylation patterns to acquire their unique phenotypes(14). Characterization of cytosine methylation patterning in tumors may thus provide important insights into how gene expression is perturbed in different tumor types.
Identification of epigenetically modified genes may be highly informative, since deregulation of key signaling or transcriptional regulatory genes can alter entire downstream pathways and have significant biological effects. For example aberrant epigenetic silencing of WNT signaling suppressor genes can result in WNT pathway hyperactivity in colon cancers(15). In acute leukemia a number of genetic lesions encode for proteins that can potentially indirectly or directly alter epigenetic regulatory states(16, 17). Hence combined genetic, transcriptional and epigenetic profiling studies may provide a mechanistic link between genetically altered proteins, their impact on chromatin, and their associated transcriptional profiles.
Several lines of investigation suggest that epigenomic programming is globally disrupted in B-ALL. For example, many of the common MLL fusion proteins can recruit the DOT1L histone methyltransferase which dimethylates H3K79, an event that is associated with transcriptional activation and elongation(18-23). Infants with MLLr B-ALL were shown to feature prominent cytosine hypermethylation of many genes, and to be susceptible to DNA methyltransferase inhibitors in vitro(24, 25). Other studies generally in limited numbers of B-ALLs and CpGs identified aberrantly methylated genes in B-ALL(26-29). Differential cytosine methylation of CpG islands was noted in subtypes of childhood B-ALL such as those with high hyperdiploidy and t(12;21)(30). In order to better understand the contribution of aberrant epigenetic gene regulation to the pathogenesis of adult B-ALL, we performed integrative genome-wide cytosine methylation and transcriptional profiling of a large cohort of adult B-ALL patients all enrolled in a single multicenter phase III clinical trial (ECOG E2993). We focused primarily on B-ALL subtypes associated with poor outcome. When coupled with ChIP sequencing (ChIP-seq) of leukemic fusion proteins and histone modifications and computational and functional assays, the resulting DNA methylation profiles provided insight into mechanisms driving aberrant gene expression in adult B-ALLs as well as new biomarkers and therapeutic targets.
Results
Specific promoter DNA methylation patterning in genetically defined B-ALL subtypes
We reasoned that cytosine methylation patterning would provide biologically and clinically significant information regarding B-ALL in adult patients. Therefore we performed HELP DNA methylation assays(31) on a cohort of 215 newly diagnosed adult patients with B-ALL with available diagnostic specimens enrolled in the ECOG E2993 multicenter phase III clinical trial (Table S1-2 for detailed patient description). Eighty-three of these patients featured BCR-ABL1 translocations, seven had E2A-PBX1 fusions and twenty eight harbored MLL rearrangements. We focused our attention principally on these B-ALLs because of their defined genetic background and association with poor clinical outcome. We also profiled normal pre-B cells (CD19+ and VpreB+) isolated from the bone marrows of 12 healthy adults as normal counterpart for our B-ALL tumor samples (Table S2). HELP was performed using a customized microarray design covering >50,000 CpGs annotated to 14,000 gene promoters and our standard quality control and normalization algorithms(31-33). We performed technical validation using base-pair resolution quantitative DNA methylation assessment by MassArray EpiTyper to confirm the accuracy of methylation values derived from HELP arrays. This procedure indicated that our HELP-derived DNA methylation values were highly concordant (correlation coefficient |r| = 0.87, Fig. S1, Table S3), and that based on the regression line in Fig. S1, a log ratio difference (dx) of 1 or 1.5 from HELP data corresponds to a methylation difference of 20% or 30% respectively by MassArray. Consistent with previous reports(14, 33), these data confirm that HELP allows DNA methylation to be accurately assessed as a continuous variable.
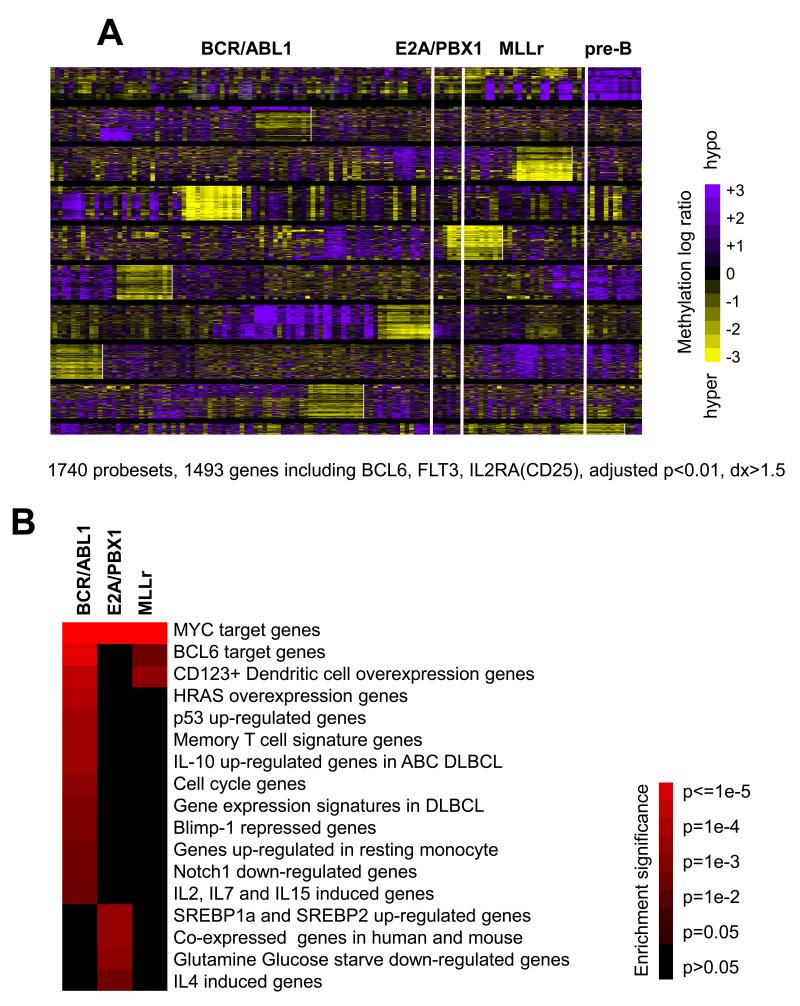
We next performed a supervised analysis comparing each of these three cytogenetically defined B-ALL subtypes to normal pre-B cells. This procedure identified a total of 1,740 probesets differentially methylated in at least one of the three B-ALL subtypes as compared to pre-B cell controls with DNA methylation difference >30% (dx>1.5 by HELP) and adjusted p-value<0.01 (ANOVA followed by Dunnett’s test(33, 34) and Benjamini-Hochberg (BH) correction(35)). These probesets corresponded to 1,493 gene promoters, with a general trend toward genes being hypomethylated in these B-ALL subtypes compared to normal pre-B cells (Fig. 1A, Table S4). Among hypomethylated probesets, 13.7% overlap with CpG islands, 55.3% with CpG shores (the 2kb flanking CpG islands(36)) and 31% outside of CpG islands or shores. B-ALL DNA methylation signatures were queried for their association with gene sets relevant to lymphoid tumor biology(37). All three B-ALL subtypes were significantly enriched with MYC target genes (p<1e-3, p<1e-11 and p<1e-7 respectively for BCR-ABL1, E2A-PBX1 and MLLr, Fisher’s exact test with BH correction, Fig. 1B, Table S5-6), suggesting MYC transcriptional networks are disrupted epigenetically in all three leukemia subtypes consistent with the critical role of Myc in hematopoietic neoplasms. BCR-ABL1 and MLLr DNA methylation signatures were enriched in BCL6 target genes (p=0.002 and p=0.049 respectively). BCR-ABL1 B-ALL also featured epigenetic deregulation of P53 induced genes (p=0.012), IL10 responsive genes (p=0.013), cell cycle genes (p=0.022), and others. E2A-PBX1 methylation signature involved gene sets associated with glutamine and glucose starvation (p=0.021), SREBP-1 and 2 activity (p=0.016), and T cell cytokines (p=0.042) (Fig. 1B, Table S5-6). Detailed explanation and references for those signatures were provided in Table S5. A similar analysis using Gene Ontology (GO) terms enriched for signal transduction, cell proliferation and gene regulation in all three subtypes (Table S5-6). These data indicate that cytogenetically distinct forms of adult B-ALL display specific and functionally defined DNA methylation signatures as compared to normal pre-B cells.
Figure 1. ALL subtypes display aberrant DNA methylation patterns as compared to normal pre-B cells.
(a) Heatmap representation of promoter DNA methylation levels of differentially methylated genes in BCR-ABL1 (n=83), E2A-PBX1 (n=7) and MLLr (n=28) B-ALLs vs. normal pre-B samples (n=12). 1740 probesets (or 1493 gene promoters) were identified with adjusted p-value<0.01 and methylation difference >30% (i.e. dx>1.5 on HELP). Each row of the heatmap represents one probe set on the HELP array, and each column represents an ALL patient or a healthy bone marrow. The color scale bar represents the methylation levels with purple for hypomethylation and yellow for hypermethylation. (b) The heatmap represents enrichment of B-cell and lymphoid specific gene sets with the DNA methylation signatures of the three B-ALL subtypes vs. normal pre-B cells. The statistical significance is provided in the color key. P values were calculated by Fisher’s exact test with BH correction.
BCR-ABL1 ALL DNA methylation signature features dysregulation of IL2RA
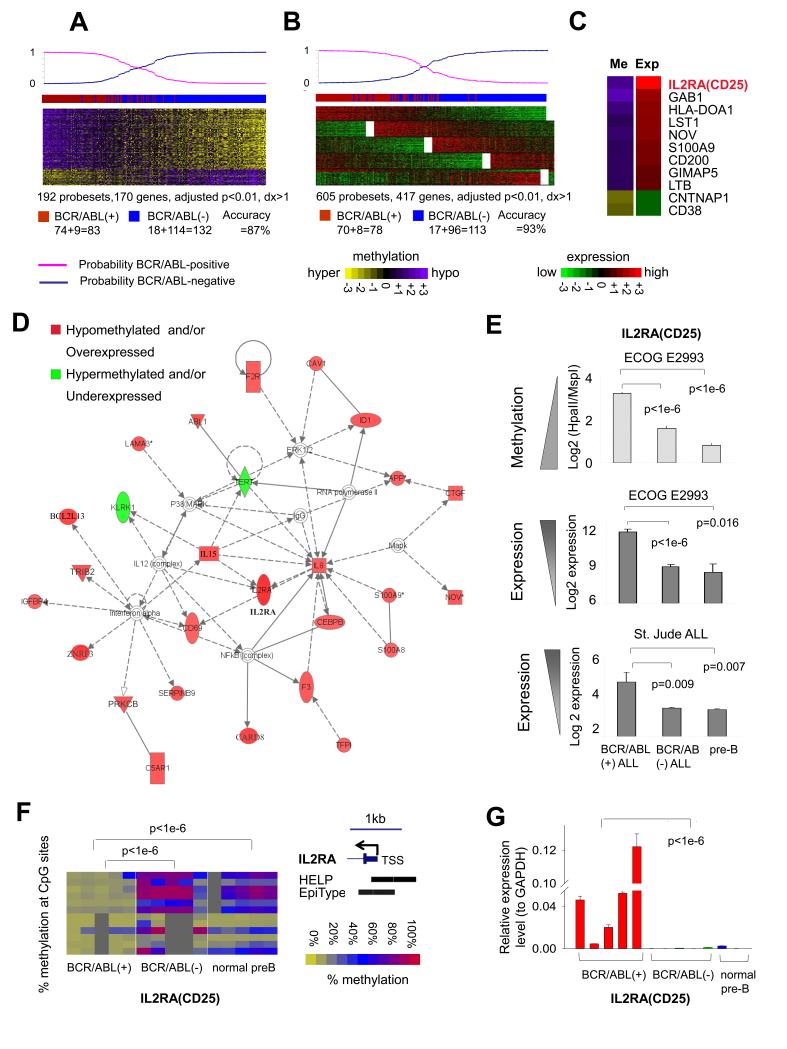
In order to investigate epigenetic programming linked to BCR-ABL1 signaling, we sought to identify genes differentially methylated in BCR-ABL1-positive ALLs. We performed a supervised analysis comparing DNA methylation profiles of the 83 BCR-ABL1-positive to the 132 BCR-ABL1-negative patients using a student t-test with BH correction for multiple testing. This yielded a set of 192 probesets (170 unique gene promoters) with methylation difference >20% (dx >1 by HELP) and adjusted p-value<0.01 (Fig. 2A, Table S7A). Gene expression profiling was also performed in these E2993 patients to define potential links between DNA methylation and transcriptional regulation. Supervised analysis of gene expression profiling in the same patients yielded 605 probesets (417 unique genes) with an adjusted p-value <0.01 and log2 fold change >1 (Fig. 2B, Table S7B). There was a tendency for DNA methylation to be inversely correlated with gene expression (i.e., the average expression fold changes (log2 fold) of BCR/ABL1-positive vs. –negative ALLs in the hypomethylated signature genes is higher than that in the hypermethylated signature genes, p=0.013, one-tailed Wilcoxon test). More than 80% of the genes in the methylation signature were hypomethylated and 80% of the genes were overexpressed in BCR-ABL1-positive B-ALLs, suggesting strong gene activation functions downstream of the fusion protein. Using a Bayesian predictor(38) and leave-one-out cross validation (LOOCV), the methylation and expression signatures re-classified 87% and 93% respectively of these B-ALL cases according to BCR-ABL1 status (Fig. 2A-B). To identify a core set of highly dysregulated genes, we examined the overlap between the DNA methylation and gene expression signatures. This analysis yielded 13 genes, 11 of which featured inversely correlated methylation and expression levels (Fig. 2C). Among the 11 core signature genes, the IL2RA gene (interleukin receptor 2 alpha chain), which encodes the CD25 protein, featured the highest overexpression and the second highest hypomethylation in BCR-ABL1 ALL (Fig. 2C). Differential expression of some of these genes were also captured in a previous gene expression profiling effort performed in 54 E2993 B-ALL patients(6). Differential expression and methylation was confirmed for all 11 genes by QPCR and MASSArry EpiTyper (Fig. S2A-B, Table S8-9). The DNA methylation and gene expression microarray data are available at GEO database (SueprSeries: GSE34941; gene expression: GSE34861; DNA methylation: GSE34937).
Figure 2. BCR-ABL1 ALL feature DNA methylation and expression signatures centered around IL2RA(CD25).
(a) Methylation signature and (b) expression signature of BCR-ABL1-positive vs. BCR-ABL1-negative B-ALL. Each row represents one probeset and each column one patient. The color scale bars represent the methylation or expression levels. The probability of signatures to re-classify cases according to BCR-ABL1 status is shown on the top. The actual case labels: Red, BCR-ABL1-positive; Blue, BCR-ABL1-negative. The numbers of correctly classified and misclassified cases are given below the heatmap. (c) The difference of mean methylation or expression values in BCR-ABL1-positive vs. -negative B-ALL is depicted by the color scale on the 11 core genes overlapping between methylation and expression signatures. (d) The gene network most highly enriched by methylation and expression signatures in BCR-ABL1 B-ALLs is centered around IL2RA(CD25). (e) The degree of IL2RA hypomethylation and expression from E2993 cases and from an independent cohort of 132 St. Jude B-ALL patients(4, 39) in BCR-ABL1-positive vs. BCR-ABL1-negative and vs. normal pre-B cells. (f) MassArray and (g) QPCR validation for IL2RA(CD25). The color key represents % methylation of each promoter CpG (rows). Columns represent individual cases. Location of HELP probesets and EpiTyper primers on IL2RA are shown (UCSC genome browser hg19).
Pathway analysis combining the full DNA methylation and gene expression signatures from Fig. 2A and 2B identified the heavy involvement of one particular gene network, centered around IL2RA and other cytokines (Fig. 2D). Collectively, these analyses pointed towards IL2RA as a potentially significant gene in BCR-ABL1-positive B-ALL. Indeed we observed that that IL2RA is significantly more hypomethylated in BCR-ABL1-positive B-ALL not only vs. BCR-ABL1-negative B-ALL but also vs. normal pre-B cells (p<1e-6 for both, t-test, Fig. 2E top panel). IL2RA mRNA in gene expression profiling of the same patients was also significantly more abundant in BCR-ABL1-positive vs. BCR-ABL1 negative B-ALL or normal pre-B cells (Fig. 2E middle panel). The same differential expression of IL2RA was detected upon examining an independent set of 132 B-ALL gene expression profiles(4, 39) (Fig. 2E bottom panel). Significant hypomethylation and upregulation of IL2RA in BCR-ABL1-positive ALL was validated by MassArray EpiTyper and QPCR on randomly selected E2993 patient samples with available DNA (p<1e-6 for all, one-tailed t-test, Fig. 2F-G).
CD25 positivity confers poor clinical outcomes
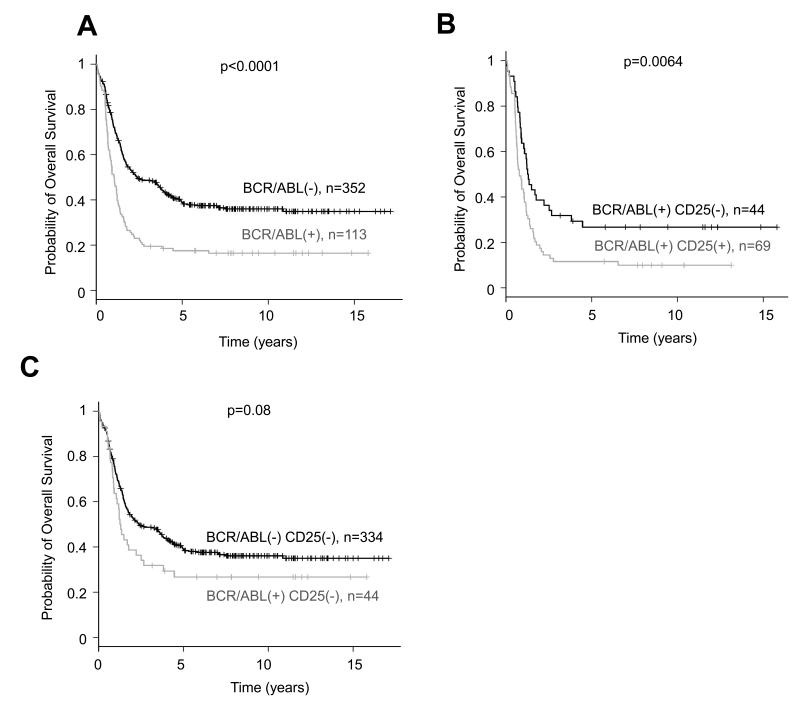
BCR-ABL1 translocation is an indicator of unfavorable outcome in adults with B-ALL(2). Examination of a total of 465 B-ALL patients enrolled in the E2993 trial including those profiled with microarrays plus additional patients with complete molecular and clinical annotation (median age 37 years, IQR: 25-47 years; median follow-up 7.9 years, IQR: 5-11 years) confirmed the inferior overall survival (OS) of BCR-ABL1-positive (n=113) vs. BCR-ABL1-negative B-ALL cases (n=352, logrank p<0.0001, Fig. 3A). The subset of Imatinib-treated B-ALL patients was excluded from the survival analysis. Consistent with a previous report(40), CD25 positivity (measured by flow cytometry) was strongly associated with BCR-ABL1 (61.1% BCR-ABL1-positive patients are also CD25 positive, 69 out of 113), but was rare in BCR-ABL1-negative patients (only 5.1% BCR-ABL1-negative patients are CD25 positive, 18 out of 352).
Figure 3. CD25 expression is associated with unfavorable clinical outcome.
(a) BCR-ABL1-positive B-ALL (n=113, median OS: 1.0 years, 95% CI: 0.8 to 1.2) have a worse OS than the BCR-ABL1-negative B-ALL (n=352, median OS: 2.4 years, 95% CI: 1.8 to 3.8) in the E2993 cases (p<0.0001, logrank test). (b) CD25 positivity further stratifies BCR-ABL1-positive B-ALL into two groups with significantly different clinical outcome: CD25-positive patients have significantly inferior OS (n=69, median OS: 0.8 years, 95% CI: 0.6 to 1.1) than the CD25-negative patients (n=44, median OS: 1.3 years, 95% CI: 0.9 to 2.5, p=0.0064, logrank test). (c) The CD25-negative BCR-ABL1-positive patients (n=44) didn’t have significantly different OS than the BCR-ABL1-negative CD25-negative patients (n=334, p=0.08, logrank test).
This difference was highly significant (p<1e-6 Fisher’s exact test, Fig. S3). When comparing expression profiles of CD25-positive vs. CD25-negative cases among BCR-ABL1-positive B-ALL, IL2RA was the most differentially expressed gene (>6 fold overexpressed in CD25-positive cases, p=0.005, t-test) and also notably hypomethylated (p=0.01, t-test, Fig. S4A). The overexpression and hypomethylation of IL2RA was validated by QPCR and MassArray (Fig. S4B). Those data suggest IL2RA (CD25) might be an important factor in BCR-ABL1-positive ALL. Indeed CD25 expression was clinically relevant, as BCR-ABL1-positive patients can be further stratified into two groups with significantly different clinical outcome according to CD25 positivity. CD25-positive patients have significantly worse OS (median OS time 0.8 years, 95% CI: 0.6 to 1.1, n=69,) than the CD25-negative patients (median OS time 1.3 years, 95% CI: 0.90 to 2.5, n=44, logrank p=0.0064, Fig. 3B). Although the CD25-negative BCR-ABL1-positive patients tended to display worse OS than the CD25-negative BCR-ABL1-negative patients, the difference did not reach statistically significance (logrank p=0.08, Fig. 3C). Collectively the data indicate that hypomethylation and corresponding overexpression of CD25 is strongly associated with BCR-ABL1 fusion protein and may be linked with processes that account at least in part for a particularly unfavorable clinical outcome in BCR-ABL1 B-ALL. CD25 can serve as a new prognostic biomarker for risk stratification in B-ALL.
Deregulated DNA methylation associated with binding of E2A-PBX1 fusion protein
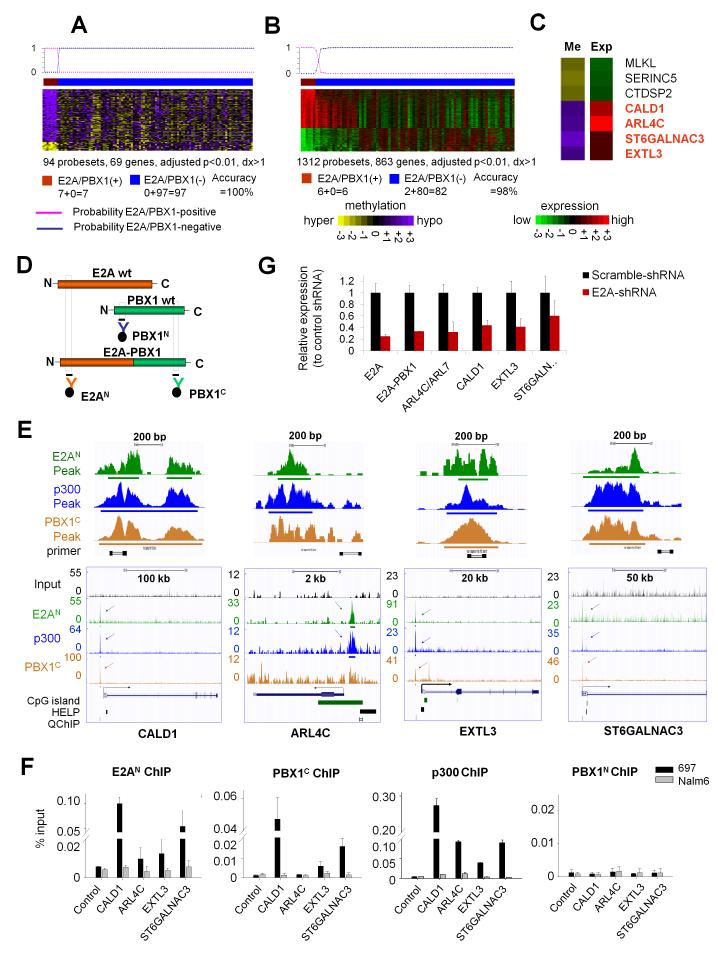
We next examined whether E2A-PBX1 translocations were also associated with specific perturbations of DNA methylation patterning. Supervised analysis comparing 7 E2A-PBX1-positive vs. 97 E2A-PBX1-negative ALLs identified 94 differentially methylated probesets (69 genes) with adjusted p-value<0.01 and methylation difference >20% (Fig. 4A, Table S10A) and 1,312 differentially expressed probesets (863 genes) with adjusted p-value<0.01 and log2 fold change >1 (Fig. 4B, Table S10B). Most genes in the methylation signatures (58 out of 69) were hypomethylated and the hypomethylation was associated with overexpression (p<0.01, one-tailed Wilcoxon test by comparing the means of the log2 expression fold changes (E2A-PBX1-positive vs. -negative cases) in the hypo- and hyper-methylated gene groups). When applied to a Bayesian predictor(38) using LOOCV, the DNA methylation and gene expression signatures re-classified E2A-PBX1 ALL with 100% and 98% accuracy, respectively. Eight genes overlapped between methylation and expression signatures and seven showed inverse correlation, including MLKL, SERINC5 and CTDSP2 (hypermethylated and underexpressed), and CALD1, ARL4C, ST6GALNAC3 and EXTL3 (hypomethylated and overexpressed, Fig. 4C). E2A-PBX1 is reported to act as an aberrant transcriptional activator that cooperates with the p300 histone acetyltransferase(41). We wondered whether the fusion protein itself might be linked to hypomethylation in these B-ALLs. We therefore performed ChIP-seq using antibodies recognizing the N terminus of E2A (E2AN), the C terminus of PBX1 (PBX1C), as well as p300 in an E2A-PBX1-positive B-ALL cell line, 697 cells (Fig. 4D, Table S11). Global analysis of these datasets identified 3,358 E2AN peaks, located predominantly at intronic and upstream regulatory regions (83%: broken down into 41% intronic, 23% distal at 2-50kb upstream and 19% intergenic >50kb from RefSeq TSS) vs. only 11% at promoters (ChIPseeqer software(42)). The 19,108 PBX1C peaks were located at a much higher frequency at promoters (36%), but where also at putative regulatory regions (28% introns, 17% distal and 13% intergenic) (Suppl. Fig. S5A, Table S14). Much of the intergenic/intronic PBX1C sites may be accounted for by E2A-PBX1 fusion protein, since there was an 83% overlap between E2AN sites with PBX1 (2,971 peaks, Z-score: 741.57, p-value: <1e-10, ChIPseeqer software(42), Suppl. Fig. S5B). P300 ChIP-seq identified 15,501 binding sites, which similar to E2AN were preferentially to regulatory regions (85%, with 43% in introns, 22% distal and 20% intergenic). Remarkably, 96% of the putative E2A/PBX1 fusion protein sites overlapped with p300 (2,684 peaks, Z-score: 960.58, p-value: <1e-10, ChIPseeqer software(42)), consistent with the known complex formation between these proteins (Suppl. Fig. S5B). Fully 87% of these joint binding sites were located at intronic/distal and intergenic regions and only 8% at promoters (Suppl. Fig. S5C). Altogether the data place a minority of E2A-PBX1-p300 complexes at traditional promoters, and the majority at both intra- and intergenic regulatory regions.
Figure 4. Specific DNA methylation and expression signatures associated with binding of E2A-PBX1 fusion protein.
(a) Methylation signature, (b) expression signature and (c) overlapping between methylation and expression signatures of E2A-PBX1-positive vs. E2A-PBX1-negative ALL. Rows are probesets and columns are patients. The color scale bars represent the methylation or expression levels. The probability of signatures to re-classify E2A-PBX1 cases is shown on the top. (c) The difference of mean methylation and expression values in E2A-PBX1-positive vs. E2A-PBX1-negative B-ALL is depicted by the color scale. (d) Graphical representations of ChIP for E2A (orange), PBX1 (green) and E2A-PBX1 fusion proteins. (e) ChIP-seq tracks and (f) QChIP results for E2AN (green), PBX1C (orange) and p300 (blue) antibodies vs. input (black) in 697 cells on the four hypomethylation signature genes in (c). (e) Y axis: number of reads for peak summits normalized by the total number of reads per track (set to 1Gbases for each track). Locations of CpG islands, HELP probesets, QChIP primers, and an enlarged view of binding peaks are shown. (g) QPCR was performed in 697 cells transfected with E2A-shRNA or Scramble-shRNA. Y axis: relative expression normalized to the scramble-shRNA. Data represent means ± s.e.m. (n=3).
Although promoter sites were the minority of fusion protein binding sites, we focused on these sites in order to more firmly link them to our studies on promoter DNA methylation. Focusing down on E2A-PBX1 signature genes, all four hypomethylated and upregulated genes in the core signature (Fig. 4C) showed enriched binding of E2AN, PBX1C and p300 in their promoter regions (Fig. 4E). Ten additional hypomethylation signature genes were enriched by E2AN, PBX1C and p300 antibodies, including ACOXL, BLK, COMT, MAST4, MYBPH, SLAMF1, SMPD3, TERT, TNS4 and WAPAL. A total of 14 genes overlap between the E2A-PBX1 hypomethylation signature (n=58 genes) and those identified as putative E2A-PBX1 and p300 direct targets (n=2,684 overlapping peaks annotated to n=1,642 genes). This level of overlap was indicative of overrepresentation of E2A/PBX1 target genes among hypomethylated genes (p=0.006, two-tailed Chi-square test with Yates’ correction). We further validated these targets by performing QChIP with E2AN, PBX1C and p300 antibodies (as positive ChIP’s) and PBX1N antibody (as a negative control ChIP) in 697 and Nalm6 cells (Nalm6 is an E2A-PBX1-negative B-ALL cell line). PBX1N QChIP serves as a negative control since PBX1N binds to only wild type PBX1 but not E2A-PBX1 fusion and wild type PBX1 is not expressed in lymphoid precursor B cells(43), therefore we would expect no PBX1N binding enrichment on those loci in neither 697 nor Nalm6 cells. This procedure confirmed binding enrichment of E2AN, PBX1C and p300 to the four core hypomethylation signature genes in 697 but not Nalm6 cells, and no binding enrichment of PBX1N to those loci in either cell type (Fig. 4F, Table S12). In order to determine whether these genes are directly controlled by E2A-PBX1, we performed lentiviral shRNA knockdown in 697 cells which targeted the middle coding region (AA 340-348) of E2A (E12/E47) and therefore knockdown both wild type E2A and E2A-PBX1 fusion. Fig. 4G shows the shRNA effectively knocked down E2A, E2A-PBX1, as well as the four core signature genes. Collectively these data identify a set of genes specifically deregulated epigenetically by E2A-PBX1 fusion, and moreover, directly implicate the fusion protein as driving this epigenetic and transcriptional signature.
Adult MLLr B-ALL displays a unique DNA methylation signature with predominant hypomethylation
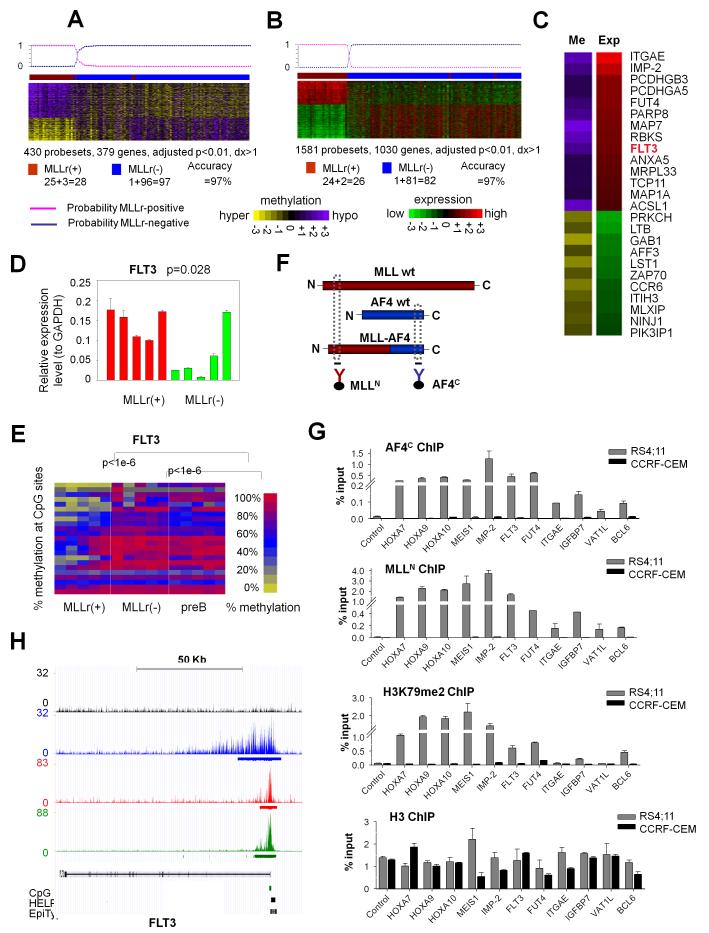
Chromosomal translocations involving MLL are frequent in B-ALL, can involve >50 partner genes, and are generally associated with a poor clinical outcome(44). Among the 28 MLLr cases in our cohort, there were 20 MLL-AF4, 6 MLL-ENL, 1 MLL-AF9 and 1 MLL-EPS15 cases. Unsupervised analysis of DNA methylation and gene expression profiles using hierarchical clustering showed that MLLr cases do not segregate according to fusion partners (data not shown). Moreover a supervised analysis between the 20 MLL-AF4 and the other eight MLLr cases showed no genes differentially methylated or expressed with adjusted p-value<0.01 (data not shown). Many of the common MLL fusion partners such as AF9, ENL and AF4 form part of transcriptional elongation complexes, and directly or indirectly recruit the DOT1L histone 3 lysine 79 (H3K79) methyltransferase(21, 23). This biochemical commonality may explain why these MLLr DNA methylation profiles are highly similar to each other. Supervised analysis comparing the 28 MLLr vs. the other 97 MLLr-negative ALLs identified 430 differentially methylated probesets (379 genes) with adjusted p-value<0.01 and methylation difference >20% (Fig. 5A, Table S13A) and 1,581 differentially expressed probesets (1,030 genes) with adjusted p-value<0.01 and log2 fold change >1 (Fig. 5B, Table S13B). Sixty percent (231 out of 379) differentially methylated genes were hypomethylated and displayed higher expression levels than the hypermethylated genes in MLLr B-ALL (p<1e-6, one-tailed Wilcoxon test by comparing the means of the log2 expression fold changes of MLLr-positive vs. -negative cases in the hypo- and hyper-methylated gene groups). When applying a Bayesian predictor(38) to these cases, the methylation and expression signatures re-classified MLLr cases with 97% and 97% accuracy respectively (Fig. 5A-B). A core set of 32 genes overlapped between the methylation and expression signatures, among which 25 showed inverse correlation between methylation and expression. Those genes include ITGAE, IGF2BP2(IMP-2), PCDHGB3, PCDHGA5, FUT4, PARP8, MAP7, RBKS, FLT3, ANXA5, MRPL33, TCP11, MAP1A and ACSL1 as hypomethylated and overexpressed, and PRKCH, LTB, GAB1, AFF3, LST1, ZAP70, CCR6, ITIH3, MLXIP, NINJ1 and PIK3IP1 as hypermethylated and downregulated in MLLr B-ALL (Fig. 5C). MassArray EpiTyper and QPCR confirmed the differential methylation and expression on fourteen randomly selected signature genes from this list (Fig. S6A-B). One of the core genes was FLT3 (hypomethylated and overexpressed in MLLr B-ALL, Fig. 5D-E), which encodes a class III receptor tyrosine kinase that plays key roles in proliferation of hematopoietic cells(45). FLT3 was previously shown to be highly expressed in MLLr leukemias and a potential therapeutic target in this B-ALL subtype(46, 47). Aberrant DNA hypomethylation may thus contribute to or facilitate increased expression of FLT3 in MLLr B-ALL.
Figure 5. DNA methylation and expression signatures are linked to MLL fusion protein binding patterns.
(a) Methylation signature (b) expression signature and (c) overlapping genes between methylation and expression signatures of MLLr vs. non-MLLr B-ALL. Rows for probesets and columns for patients. The probability of a Baysian predictor to re-classify MLLr ALL is shown on top. The numbers of correctly classified and mis-classified cases are given below the heatmap. (c) The color scale depicts the difference of mean methylation or expression values in MLLr vs. non-MLLr cases. (d) QPCR and (e) MassArray validation of FLT3 in five MLLr, five non-MLLr B-ALL and five normal pre-B samples. Data presents means ± s.e.m. (n=3). P values from one-tailed t-test. The heatmap represents the % methylation of CpGs (rows) in samples (columns). (f) Graphical representation of ChIP for MLL (red), AF4 (blue) and MLL-AF4 fusion. (g) QChIP using MLLN, AF4C, H3K79me2 or H3 antibodies in RS4;11 and CCRF-CEM cells, enriching for four positive control and seven MLLr hypomethylated and overexpressed loci. Data represent means ± s.e.m. (n=3). (h) FLT3 locus ChIP-seq enrichment profiles in RS4;11 cells. Locations of TSS, CpG islands, HELP probesets, and MASSArray Epityper primers are shown below the gene model in the UCSC genome browser view.
MLL-AF4 binding is associated with DNA hypomethylation and overexpression of genes, several with known or suspected oncogenic functions
Because MLL fusion proteins are chromatin modifiers and transcriptional activators, we hypothesized that genes aberrantly hypomethylated and highly expressed in MLLr cases might be direct targets of MLL fusions. To determine if this is the case we performed QChIP using MLL N-terminal antibody (MLLN), AF4 C-terminal antibody (AF4C), H3K79me2 antibody (as a chromatin mark induced by MLL fusions through recruitment of DOT1L) and histone H3 antibody (as a negative control) in a MLL-AF4-positive B-ALL cell line RS4;11 and a MLL-wild type ALL cell line CCRF-CEM (Fig. 5F, Table S11). We assessed enrichment at promoters of i) four positive control genes: HOXA7, HOXA9, HOXA10 and MEIS1 (Figs 5G, S7, S8A-B); ii) four genes from the core signature in Fig. 5B: FLT3, IGF2BP2/IMP-2, FUT4 and ITGAE; iii) two randomly selected genes from among the top 100 most highly overexpressed genes (IGFBP7 and VAT1L/KIAA1576), which were also hypomethylated although did not reach the threshold for the signature in Fig. 5A: IGFBP7 (dx=1.18, p=0.091) and VAT1L (dx=0.94, p=0.0002); and iv) BCL6, which was of interest because BCL6 target genes were one of the few pathways differentially methylated in MLLr patients (Fig. 1B), and BCL6 was differentially methylated in MLLr ALL vs. normal preB cells in Fig. 1A and Table S4 (dx=1.59, p= 0.00001). We observed enrichment for MLLN and AF4C at all seven of these loci and the four positive controls in RS4;11 but not CCRF-CEM cells, consistent with binding of the MLL-AF4 fusion protein (Fig. 5G). In accordance with this notion we also noted increased H3K79me2 signal in RS4;11 but not CCRF-CEM cells at nine of those eleven loci (except at ITGAE and VAT1L), and histone 3 enrichment was relatively even in both cell lines (Fig. 5G). Similar enrichment for MLLN and AF4C was observed in a second MLL-AF4–positive B-ALL cell line, SEM, at all those eleven loci (Fig. S8). We next performed ChIP-seq with MLLN, AF4C and H3K79me2 antibodies in RS4;11 cells (GEO accession numbers: GSE38403). MLLN ChIP-seq identified 3,312 binding peaks, annotated to 1,928 promoters (promoter regions defined as 2kb up- and down-stream to TSS); AF4C ChIP-seq identified 6,253 peaks (1,387 promoters); and H3K79me2 ChIP-seq identified 28,476 peaks (5,395 promoters, Tables S14-15). 603 gene promoters showed binding overlap with all three antibodies (MLLN, AF4C and H3K79me2) and were hence identified as MLL fusion direct targets by ChIP-seq data (Table S15). These 603 MLL-fusion target genes showed higher level of hypomethyation and expression than the non-target genes in the MLLr B-ALL cases (p<1e-6 for both, one-tailed Wilcoxon test, Fig. S9A-B), indicating MLL-AF4 binding is associated with DNA hypomethylation and overexpression of the target genes. Nine of the eleven loci queried by QChIP were also in this 603 gene list (except ITGAE and VAT1L). ChIP-seq read densities at the FLT3 locus are shown as an example in Fig. 5H. Our results showed that the hypomethylation signatures are linked with MLL fusion protein binding and H3K79me2, which induce overexpression of these genes. These are first examples of DNA methylation signatures directly mechanistically linked to binding patterns of aberrant fusion oncoproteins. Our data also revealed that FLT3 is in fact a direct target of MLL fusion proteins in B-ALL, and consequent hypomethylation and H3K79 dimethylation of the FLT3 promoter in these tumors may contribute to direct transcriptional activation of this oncogenic tyrosine kinase.
BCL6 is a transcriptional target of MLL fusion proteins
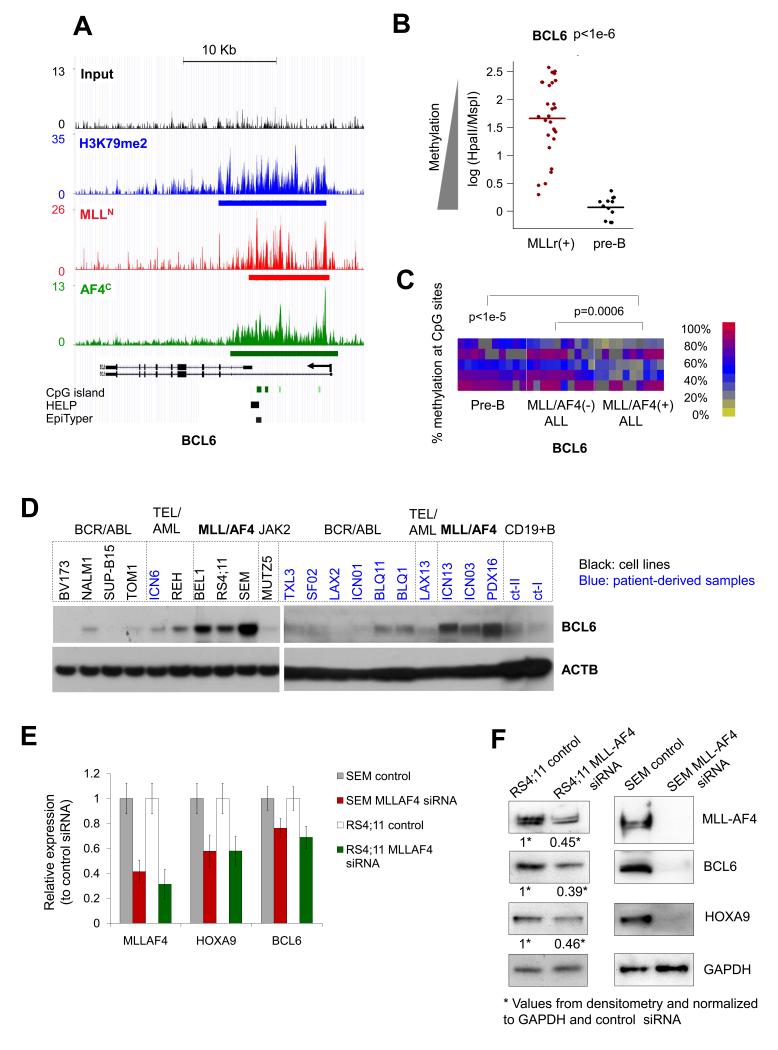
Among novel MLLr fusion targets identified in this study, BCL6 captured our particular attention because of its known role as a potent oncoprotein in B-cell lymphomas. As noted above, direct binding of MLL-AF4 as well as increased H3K79me2 was observed at the BCL6 locus by QChIP (Fig. 5G). ChIP-seq by MLLN, AF4C and H3K79me2 antibodies also confirmed enriched binding at the BCL6 locus (Fig. 6A). We showed that hypomethylation of BCL6 is indeed an aberrant feature of MLLr by comparing the degree of cytosine methylation in MLLr B-ALLs with normal pre-B cells as captured by our HELP assays (p<1e-6, t-test, Fig. 6B). Hypomethylation of BCL6 in MLLr vs. non-MLLr B-ALL and normal pre-B cells was again confirmed by MassArray Epityper (p<1e-3 and p<1e-5 respectively, t-test, Fig. 6C). Immunoblots using BCL6 antibody in a panel of 9 B-ALL cell lines, 11 primary B-ALL patient specimens and 2 normal pre-B samples revealed that expression of BCL6 protein was elevated in MLLr as compared to other B-ALL subtypes and normal pre-B-cells (Fig. 6D). BCL6 mRNA abundance was largely consistent with protein in these cells (Suppl. Fig. S10). Finally, in order to determine whether MLL fusion protein not only binds, but also induces BCL6 expression, we performed MLL-AF4 siRNA knockdown using fusion transcript specific siRNA sequences(48) in RS4;11 and SEM cells. The siRNAs effectively downregulated MLL-AF4 transcripts and proteins within 48hs, and this was accompanied by a reduction in expression of HOXA9 (a known MLL fusion target) and BCL6 at both mRNA and protein levels (Fig. 6E-F). MLL fusion proteins thus bind to the BCL6 promoter and contribute to its upregulation in MLLr B-ALL.
Figure 6. BCL6 is upregulated in MLL-AF4 ALL with its promoter hypomethylated and bound by MLL-AF4 fusion.
(a) ChIP-seq profiles at BCL6 enriched by MLLN (red), AF4C (green) and H3K79me2 (blue) antibodies vs. input (black) in RS4;11 cells. The locations of CpG islands, HELP probesets and MassArray Epityper primers are shown. (b) DNA methylation values on BCL6 derived from HELP assays in MLLr (n=28) vs. normal pre-B (n=12) samples. P value from t-test. (c) Heatmap representation of CpG methylation (rows) at the BCL6 locus measured by MassArray on 10 MLL-AF4-positive, 10 MLL-AF4-negative, and 10 normal pre-B samples (columns). The color key indicates percent DNA methylation. P values from t-test comparing the average methylation level of CpGs in each group. (d) BCL6 western blots performed in ALL cell lines (black labels) and primary specimens (blue labels). (e-f) RS4;11 and SEM cells were transduced with MLL-AF4 fusion specific siRNA as described in (48) or non-targeted control siRNA, followed 48 hrs later by (e) QPCR or (f) immunoblots for MLL-AF4, HOXA9, and BCL6. (e) Y axis represents the relative expression of MLL-AF4 siRNA knockdown normalized to the control siRNA. Data represent means ± s.e.m. from six independent knockdown experiments. (f) RS4;11 western blot bands were quantified relative to GAPDH using ImageJ(81).
BCL6 is a therapeutic target in MLLr B-ALL
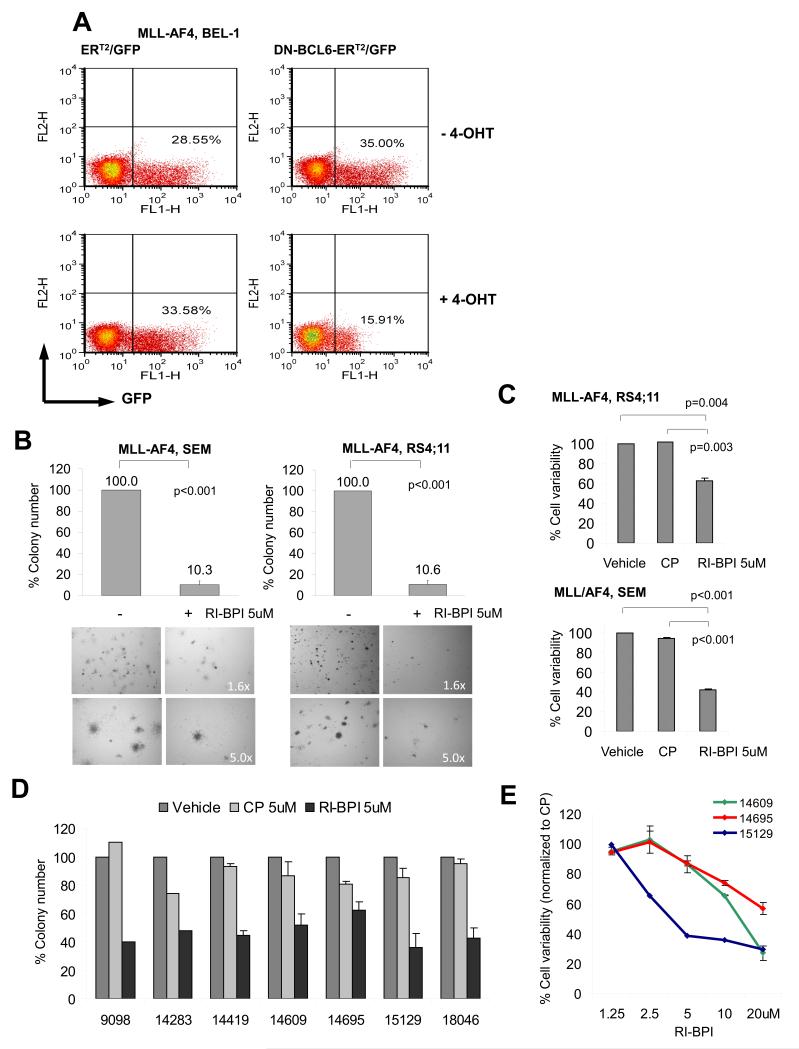
Constitutive expression of BCL6 is known to maintain the proliferation and survival of B-cell lymphomas(49). The fact that BCL6 is hypomethylated, overexpressed and a direct transcriptional target of MLL fusion proteins suggests that it might contribute to the proliferation and survival of MLLr B-ALLs. To determine if this is the case we transduced MLL-AF4 B-ALL cells (BEL-1) with a retrovirus expressing an 4OHT-inducible dominant-negative form of BCL6 consisting of its Zinc-finger domain and also expressing GFP through an internal ribosomal entry site (DNBCL6-ERT2–IRES-GFP)(50) or ERT2–IRES-GFP control vectors. We observed that while the percent of control ER-GFP transduced cells remained constant after three days exposure to 4-hydroxytamoxifen (4-OHT), there was a 55% decrease after 4-OHT induction in GFP-positive cells transduced with dominant negative BCL6 (15.91% vs. 35.00%, Fig. 7A), suggesting BCL6 is important in maintaining cell proliferation in MLLr B-ALL. Specific inhibitors of BCL6 were developed to block its transcriptional repressor activity(49, 51, 52) including the widely used peptidomimetic inhibitor RI-BPI, which specifically blocks BCL6 by preventing its binding to co-repressors(52). We tested the ability of two MLL-AF4-positive B-ALL cell lines (RS4;11 and SEM) to form colonies after exposure to 5 μM RI-BPI or vehicle. RI-BPI resulted in ~90% reduction in colonies in both SEM and RS4;11 cells (Fig. 7B, p<0.001 for both, t-test). In contrast RI-BPI treatment of two MLLr wild type ALL cell lines (Nalm6 and REH) did not suppress colony formation (Suppl. Fig. S11A-B), suggesting BCL6 dependence is a feature of MLLr. Similarly, 5 μM RI-BPI induced a ~40% reduction in viability of RS4;11 and a ~60% reduction in SEM cells after 48 hours as compared to vehicle (p=0.003 and p<0.001 respectively) or a control peptide (CP) (p=0.004 and p<0.001 respectively, t-test, Fig. 7C). Seven viably frozen primary human MLLr B-ALL specimens from the E2993 clinical trial were thawed and placed in liquid culture. Expression of BCL6 was verified in each of these cases by QPCR (Fig. S12). The cells were exposed to 5μM RI-BPI, 5μM CP or vehicle for 48 hours and assayed for colony forming potential by plating cells in methylcellulose in the presence of IL7. RI-BPI caused a 30-70% reduction in colony formation of MLLr specimens (Fig. 7D). Three patient specimens had sufficient cells to perform dose-response viability assays by flow cytometry using annexin V and 7AAD co-staining. Cells were treated with RI-BPI or CP at doses ranging from 1.25 μM to 20 μM to cover the range of IC50s reported in BCL6 dependent lymphomas(52). In all three cases, RI-BPI, but not CP, induced apoptosis of these primary human MLLr B-ALL within a similar dose range as B-cell lymphomas(52) (Fig. 7E). In contrast, when we ran cell viability experiments on three independent B-ALL primary samples (not from ECOG) with the phenotype labels blinded during the experiments, the RI-BPI suppression was only observed in the two MLLr cases but not the MLLr wild type B-ALL (Suppl. Fig. S13). These data suggest that BCL6 plays a crucial role in maintaining the survival and proliferation of MLLr B-ALL cells and is a bona fide therapeutic target. Collectively the integration of DNA methylation patterning studies in tandem with gene expression profiling and ChIP-seq provide an enhanced ability to understand the mechanism of action of oncogenic fusion proteins, establish putative prognostic biomarkers and identify new therapeutic targets in adult patients with B-ALL.
Figure 7. BCL6 targeted therapy kills MLLr B-ALL cells.
(a) BEL-1 cells (MLL-AF4 positive) were transduced with 4-OHT-inducible dominant-negative BCL6 (DN-BCL6-ERT2) or ERT2 control vectors (expressing GFP) and exposed or not exposed to 4-OHT for three days. Percentage of GFP-positive cells is indicated in the flow cytometry plots before and after 4-OHT for each construct. (b) SEM and RS4:11 cells and (d) seven E2993 MLL-AF4 patient samples were exposed to 5 μM RI-BPI, 5μM CP or vehicle for 48 hrs, followed by colony formation assay. Y axis shows percent colony formation of RI-BPI treated cells vs. vehicle. Experiments were performed in triplicates. (c) RS4:11 and SEM cells were exposed to 5μM RI-BPI, 5μM CP or vehicle for 48 hrs, followed by flow cytometry for annexin V/7AAD. Percent viability vs. vehicle is shown in Y axis. Experiments were performed in triplicates. (e) Three E2993 MLL-AF4 patient samples were treated with increasing concentrations of RI-BPI or CP for 48 hrs, and viability determined by annexin V/7AAD flow cytometry. Percent viability relative to CP is shown in Y axis. Data represent means ± s.e.m.(n=3). Student’s t-test was used to calculate p-values.
Discussion
The focus of this study was to identify promoter DNA methylation profiles associated with unfavorable B-ALL subtypes in adult patients and determine their functional and clinical significance. This effort was facilitated by profiling a large and unique cohort of clinically well-annotated adult B-ALL patients enrolled in a single multicenter phase III clinical trial (E2993). We observed that aberrant epigenetic regulation occurs universally and is distributed to specific gene sets in genetically defined B-ALL subtypes associated with poor outcomes. Integrative analysis of epigenetic and transcriptional profiles identified core gene signatures, associated with the mechanism of action of aberrant fusion proteins and pointing to key molecular mechanisms and potential therapeutic targets. We also performed unsupervised hierarchical clustering of the 97 normal karyotype B-ALLs in our cohort, which yielded two robust clusters (1-Pearson, Ward’s method, 1000x bootstrapping, data not shown). A supervised analysis revealed 1,512 genes differentially methylated (methylation difference>20%, adjusted p-value<0.01, Student’s t-test) between the two clusters, enriched in hematopoietic cell quiescence genes(53) (BH=0.03) as well as MYC (BH=0.02), and IKAROS target genes(54) (BH=0.03). These data suggest differences in the underlying pathogenesis of these cases, and point towards a rationale for further study of mechanisms driving aberrant gene regulation in translocation-negative patients.
In the case of BCR-ABL1-positive B-ALL, we identified the most deregulated gene network captured by integrative analysis of DNA methylation and gene expression profiles was centered around IL2RA, which is the most hypomethylated and overexpressed gene in BCR-ABL1-positive B-ALL. In general IL2RA(CD25) positivity was strongly associated with BCR-ABL1 translocation in this large cohort of B-ALL patients, which confirms a previous report on a smaller set of E2993 patients(55). BCR-ABL1-positive ALL patients experienced significantly worse outcome than other B-ALL patients in the E2993 clinical trial. However much of the difference appears related to CD25 expression since it was the subset of BCR-ABL1-positive CD25-positive B-ALL that featured the lowest OS, whereas BCR-ABL1-positive CD25-negative cases were not statistically worse than BCR-ABL1-negative B-ALLs. Further research will be required to determine how and why IL2RA and BCR-ABL1 are functionally linked. It is notable that CD25 expression signature in our B-ALL (data not shown) was significantly associated with a leukemia stem cell (LSC) signature(56) (BH adjusted p-value <1e-5) and a hematopoietic stem cell (HSC) signature(57) in leukemia (BH adjusted p-value <1e-6, Fisher’s exact test). Since LSCs are generally considered to be more chemo-resistant(58), it is possible that CD25 positivity might be an indicator of an enlarged LSC-like fraction. The IL2RA network also featured aberrant regulation of numerous cytokines and interleukins. Notably paracrine cytokine signaling by tumor cell microenvironment was recently shown to also confer chemo-resistance(59). Along these lines, quiescent AML leukemia stem cells localized to the endosteal niche within the bone marrow strongly expressed CD25(60). These endosteal localized AML stem cells were also particularly chemotherapy resistant(61), providing another possible link between CD25 positivity and poor outcome. Collectively these data raise the possibility that CD25 positivity may be linked in some way to cross-talk between BCR-ABL1-positive B-ALL cells and the bone marrow microenvironment. Along these lines we noted that when placed in in vitro culture, CD25 levels on B-ALL cells markedly decline (data not shown). Importantly, expression of CD25 provides an opportunity for therapeutic targeting. Monoclonal antibodies against CD25 such as RFT5.dgA, which is conjugated to deglycosylated ricin A-chain, have specific activity against CD25-positive cells in vitro and in vivo, and are currently in clinical trials for CD25-positive lymphoid malignancies(62, 63). Using such antibodies, it seems reasonable to attempt to eradicate CD25-positive B-ALL cells in patients in order to determine whether these cells contribute to chemo-resistance and relapse. CD25-immunotoxins are also able to kill infrequently dividing cells, and hence may be able to eradicate more quiescent cells such as LSCs. Meanwhile, the underlying cause of aberrant hypomethylation and upregulation of CD25 needs to be further investigated, and may provide clues to explain the etiology of CD25-positive B-ALL.
DNA methylation profiling coupled with ChIP-seq suggested that hypomethylation signatures in B-ALLs with t(1;19) and MLL rearrangements may be functionally linked to the function of these fusion oncoproteins. In E2A-PBX1 B-ALL, 14 out of the 58 hypomethylation signature genes were also identified as E2A-PBX1 fusion protein complex targets by ChIP-seq using E2AN, PBX1C and p300 antibodies. E2A-PBX1 recruits p300 and perhaps other co-activators to the target genes, which could potentially lead to loss of regulatory cytosine methylation and may epigenetically facilitate transcriptional activation. A number of these hypomethylated genes appear functionally relevant to leukemogenesis including glutamine metabolic pathways and factors such as BLK (B lymphocyte tyrosine kinase), which is a SRC kinase that can be targeted by tyrosine kinase inhibitors such as desatinib(64). In MLLr B-ALL, the MLL-AF4 fusion targets identified by the overlap of three ChIP-seq datasets (MLLN, AF4C and H3K79me2) also showed increased hypomethyation and gene expression compared to non-target genes. All MLL fusions retain the wild type MLL CXXC motif, which mediates direct binding to unmethylated CpG sequences(65). Binding of wild type MLL to CpG-rich regions in the HOXA9 locus has been shown to prevent these genomic regions from becoming methylated(66). MLL depletion resulted in gain of cytosine methylation at these HOXA9 sites and conversely re-expression of MLL rescued the locus back to a hypomethylated state(66). A similar effect was observed with MLL-AF4 and MLL-AF9(65, 66). In fact mutation of the CXXC motifs that disrupt binding to hypomethylated CpGs abrogate the leukemogenic functions of MLL fusion proteins(65), suggesting that maintenance of hypomethylation is a critical component of MLLr leukemogenesis. Our data suggest that this CXXC mechanism may extend to additional genes actively regulated by MLL fusion proteins (an illustration cartoon for a potential relationship between MLL-AF4 binding and DNA hypomethylation was shown in Suppl. Fig. S14A-B). Recent data from our group also suggest a link between HOXA9 and enhancer hypomethylation in myeloid MLLr leukemias(67). At the current time there is no evidence for direct biochemical cross-talk between DNMTs and MLL fusion proteins. However, given the previously described cooperation between Polycomb complexes and DNA methyltransferases(68) and the generally antagonistic actions of MLL/trithorax proteins with Polycomb(69), it remains possible that competition between MLL and DNMTs might occur in at least an indirect manner (Suppl. Fig. S14). Additional studies will be needed to explore whether aberrant DNA hypomethylation of loci occurring during leukemogenesis facilitates the recruitment of MLL fusions and induces abnormal transcriptional activation, or whether MLL fusion proteins in some way contribute or cause the initial aberrant hypomethylation of these loci. Whichever the case it seems clear that MLL binding to hypomethylated DNA is important to its leukemic functions, and perhaps facilitates assembly of MLL fusion complexes with DOT1L, thus accounting for the association we observed between DNA hypomethylation, MLL fusion protein binding, and H3K79me2. Perhaps CXXC induced DNA hypomethylation and DOT1L work together or form a positive feedback loop to highly upregulate certain gene sets important for leukemogenesis in MLLr cells, including not only HOX genes but also factors such as FLT3 and BCL6. In fact FLT3 was one of the most hypomethylated and highly expressed genes in MLLr B-ALL. FLT3 expression in MLLr leukemia was previously reported(70) as functionally relevant since FLT3 inhibitors could suppress the growth of MLLr cells(46). We show herein that FLT3 is in fact a bona fide direct target gene of MLL-AF4 and perhaps an example of a leukemogenic gene in which MLL fusion protein associated DNA hypomethylation and H3K79me2 epigenetically deregulate its expression.
Integrative epigenomic analysis allowed us to identify the BCL6 transcriptional repressor as a therapeutic target in MLLr B-ALL. The BCL6 locus displayed all the hallmarks of MLL fusion protein mediated epigenetic deregulation including direct MLL-AF4 binding, DNA hypomethylation, H3K79 dimethylation and transcriptional upregulation. BCL6 is required for germinal center B-cells to undergo proliferation and immunoglobulin affinity maturation. BCL6 mediates these effects by suppressing replication and DNA damage checkpoints(71-73). More recently BCL6 was shown to play a similar role in maintaining survival of pre-B-cells(74). Specifically, pre-B cells transiently express BCL6 during the transition from IL7 dependent to IL7 independent stages, whereupon BCL6 protects cells from genotoxic stress during immunoglobulin light chain recombination(74). These actions link BCL6 to processes associated with development of B-ALL. In the same way that constitutive expression of BCL6 in germinal center cells contributes to formation of diffuse large B cell lymphomas(75), it is reasonable to hypothesize that MLL fusion driven constitutive expression of BCL6 might also contribute to B-ALL leukemogenesis. Importantly, our data indicate that MLLr B-ALLs are addicted to BCL6 and require its presence to maintain proliferation and survival. This scenario is quite different than the recently described role of BCL6 as an imatinib-treatment inducible factor that protects BCR-ABL1-positive B-ALL cells from the toxic effects of ABL kinase blockade(76). This current study expands the paradigm of BCL6 targeted therapy to an aggressive form of B-ALL and supports the rationale for developing clinical trials of BCL6 inhibitors specifically for MLLr B-ALL. Altogether, it appears that MLL fusion proteins drive expression of FLT3, BCL6 and other key downstream target genes through the dual epigenetic activating effects of inducing and maintaining aberrant DNA hypomethylation and aberrant H3K79 dimethylation.
A small previous study combining DNA methylation profiling, gene expression profiling and ChIP-on-chip in five leukemia patients illustrated the proof of principle that triangulating between these platforms allowed more depth characterization of gene deregulation than single platforms alone(77). In manner similar to recent studies in AML showing that DNA methylation patterns can be used to identify new pathogenic mechanisms(14), this current analysis of adult B-ALL patients enrolled in ECOG E2993 phase III clinical study demonstrates the power of the integrative epigenomic approach to identify disease mechanisms and new prognostic biomarkers and therapeutic targets. These data point the way for future functional studies to explore in depth how BCR-ABL1, E2A-PBX1 and MLL fusions directly or indirectly alter DNA methylation during malignant transformation, and to test the role of IL2RA signaling in mediating chemotherapy resistant disease. Most importantly we expect these studies to trigger therapeutic trials of BCL6 inhibitors in MLL leukemia and the application of CD25 as a putative biomarker for risk stratification in B-ALL.
Materials and Methods
Patient samples, human cells and cell lines
Pretreatment bone marrow (BM) or peripheral blood (PB) samples were obtained at diagnosis (before any treatment) from 215 patients enrolled in the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) E2993 phase III trial in adult B-lineage ALL. Clinical details, including age, white blood cell count (WBC) and survival time were collected by the Clinical Trial Service Unit (CTSU), University of Oxford, United Kingdom. All patient specimens used in this study were accrued by ECOG. The diagnosis was established by central morphology review and confirmed by multiparameter flow cytometry and RT-PCR assays in ECOG’s Leukemia Translational Studies Laboratory. Cytogenetic information was reviewed by the ECOG Cytogenetics Committee. The study was approved by the institutional review board of each treatment center. Normal human pre-B or pro–B cells (CD19+ and VpreB+) were sorted from the bone marrows of 12 healthy donors by flow cytometry using antibodies (BD Biosciences) and a FACSVantage SE cell sorter (BD Biosciences). The use of human tissue was in agreement with research ethics board of Weill Cornell Medical College. Patient characteristics are given in Table S1-2. The human ALL cell lines RS4;11, SEM, BEL-1, CCRF-CEM, 697, Nalm6, REH, BV173, NALM1, SUP-B15, TOM1 and MUTZ5 were obtained fresh from DSMZ (Braunschweig, Germany). The expression of aberrant fusion proteins was verified by Western Blot and QPCR.
Array-based methylation analysis using HELP
The HELP (HpaII tiny fragment enrichment by ligation mediated PCR) assay was performed as previously published(31, 33, 78) (Supplementary information). HELP data quality control (QC) and analysis was performed as described previously(32) using R software(79). Basically, raw data (.pair) files were generated using NimbleScan software. Signal intensities at each HpaII amplifiable fragment were calculated as a robust (25% trimmed) mean of their component probe-level signal intensities. Any fragments found within the level of background MspI signal intensity, measured as 2.5 mean-absolute-deviation (MAD) above the median of random probe signals, were considered as “failed” probes and removed. After QC processing, a median normalization was performed on each array by subtracting the median log-ratio (HpaII/MspI), resulting in median log-ratio of 0 for each array.
HELP array data analysis
Identification of the aberrantly methylated genes for each B-ALL subtype compared to the normal pre-B cells was performed using an ANOVA test followed by Dunnett’s post hoc test(34) using the normal pre-B samples as the reference group, and multiple testing corrected by the Benjamini and Hochberg (BH) method(35). R Package ‘multcomp’ Version 1.2-6 was used(34). The comparison between the positive and negative groups of BCR-ABL1, E2A-PBX1 and MLLr was determined by Student’s t-test and the BH adjusted p-values. As methylation difference between ALL and normal pre-B cells are more apparent than the methylation difference among the ALL subtypes, to capture and prioritize the top differentially methylated genes we used a higher threshold dx>1.5 (which corresponds to 30% methylation difference) in the ALL vs. normal pre-B comparison, and a lower threshold dx>1 (which corresponds to 20% methylation difference) in the ALL subtype comparison. Based on the linear regression line in MassArray EpiTyper validation (Fig. S1, equation: y = −4.9484x + 3.3232, where x is MassArray readout and y is HELP readout), a log2 ratio difference (dx) of 1 and 1.5 from HELP data corresponds to methylation difference of 20% and 30%, respectively.
Single locus quantitative DNA methylation assays
MassArray EpiTyper assays (Sequenom, CA) were performed on bisulfite-converted DNA, as previously described(80). The primers were designed using Sequenom EpiDesigner beta software (Sequenom, Inc) and data were analyzed using EpiTYPER software version 1.0 (Sequenom, Inc). Note that MassArray and QPCR validation studies were performed in different sets of randomly selected specimens with available DNA and RNA, from among the cohort of 215 profiled E2993 patients.
Cell culture
The human B-ALL cell lines were maintained in Roswell Park Memorial Institute medium (RPMI-1640, Invitrogen, CA) with 10% fetal bovine serum (FBS), 100 IU/mL penicillin and 100 μg/mL streptomycin. Primary B-ALL cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Invitrogen, CA) with 10%FBS, LDL, 1X ß-ME, 2 mM L-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, rhFLT3 (20ng/mL), rhSCF (50ng/mL), IL3, (20ng/mL), IL6 (20ng/mL) and IL7 (10ng/mL) (Invitrogen, CA). Cell cultures were kept at 37°C in a humidified incubator under a 5% CO2 atmosphere.
MLL-AF4 and E2A-PBX1 knockdown
The MLL-AF4 knockdown in RS4;11 and SEM cells was carried out as described in Thomas et al.(48) (Supplementary information). Synthetic sense and antisense oligoribonucleotides specific for the MLL-AF4 fusion sites were synthesized by Dharmacon RNAi Technologies (Lafayette, CO). RNA was extracted 48 hrs after transduction and the levels of genes were determined by QPCR. The E2A-PBX1 knockdown in 697 cells was carried out using an E2A-shRNA that targets the middle coding region (AA 340-348) of E2A (E12/E47), and therefore affects both the E2A and E2A fusions (E2A-PBX1a and E2A-PBX1b). The shRNA vectors were obtained from Open Biosystems (Lafayette, CO), clone ID: TRCN0000017534. The lentivirus was prepared as described in the Addgene website (Addgene, Cambridge MA). RNA was extracted 96 hrs post-infection and transcript abundance determined by QPCR.
Western Blot
For BCL6 Western blot, rabbit antibody raised against BCL6-N3 (sc-858) and anti-Actin-HRP conjugated (sc-1615) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). For the Western blot experiments in MLL-AF4 siRNA knockdown, SEM and RS4;11 cells were treated with control and MLL-AF4 siRNAs as described. 4 × 106 cells were collected and whole cell extracts were made via resuspension in 40 μl of a modified RIPA buffer (50mM Hepes-KOH, pH 7.6, 500mM LiCl, 1mM EDTA, 1% NP-40, 0.7% Na-Deoxycholate, ddH2O) for 30 minutes at 4°. Whole cell extracts were split in half and loaded onto either a 3-8% Tris Acetate gel (NuPAGE, Life Sciences) or a 4-12% Bis-Tris gel (NuPAGE, Life Sciences). Gels were blotted onto PVDF membranes and probed with BCL6 antibodies (as described), αHOXA9 (Millipore, 07-178), αAF4-C (Abcam, ab31812) or αGAPDH (Bethyl, A300-641A). RS4;11 western blot bands were quantified relative to GAPDH using a standard protocol on ImageJ(81).
Cell viability assay
One hundred thousand MLL-AF4 ALL cells from the cell lines or 200,000 primary cells were seeded in a volume of 100 uL medium (as described in “Cell culture”) per well. RI-BPI or control peptides (CP) were diluted in medium and added at the indicated concentration in a total culture volume of 120 uL. After culturing for 48 hours, cell viability was measured by flow cytometry using annexin V–fluorescein isothiocyanate (FITC; BD Biosciences) and 7-aminoactinomycin (7-AAD, Molecular Probes-Invitrogen) to detect phosphatidylserine exposition and cell permeability, respectively. At least 50,000 events were recorded per condition on an LSR II flow cytometer (BD Biosciences). Data analysis was conducted using FlowJo 8.2 software for Mac OS X (TreeStar). Cells that were negative for annexin V and 7-AAD were scored as viable. Fold changes were calculated using baseline values of untreated or the CP-treated cells as a reference.
Colony-forming assay
MLL-AF4 ALL cell lines were plated with 10,000 cells per plate in semisolid agar in the presence of 5 μM retro-inverso BCL6 peptide-inhibitor (RI-BPI) or vehicle. Colonies were counted after ten days. For MLL-AF4 B-ALL primary samples, 25,000 cells per sample were treated with RI-BPI, CP or vehicle for 48 hours and plated in M3434 methylcult (Stem Cell Technologies) with IL7 (10ng/mL) (Invitrogen). Colonies were counted after 14 days.
Chromatin Immunoprecipitation (ChIP) and QChIP
RS4;11, SEM, CCRF-CEM, 697 and Nalm6 cells were fixed with 1% formaldehyde, lysed and sonicated to generate fragments less than 500bp. Sonicated lysates were incubated with antibodies overnight and after increasing stringency washes immunocomplexes were recovered and DNA was isolated. The ChIP protocol was performed as in Milne et al.(82). A fraction of the ChIP products was used as template in QPCR reactions using TaqManTM or SYBR GreenTM and a 7900 ABI qRT-QPCR machine (Applied Biosystems, CA). The QChIP results was quantified relative to inputs as explained in detail in Milne et al.(82).
ChIP sequencing (ChIP-seq)
See supplementary information for details. Peaks from ChIP-seq data were called using the ChIPseeqer program(42) with parameters indicated in Table S14.
Survival analysis
Kaplan-Meier survival analysis was used to estimate overall survival (OS) and logrank test was used to compare survival differences between patient groups. R package “survival” version 2.35-8(79) and “proc lifetest” in SAS was used for the survival analysis.
Supplementary Material
Statement of Significance.
We performed the first integrative epigenomic study in adult B-ALL, as a correlative study to the ECOG E2993 phase III clinical trial. This study links for the first time the direct actions of oncogenic fusion proteins with disruption of epigenetic regulation mediated by cytosine methylation. We identify a novel clinically actionable biomarker in B-ALL: IL2RA, which is linked with BCR-ABL1 and an inflammatory signaling network associated with chemotherapy resistance. We show that BCL6 is a novel MLL fusion protein target that is required to maintain the proliferation and survival of primary human adult MLLr cells and provide the basis for a clinical trial with BCL6 inhibitors for MLLr patients.
Acknowledgements
AM is supported by the Chemotherapy Foundation, Burroughs Wellcome Foundation, and the Sackler Center for Biomedical and Physical Sciences at the Weill Cornell Medical College. OE is supported in part by the NSF CAREER grant DBI 1054964. AM and MM are supported by LLS TRP 6097-10. AM, RGR, CDA and TAM were supported by LLS SCOR 7132-08. TAM was supported by the Medical Research Council UK. RGR was supported by Starr Cancer Consortium and NIH grants. WYC and DB were supported by LLS fellowship 5230-09 and 5089-10. We thank Dr. Peter H Wiernik for his support of the E2993 clinical trial and correlative studies as former chair of the ECOG leukemia committee, and Dr. Sue Richards and the MRC/ECOG trial management group supported by MRC grant (G8223452) for maintaining the trial database for ECOG. We are grateful for the assistance of the Weill Cornell Epigenomics Core Facility.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–78. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23(26):6306–15. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 3.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 4.Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102(8):2951–9. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 5.Chiaretti S, Li X, Gentleman R, Vitale A, Wang KS, Mandelli F, et al. Gene expression profiles of B-lineage adult acute lymphocytic leukemia reveal genetic patterns that identify lineage derivation and distinct mechanisms of transformation. Clin Cancer Res. 2005;11(20):7209–19. doi: 10.1158/1078-0432.CCR-04-2165. [DOI] [PubMed] [Google Scholar]
- 6.Juric D, Lacayo NJ, Ramsey MC, Racevskis J, Wiernik PH, Rowe JM, et al. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J Clin Oncol. 2007;25(11):1341–9. doi: 10.1200/JCO.2006.09.3534. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 9.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–15. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 11.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–9. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28(8):848–55. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36(4):417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 16.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295(5557):1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 17.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14(5):355–68. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–68. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, et al. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37(3):429–37. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17(2):198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stumpel DJ, Schneider P, van Roon EH, Boer JM, de Lorenzo P, Valsecchi MG, et al. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood. 2009;114(27):5490–8. doi: 10.1182/blood-2009-06-227660. [DOI] [PubMed] [Google Scholar]
- 25.Schafer E, Irizarry R, Negi S, McIntyre E, Small D, Figueroa ME, et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. Blood. 2010;115(23):4798–809. doi: 10.1182/blood-2009-09-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118(19):5218–26. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, et al. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67(6):2617–25. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 28.Kuang SQ, Tong WG, Yang H, Lin W, Lee MK, Fang ZH, et al. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22(8):1529–38. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- 29.Kuang SQ, Bai H, Fang ZH, Lopez G, Yang H, Tong W, et al. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood. 2010;115(12):2412–9. doi: 10.1182/blood-2009-05-222208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidsson J, Lilljebjorn H, Andersson A, Veerla S, Heldrup J, Behrendtz M, et al. The DNA methylome of pediatric acute lymphoblastic leukemia. Hum Mol Genet. 2009;18(21):4054–65. doi: 10.1093/hmg/ddp354. [DOI] [PubMed] [Google Scholar]
- 31.Figueroa ME, Melnick A, Greally JM. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- 32.Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, et al. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24(9):1161–7. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346–63. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 36.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41(2):246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 38.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100(17):9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zelm MC, van der Burg M, de Ridder D, Barendregt BH, de Haas EF, Reinders MJ, et al. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175(9):5912–22. doi: 10.4049/jimmunol.175.9.5912. [DOI] [PubMed] [Google Scholar]
- 40.Paietta E, Racevskis J, Neuberg D, Rowe JM, Goldstone AH, PH W. Expression of CD25 (interleukin-2 receptor alpha chain) in adult acute lymphoblastic leukemia predicts for the presence of BCR/ABL fusion transcripts: results of a preliminary laboratory analysis of ECOG/MRC Intergroup Study E2993. Eastern Cooperative Oncology Group/Medical Research Council. Leukemia. 1997;11(11):1887–90. doi: 10.1038/sj.leu.2400836. [DOI] [PubMed] [Google Scholar]
- 41.Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, et al. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J Biol Chem. 2004;279(53):55362–71. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- 42.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC Bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aspland SE, Bendall HH, Murre C. The role of E2A-PBX1 in leukemogenesis. Oncogene. 2001;20(40):5708–17. doi: 10.1038/sj.onc.1204592. [DOI] [PubMed] [Google Scholar]
- 44.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 45.Sanz M, Burnett A, Lo-Coco F, Lowenberg B. FLT3 inhibition as a targeted therapy for acute myeloid leukemia. Curr Opin Oncol. 2009;21(6):594–600. doi: 10.1097/CCO.0b013e32833118fd. [DOI] [PubMed] [Google Scholar]
- 46.Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3(2):173–83. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 47.Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, et al. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103(9):3544–6. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 48.Thomas M, Gessner A, Vornlocher HP, Hadwiger P, Greil J, Heidenreich O. Targeting MLL-AF4 with short interfering RNAs inhibits clonogenicity and engraftment of t(4;11)-positive human leukemic cells. Blood. 2005;106(10):3559–66. doi: 10.1182/blood-2005-03-1283. [DOI] [PubMed] [Google Scholar]
- 49.Polo JM, Dell’Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10(12):1329–35. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 50.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13(2):199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 51.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17(4):400–11. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113(15):3397–405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101(16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paietta E, Racevskis J, Neuberg D, Rowe JM, Goldstone AH, Wiernik PH. Expression of CD25 (interleukin-2 receptor alpha chain) in adult acute lymphoblastic leukemia predicts for the presence of BCR/ABL fusion transcripts: results of a preliminary laboratory analysis of ECOG/MRC Intergroup Study E2993. Eastern Cooperative Oncology Group/Medical Research Council. Leukemia. 1997;11(11):1887–90. doi: 10.1038/sj.leu.2400836. [DOI] [PubMed] [Google Scholar]
- 56.Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 304(24):2706–15. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2010;17(9):1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 58.Konopleva MY, Jordan CT. Leukemia stem cells and microenvironment: biology and therapeutic targeting. J Clin Oncol. 29(5):591–9. doi: 10.1200/JCO.2010.31.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143(3):355–66. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saito Y, Kitamura H, Hijikata A, Tomizawa-Murasawa M, Tanaka S, Takagi S, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2(17):17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–21. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 62.Schnell R, Vitetta E, Schindler J, Borchmann P, Barth S, Ghetie V, et al. Treatment of refractory Hodgkin’s lymphoma patients with an anti-CD25 ricin A-chain immunotoxin. Leukemia. 2000;14(1):129–35. doi: 10.1038/sj.leu.2401626. [DOI] [PubMed] [Google Scholar]
- 63.Schnell R, Borchmann P, Staak JO, Schindler J, Ghetie V, Vitetta ES, et al. Clinical evaluation of ricin A-chain immunotoxins in patients with Hodgkin’s lymphoma. Ann Oncol. 2003;14(5):729–36. doi: 10.1093/annonc/mdg209. [DOI] [PubMed] [Google Scholar]
- 64.Montero JC, Seoane S, Ocana A, Pandiella A. Inhibition of SRC family kinases and receptor tyrosine kinases by dasatinib: possible combinations in solid tumors. Clin Cancer Res. 17(17):5546–52. doi: 10.1158/1078-0432.CCR-10-2616. [DOI] [PubMed] [Google Scholar]
- 65.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17(1):62–8. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia ZB, et al. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci U S A. 2008;105(21):7517–22. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8(6):e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 69.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 70.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30(1):41–7. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 71.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8(7):705–14. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 72.Ranuncolo SM, Wang L, Polo JM, Dell’Oso T, Dierov J, Gaymes TJ, et al. BCL6-mediated attenuation of DNA damage sensing triggers growth arrest and senescence through a p53-dependent pathway in a cell context-dependent manner. J Biol Chem. 2008;283(33):22565–72. doi: 10.1074/jbc.M803490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–9. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 74.Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207(6):1209–21. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7(5):445–55. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 76.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473(7347):384–8. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Figueroa ME, Reimers M, Thompson RF, Ye K, Li Y, Selzer RR, et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS One. 2008;3(3):e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16(8):1046–55. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Development Core Team R: A language and environment for statistical computing. 2008 2008. http://www.R-project.org.
- 80.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(44):15785–90. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43(1 Suppl):25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 82.Milne TA, Zhao K, Hess JL. Chromatin immunoprecipitation (ChIP) for analysis of histone modifications and chromatin-associated proteins. Methods Mol Biol. 2009;538:409–23. doi: 10.1007/978-1-59745-418-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.