Abstract
Colorectal cancer (CRC), the third most commonly diagnosed type of cancer in men and women worldwide is recognized as a complex multi-pathway disease, an observation sustained by the fact that histologically identical tumors may have different outcome, including various response to therapy. Therefore, particularly in early and intermediate stage (stages II and III, respectively) CRC, there is a compelling need for biomarkers helpful of selecting patients with aggressive disease that might benefit from adjuvant and targeted therapy. Histopathological examination shows that likely other solid tumors the development and progression of human CRC is not only determined by genetically abnormal cells, but also by intricate interactions between malignant cells and the surrounding microenvironment. This has led to reconsider the features of tumor microenvironment as potential predictive and prognostic biomarkers. Among the histopathological biomarkers, tumor budding (i.e., the presence of individual cells and small clusters of tumor cells at the tumor invasive front) has received much recent attention, particularly in the setting of CRC. Although its acceptance as a reportable factor has been held back by a lack of uniformity with respect to qualitative and quantitative aspects, tumor budding is now considered as an independent adverse prognostic factor in CRC that may allow for stratification of patients into risk categories more meaningful than those defined by tumor-node-metastasis staging alone, and also potentially guide treatment decisions, especially in T2-T3 N0 (stage II) CRCs.
Keywords: Colorectal cancer, Tumor budding, Biomarker, Histopathology
INVITED COMMENTARY ON HOT ARTICLES
We read with great interest the recent article by Lugli et al[1] describing the morphology of “tumor budding” as a promising histopathological prognostic feature in colorectal cancer (CRC) and strongly recommend it to the readers.
Although in certain countries a decline in CRC incidence rate has been registered, attributed to increases in screening adhesion rates and linked detection and removal of precancerous polyps[2], CRC remains one of the most common cancers[3]. By its frequency, CRC ranks third in men and women worldwide[3]. Explained as a multi-step dynamical disease in the last two decades, CRC develops slowly over several years and progresses through cytologically distinct benign and malignant states, from single crypt lesions through adenoma, to malignant carcinoma with the potential for local invasion and distant metastasis[4,5]. According to the model of multi-step carcinogenesis, adenomatous cells accumulate a number of molecular abnormalities to eventually become fully malignant[6,7]. In spite of unifying theories, genetic and epigenetic events during the carcinogenesis process differ considerably from tumor to tumor. Thus, CRC is not a unique disease; rather it encompasses different molecular and pathological entities with a wide range of clinical behaviors[8]. At the molecular level, CRC encloses a complex array of gene alterations. Essentially, like individual fingerprints, each tumor arises and behaves in a distinctive fashion that is unlikely to be fully recapitulated by any other tumor. Nevertheless, molecular changes allow for a basic categorization of CRC, which is largely acknowledged, although likely over-simplistic. It has been demonstrated that genetic and epigenetic features, such as microsatellite instability (MSI), chromosomal instability, CpG island methylator phenotype or even global DNA hypomethylation, lead to alterations of gene function on a genome-wide scale. It is known that activation of oncogenes, including KRAS, BRAF, TGFBR, PIK3CA and TP53, affects complex intracellular signaling pathways[9,10]. The suppressor pathway is disrupted in CRC with chromosomal instability occurring in the majority of CRCs (nearly, 85%), which have a molecular profile characterized by specific chromosomal amplifications and transformations, aneuploidy, and loss of heterozygosity[8-10]. Differently, CRCs of the mutator pathway (roughly, 15%) have a defective DNA mismatch repair (MMR) system, which leads to accumulation of unrepaired mutations[9], and harbor frameshift mutations in coding mononucleotide repeats of cancer-related genes (targets)[11]. It is now accepted that MSI CRCs have a heterogeneous histological appearance, better prognosis due to a reduced metastatic potential, and a different response to 5-fluoro-uracyl[12-14].
Histopathology of CRC
Histopathological examination shows that likely other solid tumors, CRCs are infiltrated by various innate and adaptive immune cells[15-17], and that in the cancer context, epithelial cells coexist with different extracellular matrix components and non-neoplastic cell types, including fibroblasts, myofibroblasts, adipocytes, endothelial cells, pericytes, which collectively form the tumor microenvironment[18].
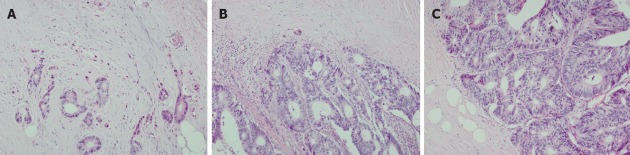
It is well known that histopathology reports usually include various features and including tumor grade, histological sub-type, state of resection margins and information on vascular and perineural invasion, but the tumor-border configuration (i.e., growth pattern) and especially tumor budding remain rarely described[1]. The term “tumor budding” denotes the presence of isolated single neoplastic cells or small clusters of cells (conventionally, up to 5 cells) scattered in the stromal compartment at the tumor invasive margin (Figure 1)[19-21].
Figure 1.
Colorectal cancer at the invasive front shows different growth patterns. Tumor budding denotes the presence of isolated single neoplastic cells or small clusters of cells scattered in the stromal compartment at the invasive tumor margin (A), although intra-tumoral budding is also reported. Tumor margin organized in larger tumor cell clusters (B) and a smooth infiltration tumor margin (C). Hematoxylin and eosin stain, objective magnification 20×.
Tumor stage as stated by the American Joint Committee on Cancer⁄International Union Against Cancer (AJCC⁄UICC) tumor-node-metastasis (TNM) system is currently considered as the most robust prognostic criterion for CRC patients. The inability of the AJCC⁄UICC staging system to accurately predict the outcome of individual patients with stage II and stage III CRC might be overcome by adding morphological, molecular or treatment-related features, that could stratify patients more accurately into different risk categories[22]. Depth local tumor infiltration (pT), loco-regional lymph-node involvement (N status), venous and lymphatic invasion, and tumor grade, are currently recognized as the main histopathological characteristics associated with worse patient outcome.
Tumor budding and CRC
Tumor budding first introduced by Jass et al[23], as a reliable histopathological hallmark to estimate the aggressiveness of rectal cancer, was initially shown to have a superior prognostic value when compared to other histopathological characteristics, including tumor differentiation and venous invasion. Tumor grading based on the nature of the advancing tumor margins, which in the scoring system proposed by Giger et al[24] divided rectal tumors into expanding type and infiltrating type, obtained wide acceptance among surgical pathologists worldwide. Although subsequent studies revealed a scarce reproducibility of Jass scoring system, several authors highlighted the potential role of tumor budding as a valid prognosticator also in tumors other than CRC, including lung cancer[25,26], invasive ampullary adenocarcinomas[27], and oesophageal and gastro-oesophageal junction cancers[28]. In CRC, tumor budding is considered as a stage IIB prognostic factor, and strictly associated with lymph-node metastasis[1]. It has been shown that the presence of “buds” at the tumor invasive front represents an independent predictor of lymph node metastasis in patients with sub-mucosal invasive or early pT1 CRCs[29]. It has also been suggested that the frequency of tumor budding increases with more advanced TNM stage[1].
Tumor budding is virtually absent in MMR-deficient cancers[30,31]. MSI CRCs have significantly more pronounced tumor infiltrating lymphocytes (i.e., CD3+ or CD8+ cells), peritumoral lymphocytes inflammation, and bundling edge (i.e., the ability of cells to adhere and to migrate) compared with microsatellite-stable CRCs, all factors contributing to the absence of tumor budding in MSI CRCs.
Tumor budding and the epithelial-mesenchymal transition
A parallel between tumor budding and the epithelial to mesenchymal transition (EMT) has also been recently proposed. This (potentially reversible) process thought to occur physiologically during embryological development (EMT subtype I), has been also associated with wound healing, tissue regeneration, organ fibrosis (EMT subtype II), and tumor invasion (EMT subtype III). Cells in EMT loss their epithelial phenotype (i.e., lack of E-Cadherin and cell polarity, expression of transcription factors including the zinc finger proteins SNAIL and SLUG, TWIST, ZEB 1/2 and SMAD) and dynamically acquire a mesenchymal phenotype (i.e., taking on a spindle-like, fusiform morphology, become motile, and start expressing mesenchymal markers including N-cadherin, fibronectin and vimentin)[32]. While the mechanisms promoting distant metastasis are extremely wide and still under intense investigation, the presence of EMT features in cells of the tumor microenvironment has been associated with an increased metastatic potential[32,33].
Assessing the tumor budding in colorectal cancer tissues
Rapidly growing insights into the cell biology of CRC and the recent developments of high-throughput technologies, gene sequencing and molecular diagnostics have led to practicable expectations for the identification of molecular biomarkers to be used in optimized and tailored treatment regiments. However, histopathological interpretation of CRC tissues remains the gold standard for cancer diagnosis. Tissue specimens, consisting of different cell types related to each other in complex spatial patterns, are important resources for both primary research efforts and validation of biological findings that are made in laboratory[34]. Working with human tissues poses several challenges to investigators, including: (1) tissue sampling (i.e., appropriate processing, histological variability, tissue heterogeneity with different areas of cancer, necrosis, inflammation and natural tissue); (2) selection of the proper preservation technique (i.e., maintenance of tissue morphology and molecular profile); (3) tissue complexity (i.e., requirement of an accurate histopathological interpretation); and (4) not least ethical and legal rules. However, an approach that integrates histopathology and molecular biology within a unique translational system is a mandatory strategy to pursue a better understanding of cancer. Such an effort can be achieved only through a more effective incorporation of pathology into clinical research, and conversely by integrating biological research into the pathological assessment, likely through efficient networks of translational researchers joining their data.
The morphology of the tumor invasive front has come into the focus of scientific studies because it appears to be intimately linked to cancer aggressiveness. Despite the established prognostic relevance of tumor budding in CRC, the reproducibility of actual methods proposed for its assessment, however, remains unstandardized, limiting its application in routine pathology practice[35]. Diagnostic reproducibility is a prerequisite for the validation of a diagnostic test and is crucial for patient care. Tumor budding promises to be a histopathological prognostic factor in CRC, and although the level of agreement needs to be improved and further investigations are compulsory to confirm any association between the rates of tumor budding detection and clinical outcome, its evaluation can be improved first by an appropriate physician training. In addition, the use of immunohistochemistry (IHC) highlights budding cells by pan-cytokeratin antibodies leading to a significant increase of tumor budding-positive cases. Single tumor cells can be more accurately detected by immunological techniques than standard hematoxylin and eosin staining, even when they appear at the tumor boundary showing glandular disruption. Under these circumstances, dissociated tumor cells should not be interpreted as budding to avoid biasing tumor budding evaluation[31].
As tumor budding has been shown as an independent prognostic factor in CRC, particularly in node-negative disease, its assessment has the potential to increase prognostic accuracy and influence treatment algorithms. When examined carefully, the majority of CRCs display some degree of budding; hence, attempts have been made at developing scoring systems to identify a prognostically significant degree of budding, commonly termed “high-grade” budding. Definitions of high-grade budding, however, vary substantially among different observers and even among different studies by the same observers[31].
Final remarks
Consensus criteria for its evaluation must be better established, to guide further research in this area and to provide the practicing pathologist with reporting guidelines. With respect to setting these criteria, studies focusing on budding should be designed to define objective cut-off for meaningful tumor budding. In mathematical terms, also tumor budding is a continuous variable; thus, a cut-off threshold should be less arbitrary as possible, and an attempt should be made to identify the budding threshold that results into relevant predictive information. Along this line, pathologist reporting on tumor budding should provide detailed information regarding the qualitative and quantitative criteria used to evaluate budding in order to allow for meaningful comparisons among different studies. Finally, the role of IHC in the evaluation of budding needs to be clarified. Although it might be impractical to perform IHC on all CRCs, there may be certain cases (i.e., in the context of a remarkable inflammatory reaction at the tumor invasive front), where it may reveal buds that are dubious when observed in standard hematoxylin and eosin stained histological sections.
It is indubitable that the substantial impediment to the adoption of tumor budding as a routinely reportable feature is the lack of a well defined, standardized and quantitative assessment. At any event, due to the forceful evidence that tumor budding is one of the most promising prognostic factors actually available, it is incumbent on the scientific community participating to the identification of CRC prognostic factors to move promptly to addressing it and removing the obstacles to its routine reporting and comparison with other predictive factors.
Footnotes
Supported by Ministero dell’Istruzione, dell’Università e della Ricerca, Target Project Oncologia 2006; Alleanza Contro il Cancro; the Italian Association for Cancer Research, grant project No. IG5256
Peer reviewer: Asmaa Gaber Abdou, Assistant Professor, Department of Pathology, Faculty of Medicine, Menofiya University, Shebein Elkom 002-048, Egypt
S- Editor Song XX L- Editor A E- Editor Li JY
References
- 1.Lugli A, Karamitopoulou E, Zlobec I. Tumour budding: a promising parameter in colorectal cancer. Br J Cancer. 2012;106:1713–1717. doi: 10.1038/bjc.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 4.Michor F, Iwasa Y, Lengauer C, Nowak MA. Dynamics of colorectal cancer. Semin Cancer Biol. 2005;15:484–493. doi: 10.1016/j.semcancer.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Jass JR. Colorectal cancer: a multipathway disease. Crit Rev Oncog. 2006;12:273–287. doi: 10.1615/critrevoncog.v12.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 6.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 8.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laghi L, Ranzani GN, Bianchi P, Mori A, Heinimann K, Orbetegli O, Spaudo MR, Luinetti O, Francisconi S, Roncalli M, et al. Frameshift mutations of human gastrin receptor gene (hGARE) in gastrointestinal cancers with microsatellite instability. Lab Invest. 2002;82:265–271. doi: 10.1038/labinvest.3780420. [DOI] [PubMed] [Google Scholar]
- 12.Hyde A, Fontaine D, Stuckless S, Green R, Pollett A, Simms M, Sipahimalani P, Parfrey P, Younghusband B. A histology-based model for predicting microsatellite instability in colorectal cancers. Am J Surg Pathol. 2010;34:1820–1829. doi: 10.1097/PAS.0b013e3181f6a912. [DOI] [PubMed] [Google Scholar]
- 13.Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 2007;13:3831–3839. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 14.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 15.Deschoolmeester V, Baay M, Van Marck E, Weyler J, Vermeulen P, Lardon F, Vermorken JB. Tumor infiltrating lymphocytes: an intriguing player in the survival of colorectal cancer patients. BMC Immunol. 2010;11:19. doi: 10.1186/1471-2172-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peddareddigari VG, Wang D, Dubois RN. The tumor microenvironment in colorectal carcinogenesis. Cancer Microenviron. 2010;3:149–166. doi: 10.1007/s12307-010-0038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 18.Disis ML. Immune regulation of cancer. J Clin Oncol. 2010;28:4531–4538. doi: 10.1200/JCO.2009.27.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puppa G, Senore C, Sheahan K, Vieth M, Lugli A, Zlobec I, Pecori S, Wang LM, Langner C, Mitomi H, et al. Diagnostic reproducibility of tumour budding in colorectal cancer: a multicentre, multinational study using virtual microscopy. Histopathology. 2012:Jul 5; Epub ahead of print. doi: 10.1111/j.1365-2559.2012.04270.x. [DOI] [PubMed] [Google Scholar]
- 20.Prall F. Tumour budding in colorectal carcinoma. Histopathology. 2007;50:151–162. doi: 10.1111/j.1365-2559.2006.02551.x. [DOI] [PubMed] [Google Scholar]
- 21.Zlobec I, Lugli A. Invasive front of colorectal cancer: dynamic interface of pro-/anti-tumor factors. World J Gastroenterol. 2009;15:5898–5906. doi: 10.3748/wjg.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puppa G, Sonzogni A, Colombari R, Pelosi G. TNM staging system of colorectal carcinoma: a critical appraisal of challenging issues. Arch Pathol Lab Med. 2010;134:837–852. doi: 10.5858/134.6.837. [DOI] [PubMed] [Google Scholar]
- 23.Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet. 1987;1:1303–1306. doi: 10.1016/s0140-6736(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 24.Giger OT, Comtesse SC, Lugli A, Zlobec I, Kurrer MO. Intra-tumoral budding in preoperative biopsy specimens predicts lymph node and distant metastasis in patients with colorectal cancer. Mod Pathol. 2012;25:1048–1053. doi: 10.1038/modpathol.2012.56. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi Y, Ishii G, Kojima M, Yoh K, Otsuka H, Otaki Y, Aokage K, Yanagi S, Nagai K, Nishiwaki Y, et al. Histopathologic features of the tumor budding in adenocarcinoma of the lung: tumor budding as an index to predict the potential aggressiveness. J Thorac Oncol. 2010;5:1361–1368. doi: 10.1097/JTO.0b013e3181eaf2f3. [DOI] [PubMed] [Google Scholar]
- 26.Taira T, Ishii G, Nagai K, Yoh K, Takahashi Y, Matsumura Y, Kojima M, Ohmatsu H, Goto K, Niho S, et al. Characterization of the immunophenotype of the tumor budding and its prognostic implications in squamous cell carcinoma of the lung. Lung Cancer. 2012;76:423–430. doi: 10.1016/j.lungcan.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Ohike N, Coban I, Kim GE, Basturk O, Tajiri T, Krasinskas A, Bandyopadhyay S, Morohoshi T, Shimada Y, Kooby DA, et al. Tumor budding as a strong prognostic indicator in invasive ampullary adenocarcinomas. Am J Surg Pathol. 2010;34:1417–1424. doi: 10.1097/PAS.0b013e3181f0b05a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown M, Sillah K, Griffiths EA, Swindell R, West CM, Page RD, Welch IM, Pritchard SA. Tumour budding and a low host inflammatory response are associated with a poor prognosis in oesophageal and gastro-oesophageal junction cancers. Histopathology. 2010;56:893–899. doi: 10.1111/j.1365-2559.2010.03559.x. [DOI] [PubMed] [Google Scholar]
- 29.Kye BH, Jung JH, Kim HJ, Kang SG, Cho HM, Kim JG. Tumor budding as a risk factor of lymph node metastasis in submucosal invasive T1 colorectal carcinoma: a retrospective study. BMC Surg. 2012;12:16. doi: 10.1186/1471-2482-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lugli A, Vlajnic T, Giger O, Karamitopoulou E, Patsouris ES, Peros G, Terracciano LM, Zlobec I. Intratumoral budding as a potential parameter of tumor progression in mismatch repair-proficient and mismatch repair-deficient colorectal cancer patients. Hum Pathol. 2011;42:1833–1840. doi: 10.1016/j.humpath.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Mitrovic B, Schaeffer DF, Riddell RH, Kirsch R. Tumor budding in colorectal carcinoma: time to take notice. Mod Pathol. 2012;25:1315–1325. doi: 10.1038/modpathol.2012.94. [DOI] [PubMed] [Google Scholar]
- 32.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui G, Shi Y, Cui J, Tang F, Florholmen J. Immune microenvironmental shift along human colorectal adenoma-carcinoma sequence: is it relevant to tumor development, biomarkers and biotherapeutic targets? Scand J Gastroenterol. 2012;47:367–377. doi: 10.3109/00365521.2011.648950. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Canales J, Eberle FC, Jaffe ES, Emmert-Buck MR. Why is it crucial to reintegrate pathology into cancer research? Bioessays. 2011;33:490–498. doi: 10.1002/bies.201100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karamitopoulou E, Lugli A, Panayiotides I, Karakitsos P, Peros G, Rallis G, Patsouris ES, Terracciano L, Zlobec I. Systematic assessment of protein phenotypes characterizing high-grade tumour budding in mismatch repair-proficient colorectal cancer. Histopathology. 2010;57:233–243. doi: 10.1111/j.1365-2559.2010.03615.x. [DOI] [PubMed] [Google Scholar]