Abstract
Herbivorous insects are among the most successful radiations of life. However, we know little about the processes underpinning the evolution of herbivory. We examined the evolution of herbivory in the fly, Scaptomyza flava, whose larvae are leaf miners on species of Brassicaceae, including the widely studied reference plant, Arabidopsis thaliana (Arabidopsis). Scaptomyza flava is phylogenetically nested within the paraphyletic genus Drosophila, and the whole genome sequences available for 12 species of Drosophila facilitated phylogenetic analysis and assembly of a transcriptome for S. flava. A time-calibrated phylogeny indicated that leaf mining in Scaptomyza evolved between 6 and 16 million years ago. Feeding assays showed that biosynthesis of glucosinolates, the major class of antiherbivore chemical defense compounds in mustard leaves, was upregulated by S. flava larval feeding. The presence of glucosinolates in wild-type (WT) Arabidopsis plants reduced S. flava larval weight gain and increased egg–adult development time relative to flies reared in glucosinolate knockout (GKO) plants. An analysis of gene expression differences in 5-day-old larvae reared on WT versus GKO plants showed a total of 341 transcripts that were differentially regulated by glucosinolate uptake in larval S. flava. Of these, approximately a third corresponded to homologs of Drosophila melanogaster genes associated with starvation, dietary toxin-, heat-, oxidation-, and aging-related stress. The upregulated transcripts exhibited elevated rates of protein evolution compared with unregulated transcripts. The remaining differentially regulated transcripts also contained a higher proportion of novel genes than the unregulated transcripts. Thus, the transition to herbivory in Scaptomyza appears to be coupled with the evolution of novel genes and the co-option of conserved stress-related genes.
Keywords: Arabidopsis, Drosophila, glucosinolates, herbivory, host specialization, transcriptome
Introduction
Phytophagous insects are among the most evolutionarily successful radiations. Herbivorous taxa comprise almost 50% of living insect species and 25% of living metazoan species (Ehrlich and Raven 1964; Bernays 1998) and are more diverse than their aphytophagous sister lineages (Mitter and Farrell 1988; Farrell 1998). Adapting to a herbivorous lifestyle is thought to have been a difficult transition for most insects, however, due to morphological and physiological challenges of consuming living plant tissues that are typically well defended against insects (Frankel 1959; Ehrlich and Raven 1964; Southwood 1973). Some antiherbivore defenses, including insecticidal compounds produced by plants, are dramatically induced within hours of herbivore attack and are tailored toward specific guilds of insect enemies (Kunitz 1945; Bostock 2005; Textor and Gershenzon 2009). In addition, these compounds are often effective against other natural enemies, such as microbes (Fan et al. 2011), and vertebrates (Magnanou et al. 2009).
The genomic changes that accompany a shift from an aphytophagous to phytophagous lifestyle are not well understood. The transition to herbivory has evolved independently many times in the insects and frequently involved adaptation to a more specialized host plant range. Indeed, most herbivorous species are specialized in host range and are only able to complete development on a limited number of species. Many evolutionary changes are likely to be required for a successful transition to herbivory, including the ability to find suitable host plants, utilize potentially nutritionally imbalanced plant tissues, and cope with host plant defenses (Bernays and Chapman 1994; Govind et al. 2010).
To study the transition to herbivory from an evolutionary perspective, we have focused on a recently derived herbivorous species that is closely related to a model genetic species. Our long-term goal is to understand how the evolution of novel genes and/or the recruitment of existing metabolic or signaling pathways have enabled herbivores to adapt to a fundamentally new niche. Most phytophagous insects are lepidopterans or coleopterans. Because the evolution of herbivory in these lineages is estimated to have occurred in the Cretaceous, 145–65 million years ago (Whalley 1977; Moran 1989; Labandeira et al. 1994; Bernays 1998; Farrell 1998), using comparative genomics to dissect the genetic bases of these transitions would be difficult. However, herbivory has also evolved many times in dipterans (Labandeira 2003), and the family Drosophilidae includes 12 species with published genome sequences and at least two genera (Scaptomyza and Scaptodrosophila) with phytophagous members (Hackman 1959; Bock and Parsons 1975; Clark et al. 2007). These provide the opportunity to analyze more recent transitions to herbivory.
The Scaptomyza lineage is nested in the paraphyletic subgenus Drosophila (O′Grady and Desalle 2008). Most of the described drosophilid species (ca., 2,000) are saprophagous, feeding externally on microbes growing on decaying vegetation (e.g., Drosophila melanogaster), ripe fruits (e.g., D. sechellia), or fruiting bodies of fungi (e.g., D. recens). Although decaying vegetation and fungi can be toxin-rich and likely present a barrier to colonization (Jaenike et al. 1983; Frank and Fogleman 1992; Jones 2001, 2005; Markow and O'Grady 2005; Dworkin and Jones 2009), it seems unlikely that host plants or fungi would mount a defense response against these insects. However, drosophilids in the genus Scaptomyza (fig. 1) have evolved the ability to feed on living angiosperm leaves, which activate inducible defenses in response to feeding damage (Hackman 1959; Whiteman et al. 2011). It is likely that a variety of preformed and inducible defense compounds were novel traits encountered by Scaptomyza and other herbivorous Drosophilidae in their transition from a saprophagous to a phytophagous lifestyle.
Fig. 1.—
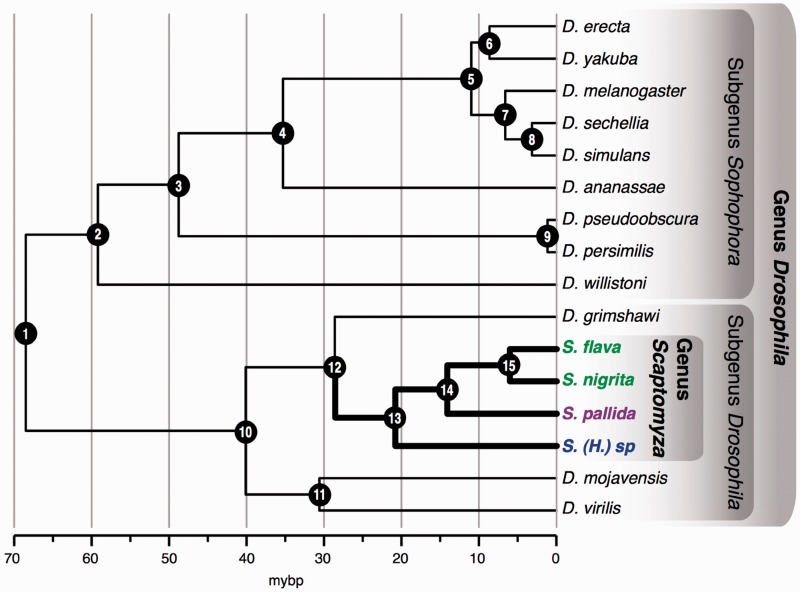
Chronogram of combined nucleotide sequence data set from 9 genes from the 12 Drosophila species with completely sequenced genomes and multiple Scaptomyza species (S. flava, S. nigrita, S. pallida, and S. [Hemiscaptomyza] sp.). Initial tree was a maximum likelihood partitioned analysis of 9 genes inferred using RAxML (−ln L = −29,522.877). See table 1 for bootstrap support values for nodes. Nodes were constrained to ages estimated from Tamura et al. (2004). The two leaf-mining Scaptomyza species are resolved as sister taxa. Scaptomyza (Parascaptomyza) pallida and Scaptomyza (Hemiscaptomyza) sp. do not mine leaves of plants and can be reared on Drosophila media, and larvae are associated with decaying vegetation. The split between the most recent common ancestor of the two leaf-mining taxa, S. flava and S. nigrita (labeled in green), was estimated at 6.31(±0.63 SD) MYBP, and we inferred this to be the most recent date for the evolution of mustard leaf mining.
The ancestral host plants of the earliest Scaptomyza herbivores are unknown, but many herbivorous Scaptomyza species are monophagous or oligophagous on plants in the order Brassicales (Hackman 1959). The larvae of S. flava feed on Arabidopsis thaliana (hereafter referred to as Arabidopsis) and other species of Brassicales in the wild (Chittenden 1902; Whiteman et al. 2011) and require living plants to complete their life cycle (Whiteman et al. 2011). Because they have been found on many members of the Brassicales and on species in a few other plant families (Caryophyllaceae and Fabaceae), we consider this species to be oligophagous (Martin 2004). We took advantage of the availability of Arabidopsis mutants blocking the synthesis of insecticidal compounds to test the hypothesis that major changes associated with the transition to feeding on living plants can be detected at the genomic level in S. flava.
We focused our attention on interactions between larval S. flava and glucosinolates, which are well characterized functionally and metabolically in Arabidopsis (Halkier and Gershenzon 2006) and which we hypothesized were a likely barrier to colonization by ancestors of S. flava. Glucosinolates are a major barrier to insect colonization of Brassicales, including economically important crops such as canola (Brassica napus), broccoli, cabbage (B. oleracea), and papaya (Carica papaya). Glucosinolates (and related compounds called camalexins [Hull et al. 2000]) are amino acid-derived thioglucosides that are present constitutively in tissues of plants in the Brassicaceae and are highly inducible after herbivore attack, increasing in concentration in leaves by up to 40-fold (Halkier and Gershenzon 2006; Textor and Gershenzon 2009). During tissue damage, glucosinolates are hydrolyzed by endogenous myrosinases and are transformed into toxic, electrophilic mustard oils (isothiocyanates) that can damage proteins and DNA (Halkier and Gershenzon 2006).
We used a variety of approaches, including genome-wide transcriptional profiling by RNA-seq, to characterize the evolutionary and functional impact of glucosinolates, a major plant antiherbivore defense, on S. flava. Our results provide a first glimpse into how the genomes of insects are shaped by the major ecological transition to herbivory from nonherbivorous ancestors. We found that new and conserved stress-related genes appear to be co-opted by S. flava in its response to host plant defenses. The powerful tools available for the genetic model organisms Drosophila and Arabidopsis should facilitate functional characterization of genes identified here as candidates, allowing us to further elucidate the molecular changes that accompany the ecological transition to herbivory and host plant specialization.
Materials and Methods
Phylogenetic Inference and Dating
To determine the relationships and times of divergence between the 12 Drosophila species with completely sequenced genomes and S. flava, we inferred a phylogeny using a codon-partitioned, concatenated data set of ∼5,000 genes from each of the 13 species, relying on genomic data for the 12 Drosophila species and transcriptome data for S. flava. To gain further insight into the timing of the evolution of herbivory, we also sequenced nine genes (seven nuclear and two mitochondrial) from an expanded taxonomic sample of leaf-mining and saprophagous Scaptomyza species and estimated phylogenetic relationships among these species and other drosophilids. In both analyses, we used independent evidence to calibrate nodes and estimate the timing of the origins of herbivory, and we evaluated nodes for statistical support (supplementary materials and methods, Supplementary Material online).
GUS Histochemical Assay
To determine whether feeding by S. flava larvae induces transcription of genes essential for glucosinolate biosynthesis, cyp79B2-promoter-GUS fusion Arabidopsis plants (Millet et al. 2010) were grown at 22°C and 50% relative humidity with a 16h:8h light:dark cycle. These reporter lines yield a qualitative measure of cyp79B2 gene expression. At 32 days following germination, a single third instar larva reared on Col-0 wild-type (WT) Arabidopsis plants was placed on each of two leaves per plant and allowed to feed for 12 h. At the same time, two leaves per mechanical control plant were crushed using a forceps three times mimicking the amount of area consumed by the larva at each time point. Although this treatment is unlikely to precisely mimic active feeding by S. flava larvae, we damaged roughly the same leaf area as a larva would over a 12-h period. An undamaged control was left untouched. Infiltration with GUS substrate solution and tissue fixation followed Millet et al. (2010). Images were visualized using a Leica MZ6 dissecting scope with a 0.5 × lens, and results were analyzed qualitatively.
Performance Assays on Wild-Type and Glucosinolate Knockout Arabidopsis thaliana Lines
To determine whether glucosinolates reduce growth rates of S. flava, we grew WT (Col-0) Arabidopsis, aliphatic glucosinolate mutant (myb28myb29) (aliphatic glucosinolate knock out [AGKO]), indolic glucosinolate mutant (cyp79b2cyp79b3) (abbreviated IGKO), and an indolic and aliphatic glucosinolate mutant (cyp79b2cyp79b3myb28myb29) (glucosinolate knockout [GKO]) plants to 4 weeks of age (see supplementary materials and methods, Supplementary Material online, for details). We allowed approximately 400 gravid female S. flava flies to create feeding punctures (stipples) and lay eggs in WT (N = 60 plants), AGKO (N = 60 plants), IGKO (N = 60 plants), and GKO (N = 60 plants) Arabidopsis plants for 24 h by randomly arraying five plants of each genotype in a cage and repeating this for all 240 plants. For each plant individual, we quantified the number of stipples created by female flies and egg number. We compared the intercepts and slopes of stipple number versus egg number regression lines across the four plant genotypes using an analysis of covariance (ANCOVA) approach. After hatching, larvae were allowed to feed on leaves for either 3 or 5 days. We compared average larval masses across plant genotypes for each cohort (days 3 and 5) also using an ANCOVA (using the aov function in the statistical package R v. 2.15.0) to test for effects of crowding on larval growth rates as there were multiple larvae per plant. A subset of larvae harvested at 5 days post–egg hatching were flash frozen in LN2 vapor and stored at −80°C. A subset of these 5-day-old larvae, two pools from two host plant lines (WT and GKO), were used for RNA-sequencing on the Illumina GAII platform.
In a separate experiment, the same plant lines were reared as above, and at 4 weeks, individuals of Col-0 (N = 15 plants) and GKO (N = 15 plants) were placed with approximately 50 ovipositing S. flava females for 24 h. Each plant was then isolated in a clear acrylic rearing box, placed in randomized arrays under lights as above, and watered to keep soil moisture constant, and larvae were allowed to develop freely. Beginning on day 15 post–egg hatch, we recorded the number of adult flies that emerged twice each day. Development time was calculated as the average number of days from egg hatch to adult emergence for a cohort of flies that emerged from a single plant.
Metabolic Profiling of Arabidopsis Lines Used in Herbivore Performance and Transcriptional Profiling Experiments
We conducted nontargeted metabolite profiling of polar compounds (sugars, hydroxy- and amino acids) by fractionation and gas chromatography/mass spectrometry. Glucosinolates are not detectable by this protocol and were not analyzed using other means because the levels in GKO plants have been previously characterized and shown to be undetectable (Muller et al. 2010).
We used 4-week-old WT and GKO (myb28myb29cyp79b2cyp79b3) Arabidopsis plants as above, which were grown under a 16 h light:8 h dark cycle (using Gro-Lux [Sylvania, Danvers, MA, USA] bulbs) at 22°C, 50% relative humidity, in a Conviron reach-in Adaptis growth chamber at the University of Arizona. Five plants of each genotype were flash frozen in LN2 and shipped on dry ice to the Proteomics and Metabolomics Facility at Colorado State University (http://www.pmf.colostate.edu/) for extraction and GC-MS analysis (details given in the supplementary materials and methods, Supplementary Material online).
454 Titanium GSFLX Transcriptome Sequencing
RNA Preparation, cDNA Synthesis, and Normalization
In 2008, we froze under LN2 vapor fresh eggs, larvae, pupae, and adults of S. flava from a population cage of flies derived from larvae collected from leaves of Barbarea vulgaris (Brassicaceae) in Belmont, MA, and subsequently reared on Arabidopsis Col-0 and Tsu-0 accessions. A subset of the adults was septically injured with cultures of Escherichia coli, Serattia marcescens, Pseudomonas syringae, or Staphylococcus aureus to maximize gene expression from as many loci as possible (De Gregorio et al. 2001; Lemaitre and Hoffmann 2007). Details of the RNA preparation, cDNA synthesis, and normalization can be found in the supplementary materials and methods, Supplementary Material online.
Library Preparation (DNA Processing) for 454 GS FLX
We followed previously published protocols to prepare the 454 GS FLX library (Toth et al. 2007). cDNAs were nebulized and size selected for an average size of 400–800 bp. 454 GS FLX Titanium-specific adapters, AdapterA and AdapterB, were ligated to the cDNA ends after end polishing. The adapter-ligated DNAs were then mobilized to the library preparation beads, and sst-cDNAs were captured. The number of molecules of the sst-cDNAs was calculated using the concentration and average fragment length for emPCR.
454 GSFLX Sequencing
We sequenced the library on a 454 GSFLX flow cell using titanium chemistry at the University of Illinois. The raw reads were processed as described later and combined with the Illumina-generated data to create a grand assembly.
Illumina GAII 72 bp Transcriptome Sequencing
RNA Preparation
Larvae harvested 5 days post–egg hatch were placed in tubes directly after harvesting from plant leaves, and masses were obtained for performance studies described earlier. Four larval pools were obtained, two pools from Col-0 plants (pool 1, N = 20 larvae; pool 2, N = 18 larvae) and two from GKO plants (pool 3, N = 12 larvae; pool 4, N = 12 larvae). Larvae were flash frozen in LN2 and then crushed in a Qiagen Tissue Lyser (Retsch Mixer Mill) and homogenized with 500 µl TRI REAGENT per sample, generally following the RNA extraction protocol above (for details see supplementary materials and methods, Supplementary Material online). Genomic DNA was removed using a gDNA Eliminator spin column (Qiagen, Inc.). A sample of RNA from each of the four RNA pools was run on an Agilent 2100 Bioanalyzer for (RNA Nano Chip) to determine the integrity of the RNA samples.
mRNA Isolation, cDNA Synthesis, and Library Preparation (DNA Processing) for Illumina Sequencing
The Illumina mRNA sequencing sample prep kit was used to prepare libraries for each of the four RNA pools, resulting in four RNA-seq libraries that were subjected to clustering and 72 bp sequencing on an Illumina GA II instrument, following the manufacturer’s protocol. These libraries were not normalized.
454 and Illumina Data Assembly
Quality Control and Preprocessing of Sequencing Data
The RNA-seq data consisted of a full plate of 454 sequencing (990,223 reads total, from normalized cDNA pools) and four lanes of 72 bp Illumina sequencing [73,147,138 reads total, from un-normalized cDNA pools from 5-day-old larvae raised on either WT or the GKO (cyp79b2cyp79b3myb28myb29) Arabidopsis plants]. Quality control and preprocessing details can be found in the supplementary materials and methods, Supplementary Material online.
Contig Assembly
The filtered reads were initially assembled using the software program AbySS version 1.2.1 (Birol et al. 2009; Simpson et al. 2009) as is described in detail in the supplementary materials and methods, Supplementary Material online.
Homology Assessment and Scaffolding
To identify the closest homolog of each assembled S. flava contig, we first used blastx to map contigs to proteins from D. grimshawi, with an E value cutoff of 1e−10. We retained all hits for which at least 20% of the S. flava contig hits a D. grimshawi protein; we then filtered out all hits with E values more than 10 orders of magnitude worse than the best hit. We also ran an all-against-all blastn search with an E-value cutoff of 1e−10 and using the S. flava contigs as both query and subject to identify potential paralogs that were not collapsed by our assembly procedure. For this blastn run, we kept all hits, regardless of E value, where the hit covered at least 90% of the smaller contig. We then identified assembled transcript regions (transcripts) as sets of contigs and D. grimshawi proteins that are all connected to each other via blastn or blastx hits, using an undirected graph algorithm implemented in custom Perl scripts. We then generated three different transcript sets based on our confidence in assignment. The high confidence set is based on blastx hits with at least 85% coverage; the medium confidence set is based on blastx hits with between 60% and 85% coverage; and the low confidence set is based on blastx hits with between 20% and 60% coverage. In all three sets, we flagged contigs if they 1) were less than 200 bp and a singleton, 2) had a significant blast hit to a non-Drosophila sequence in nr, but not to any Drosophila, or 3) had no blast hits with at least the minimum 20% query coverage. For all confidence sets, we built assemblies by merging adjacent or overlapping S. flava sequence that maps to a given D. grimshawi protein and filling in regions of the D. grimshawi protein not sequenced in our cDNA libraries as Ns. These merged sequences form the basis of the molecular evolutionary analysis (supplementary table S5, Supplementary Material online) and differential regulation analysis described later (supplementary table S6, Supplementary Material online).
By scaffolding contigs and merging potential S. flava paralogs identified via all-against-all blastn searches, we collapsed the original contigs to assembled transcripts. These transcripts were then filtered to remove short contigs and those with weak or complex blast hits, which served as the transcriptome against which the short-reads from Illumina sequencing of the 5-day-old larvae from WT and GKO plants were mapped to quantify differential regulation of S. flava genes.
Quantifying Differential Gene Regulation Using the Transcriptome and RNA- S eq
Here, we used RNA-seq to measure differential expression between the transcriptomes of flies raised on WT and GKO Arabidopsis. We mapped each of our four filtered Illumina RNA-seq samples against the S. flava (scap) contig set derived from all transcriptome data using Bowtie 0.12.5 (Langmead et al. 2009) with the following command line parameters: -S -n 2 -e 80 -l 25 -nomaqround –y –a –best –strata -m 1 –solexa1.3-quals –sam-nosq -p 2. We were able to uniquely map between 37.9% and 43.79% of reads in each sample to our filtered contig data set, representing 28.2 million total mapped sequences. We then counted the number of reads mapped to each contig and summed across all contigs in each transcript to generate a count of reads mapped to each transcript in each condition. We tested for differential expression of transcripts in larvae reared on the two Arabidopsis lines by first summing the reads mapped across all contigs in each transcript to generate a count of reads mapped to each transcript in each pool, and estimating log2-fold change per treatment (WT–GKO) using a negative binomial model implemented in the R/Bioconductor package DESeq (Anders and Huber 2010).
Estimating Rates of Molecular Evolution
The ratio of nonsynonymous substitutions per nonsynonymous site normalized by the number of synonymous substitutions per synonymous site (dN/dS) was calculated to estimate rates of molecular evolution in S. flava and across the subgenus Drosophila, within which the genus Scaptomyza is nested. First, we identified all one-to-one homologs present in S. flava and the three species in the subgenus Drosophila with sequenced genomes: D. grimshawi, Drosophila virilis, and Drosophila mojavensis (details provided in the supplementary materials and methods, Supplementary Material online).
To estimate dN/dS for each alignment, we used a maximum likelihood framework implemented in the codeml module of PAML version 4 (Yang 2007). The unrooted input tree (D. virilis, D. mojavensis, (D. grimshawi, S. flava #)) was used with the two-ratio branch model, which estimates one dN/dS in a specified foreground branch (S. flava in this case) and a separate dN/dS across the rest of the tree. As a quality control, we excluded from analysis all homolog sets where greater than 5% of sites were variable in the S. flava contig or where dS in S. flava was greater than 0.8158 (two times the median dS across the full S. flava data set), which may represent contig misassembly or sequence misalignment, respectively. We also excluded sets where dS in S. flava was less than 0.01 or less than 70 codons were present without gaps in all four species to avoid unreliable estimates of dN/dS due to insufficient data. Results of all these analyses are reported in supplementary table S6, Supplementary Material online.
Filtering Developmentally Regulated Genes
We mined a high-throughput gene expression data set constructed using D. melanogaster, in which Graveley et al. (2011) used Illumina RNA-seq to quantify gene expression across 30 developmental stages in isogenic (y1; cn bw1 sp1) D. melanogaster flies. Raw expression values expressed as reads per kilobase per million reads mapped (Graveley et al. 2011) were obtained from the analysis of Gelbart and Emmert (2010), which intersected coverage expression data with FlyBase exons.
We excluded data from D. melanogaster genes that had been split into multiple separate gene models as of 5 September 2011. A gene was flagged as developmentally confounded if the log2 (fold change) between 1) second instar larvae and larvae 12 h into third instar or 2) 12-h third instar larvae and larvae that had just entered the third instar puffstage, was greater than 50% of the log2 (fold expression change) between GKO- and WT-fed larvae in our experiment.
Results
Timing of the Evolution of Herbivory and Host Specialization
The date uniting the Scaptomyza + Hawaiian Drosophila using nine gene fragments (29.76 ± 2.25 millions of years before present [MYBP]) was very close to the one estimated for this node using S. flava transcriptomic data and genomic data for the 12 sequenced Drosophila species (27.06 ± 0.66 MYBP) (table 1). The divergence time for the two herbivorous taxa included in this analysis (S. flava and S. nigrita) was estimated to be 6.31 (± 0.63) MYBP, and the divergence time between this clade and S. pallida, which is not herbivorous, was estimated to be 14.70 (±1.26) MYBP. These dates give a conservative estimate of the range (6–16 MYBP) within which herbivory and mustard specialization likely evolved.
Table 1.
Ages of Nodes in Figure 1 and Supplementary Figure S1, Supplementary Material Online, Inferred by r8s Analysis
| Node | Calibration Range (MYBP) | Transcriptome Alignment, Age ± SD (MYBP) | Multiple Scaptomyza spp. | |
|---|---|---|---|---|
| Age ± SD (MYBP) | Bootstrap Support | |||
| 1 | 50.5–75.3 | 74.81 ± 1.14 | 71.19 ± 4.10 | |
| 2 | 62.55 ± 0.74 | 61.44 ± 3.90 | 100 | |
| 3 | 48.85 ± 0.97 | 50.72 ± 3.01 | 100 | |
| 4 | 35.3–53.1 | 35.34 ± 0.73 | 36.75 ± 1.97 | 100 |
| 5 | 8.80 ± 1.38 | 11.40 ± 0.84 | 100 | |
| 6 | 6.48 ± 0.80 | 8.96 ± 0.78 | 57 | |
| 7 | 4.50 ± 0.70 | 6.85 ± 0.62 | 100 | |
| 8 | 2.31 ± 0.52 | 3.25 ± 0.40 | 100 | |
| 9 | 0.56–1.14 | 1.14 ± 0.05 | 1.14 ± 0.00 | 100 |
| 10 | 34.2–51.6 | 43.26 ± 0.74 | 41.70 ± 2.92 | 100 |
| 11 | 32.04 ± 0.66 | 31.82 ± 2.57 | 34 | |
| 12 | 23.9–37.1 | 27.06 ± 0.66 | 29.76 ± 2.25 | 100 |
| 13 | – | 21.69 ± 1.76 | 100 | |
| 14 | – | 14.70 ± 1.26 | 100 | |
| 15 | – | 6.31 ± 0.63 | 100 | |
Note.—Node numbers refer to nodes labeled in figure 1 and supplementary figure S1, Supplementary Material online. Date intervals for calibrating are in the second column. Divergence dates are in millions of years before present (MYBP) followed by standard deviation of dates estimated by profiling nodes across 1,000 bootstrapped replicates. Two phylogenies were used: the first used characters from an alignment of nucleotide sequences from the 12 Drosophila genomes and Scaptomyza flava transcriptome, and the second used 9 genes from a broader sampling of drosophilids, including herbivorous and nonherbivorous Scaptomyza species. All bootstrap values for the tree generated for transcriptome alignment were 100.
Phenotypic Consequences of Arabidopsis Antiherbivore Defenses on S. flava
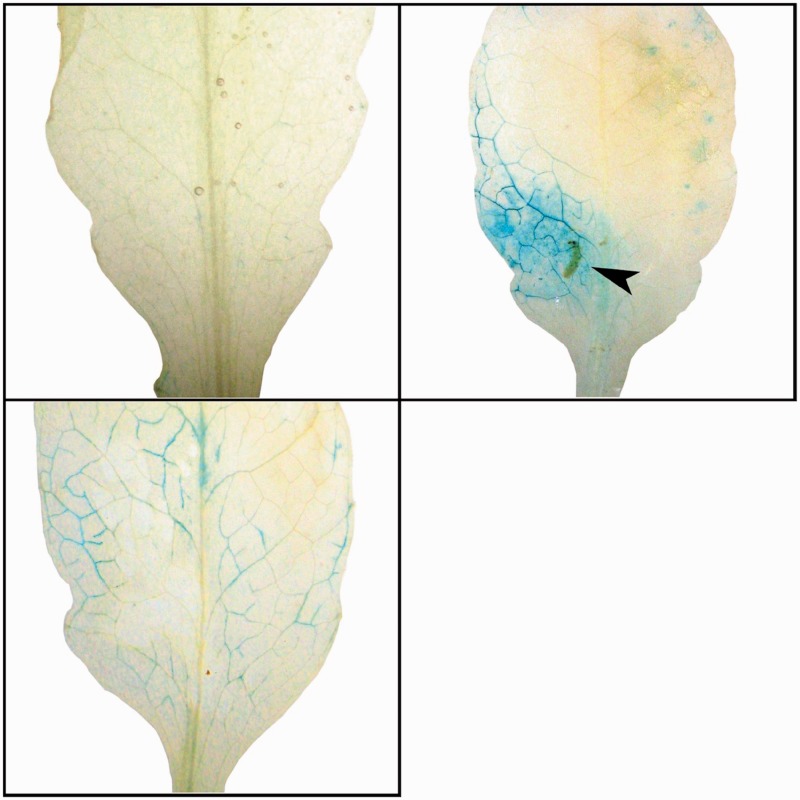
We first tested whether herbivory by S. flava larvae activated the glucosinolate biosynthetic pathway in Arabidopsis. We qualitatively assayed transcriptional activity of the Arabidopsis cyp79b2 gene in response to feeding by third instar S. flava larvae that were placed on adult Arabidopsis cyp79b2promoter:β-glucuronidase (GUS) reporter plants and allowed to form mines for 12 h. Arabidopsis cyp79b2 encodes a cytochrome P450 enzyme required for indolic glucosinolate biosynthesis (Hull et al. 2000; Muller et al. 2010). Although there was only modest transcriptional (GUS) activity in leaves that were mechanically wounded (repeatedly) with stainless steel forceps, the Arabidopsis cyp79b2 promoter drove strong transcriptional induction in leaf tissue surrounding actively feeding larvae (fig. 2). Little transcriptional activity was observed in unwounded control leaves (fig. 2).
Fig. 2.—
The indolic glucosinolate biosynthetic pathway is induced by S. flava larval feeding. Activation of the indolic glucosinolate biosynthetic cyp79b2 gene in the Arabidopsis thaliana promoter:GUS reporter line when attacked by S. flava larvae for 12 h. Strong induction occurs within and around larval mines (top right) but less so when leaves are mechanically wounded (bottom left) and induction appears absent in untouched leaves (top left).
We next tested whether glucosinolates are involved in resistance against S. flava by rearing larvae from birth on WT Arabidopsis or three Arabidopsis glucosinolate mutant lines (AGKO, IGKO, and GKO lines). We first exposed 60 plants of each of the four genotypes to S. flava females. The main purpose of this was simply to allow eggs to hatch in plants for the larval performance and transcriptional profiling experiments. However, we used it as an opportunity to test whether female flies varied in their feeding or oviposition preferences across the plant lines. We regressed egg number on stipple number and stipple number explained 54%–66% of the variation in egg number across the four host plant lines (supplementary fig. S2, Supplementary Material online). Although the slopes of these regression lines did not differ significantly among host plant lines (ANCOVA, F = 0.8844, P = 0.45), the intercepts were significantly lower for GKO (b = 1.19, t = −2.632, P = 0.0091) and AGKO (b = −1.66, t = −2.656, P = 0.0084) plants, indicating that these genotypes, which both are lacking aliphatic glucosinolates, were more heavily attacked by female flies than WT or IGKO plants (supplementary fig. S2, Supplementary Material online).
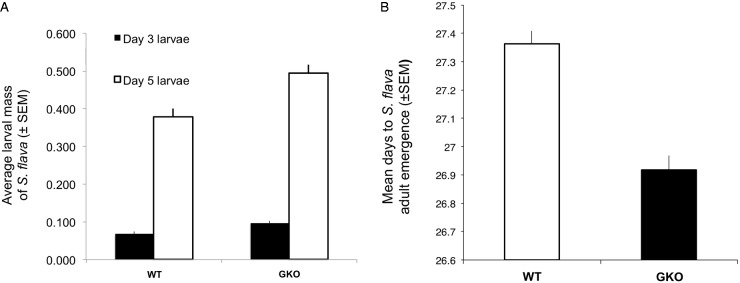
Using these same plants, 3 days after egg hatch, we extracted 311 larvae from WT (N = 18 plants), 349 larvae from AGKO plants (N = 18 plants), 285 larvae from IGKO plants (N = 18 plants), and 246 larvae from GKO (N = 18 plants) plants and recorded fresh larval mass. Five days after egg hatch, we extracted 351 larvae from WT (N = 36 plants), 364 larvae from AGKO plants (N = 36 plants), 319 larvae from IGKO plants (N = 33 plants), and 280 larvae from GKO (N = 34 plants) plants and recorded fresh larval mass as above. Results from the ANCOVA across all four host plant genotypes are presented in supplementary figure S3, Supplementary Material online. The number of larvae per plant was not a significant covariate with plant genotype (ANCOVA, plant genotype x number of larvae per plant, F1,64 = 0.777, P = 0.51). In an additive model, total number of larvae in each plant individual was not a significant factor on its own across genotypes (plant genotype + number of larvae per plant, day 3: F1,67 = 2.365, P = 0.129; day 5: F1,135 = 0.842, P = 0.36). However, plant genotype alone remained a significant factor in explaining average larval mass (F6,67 = 6.767, P = 0.0005; day 5: F3,135 = 7.462, P = 0.0001). In addition, we found no significant (α = 0.05) correlation between total number of larvae per plant with mass per larva using individual linear models for each plant line separately (supplementary fig. S3, Supplementary Material online). Post hoc tests showed that average masses of larvae reared on GKO plants were significantly larger than larvae reared on WT (fig. 3A) and AGKO plants (supplementary fig. S3, Supplementary Material online), 3 days and 5 days post–egg hatching. Similarly, egg-to-adult development time was more rapid in S. flava reared on GKO plants than in WT plants. Cohorts of adult flies emerged from GKO plants a half day earlier than flies from WT plants (P < 0.05, two-sided t-test) (fig. 3B).
Fig. 3.—
The absence of aliphatic and indolic glucosinolate biosynthesis pathways in Arabidopsis increases weight gain and decreases development time in S. flava. (A) Larvae reared from eclosion in wild-type (WT) (Col-0) or quadruple glucosinolate knockout (GKO) (cyp79b2cyp79b3myb28myb29) Arabidopsis lines that were harvested at 3 and 5 days posteclosion weighed significantly more when reared on the glucosinolate knockout. (B) S. flava reared from eggs in GKO Arabidopsis plants develop more rapidly than those reared on WT plants.
Secondary plant compounds such as glucosinolates require amino acid and carbon precursors and are potentially costly to produce (Heil 2002). Although the GKO Arabidopsis plants are phenotypically nearly identical to WT to human observers, it is possible that the leaves of GKO plants, blocked from synthesizing glucosinolates, may retain a liberated pool of nutritive precursors such as sugars and amino acids. The observed increase in S. flava growth rate in GKO relative to WT plants could be explained by a buildup of these plant primary metabolites. To test this possibility, we profiled primary metabolites in above ground tissues of 4-week-old plants of each genotype (GKO and WT, N = 5 plants each) by fractionation and gas chromatography–mass spectrometry (GC-MS). None of the 7,893 major ion features (e.g., sugars or amino acids) detected in this survey were significantly higher (P < 0.01) in GKO than WT plants (supplementary table S1, Supplementary Material online). Similarly, only five compounds with best matches to the following sugars were enriched in WT plants relative to GKO: arabinofuranose, galactofuranose, turanose, 1,6-anhydro-ß-d-glucose, and ß-dl-arabinopyranose (raw peak areas for each feature for all samples given in supplementary table S1, Supplementary Material online).
The GKO mutant we used is also deficient in an indole alkaloid phytoalexin called camalexin (3-thiazol-2'-yl-indole) (Zhao and Last 1996). Camalexin is synthesized first from tryptophan, which is converted to indole acetaldoxime by CYP79B2 and CYP79B3 (Glawischnig et al. 2004). Because the GKO is null for cyp79b2 and cyp79b3, we cannot exclude the hypothesis that S. flava performance and transcriptional phenotypes tested on the GKO mutant are due in part to the absence of camalexins. Camalexins have antimicrobial properties and could potentially have antiherbivore propteries (Rogers et al. 1996).
Sequencing and Assembly of the S. flava Transcriptome
The results described earlier suggest that glucosinolates are an important defense against S. flava and might have been a colonization barrier during or after the evolution of herbivory in the lineage. We reasoned that S. flava genes that respond transcriptionally to feeding on glucosinolate-bearing plant tissues were likely to be involved in the evolutionary transition to herbivory. We therefore used next-generation sequencing (RNA-seq) to analyze the transcriptional responses of S. flava flies reared on WT or GKO plants. We chose first to sequence a cDNA library from an RNA pool derived from mixed life stages (eggs, larvae, pupae, and adults) of an iso-female colony of S. flava using 454 Titanium FLX chemistry. We combined this assembly with reads from four Illumina GA II RNA-seq reads of 5-day-old S. flava larvae reared on either WT or GKO plants that are described later, to maximize our coverage. This resulted in a total of 104,118 preliminary contigs with an N50 of 289 (supplementary fig. S7, Supplementary Material online).
Before further analysis, we screened these 104,118 contigs for transposable elements, simple sequence repeats, and low-complexity regions using RepeatMasker with the default options and a Drosophila-specific library (RepBase: repeatmaskerlibraries-20090604.tar.gz). We removed from further consideration 8,819 contigs with either evidence for TE contamination or with greater than 10% of their sequence consisting of simple sequence repeats or low complexity regions, leaving us with a total of 95,299 contigs. These form the basis of all our subsequent analysis and are referred to as the S. flava contig set. We then used coding sequences from D. grimshawi (the closest fully sequenced sister taxon to S. flava) as scaffolds to assemble contigs using blastx (Crawford et al. 2010). By scaffolding contigs and merging potential S. flava paralogs identified via all-against-all blastn searches, we collapsed the original 95,299 contigs to 36,813 or 48,872 (depending on homology cutoffs; see supplemental materials, Supplementary Material online) assembled transcripts. These transcripts were then filtered to remove short contigs and those with weak or complex Blast hits, resulting in a total of 16,476 high-confidence transcript models, which served as the transcriptome against which the short reads from Illumina sequencing of the 5-day-old larvae from WT and GKO plants were mapped to quantify differential regulation of S. flava genes.
Transcriptional Analysis of the S. flava Response to Glucosinolates
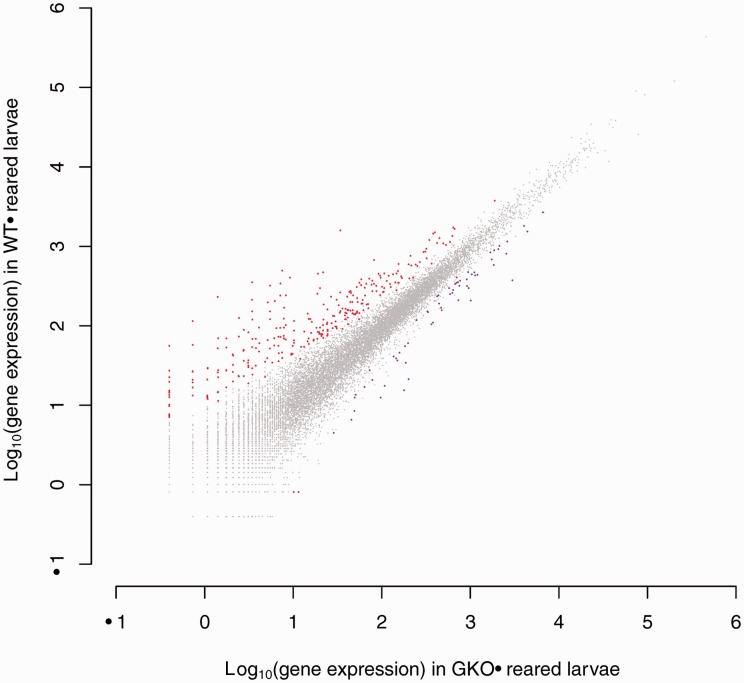
To measure differential expression between the transcriptomes of flies raised on Arabidopsis plants with and without glucosinolates, we collected 5-day-old larvae from the performance study, which were reared in WT and GKO plants. Using two pools of RNA for each treatment, we conducted an RNA-seq experiment using Illumina 75 bp sequencing. We identified a total of 341 transcripts that were differentially expressed between 5-day-old S. flava larvae reared on WT and GKO Arabidopsis (fig. 4). Of these 341 S. flava transcripts, 278 (82%) were significantly upregulated (induced) and 63 (18%) were significantly downregulated (repressed) in larvae reared on WT relative to GKO plants. Glucosinolates in the diet of S. flava thus appear to induce many more transcripts than they repress. We found one-to-one D. melanogaster orthologs for 121 S. flava transcripts: 105 of 278 induced S. flava transcripts and 16 of 63 repressed S. flava transcripts. Gene ontology (GO) categories for 76 of these 121 differentially regulated S. flava transcripts with homologous D. melanogaster genes could be inferred in AmiGO (Ashburner et al. 2000), and there was an enrichment of GO categories related to hemolymph coagulation (biological process), plasma membrane (cellular component), and structure constituent of the cuticle/chitin (molecular function) (table 2).
Fig. 4.—
Scatterplot showing differentially regulated transcripts from S. flava 5-day-old larvae raised on WT versus GKO plants. Transcripts that were significantly induced by glucosinolates in the diet of S. flava are in red, and those repressed by glucosinolates in the diet are in purple. Two RNA pools (biological replicates) for each treatment were each sequenced on single Illumina lanes (four lanes total). These sequences were mapped back to an assembled transcriptome that was scaffolded on the D. grimshawi genome. A total of 341 transcripts were differentially regulated. Larvae used for this analysis were obtained from those used in the weight-gain analysis presented in figure 3.
Table 2.
Selected Gene Ontology Enrichment for 76 of 121 Differentially Regulated Scaptomyza flava Transcripts with Homologs in Drosophila melanogaster
| GO Category | GO Term | P Value | Sample Frequency | Background Frequency | Genes |
|---|---|---|---|---|---|
| Biological process | 0050817, coagulation, metamorphosis | 2.17e-03 | 3/76 | 6/13,178 | Muc68Ca, Hml, fon |
| 0010171, body morphogenesis | 4.11e-03 | 4/76 | 22/13,178 | TwdlR, TwsdlS, TwdlW, TwdlBeta | |
| Cellular component | 0031226, intrinsic to plasma membrane | 1.95e-03 | 9/76 | 211/13,178 | sas, Osi19, Osi6, Osi7, Osi18, Osi15, Osi9, Osi2, Osi20 |
| Molecular function | 0042302, structural constituent of cuticle | 3.89e-07 | 11/76 | 146/13,178 | Cpr72Ec, CG7548, TwdlR, TwdlS, CG8543, TwldW, CG8927, CG8541, Cpr64Ab, dyl, TwldBeta |
Stress-Related Genes Are Regulated by Glucosinolates in S. flava
We evaluated the data set of 121 differentially regulated S. flava transcripts with D. melanogaster homologs in two ways. First, we compared these transcripts with the set of D. melanogaster genes activated or repressed by exposure to various stressors, including aging (Landis et al. 2004), the dietary toxin phenobarbital (Misra et al. 2011), heat (Sorensen et al. 2005), oxidation (Landis et al. 2004), and starvation (Harbison et al. 2005). Two genes induced by at least one stressor in D. melanogaster are homologous to S. flava transcripts induced by the presence of glucosinolates in the diet (table 3). This analysis suggests that there is at least some overlap between genes modulated by known physiological stressors in D. melanogaster and by the presence/absence of glucosinolates in larval host plants of S. flava.
Table 3.
Signature Transcripts Differentially Regulated by the Presence of Dietary Glucosinolates in Scaptomyza flava Whose Homologs are Differentially Regulated in the Same Direction by Environmental Stressors in Drosophila melanogaster and/or Those That Were Not Correlated with Development in Larvae Reared in GKO versus WT plants (developmental expression changes explain <50% of the log2 fold change)
| S. flava Assembled Transcript Region | Log2 Fold Change in Expression in S. flava Reared on WT vs. GKO Plants | D. melanogaster Homolog | FlyBase Potential Molecular Function (MF) and Biological Process (BP) | Stressors in D. melanogaster Causing Similar Changes |
|---|---|---|---|---|
| SF10967 | Induced | Ance-3 | MF, peptidyl-dipeptidase; BP, proteolysis | Age |
| SF777 | Induced | Thor | MF, translational regulator; BP, response to starvation; immune response; determination of adult lifespan; response to oxidative stress; regulation of cell growth; triglyceride metabolic process; negative regulation of cell size; response to stress; antibacterial humoral response; regulation of mitochondrial translation | Starvation, phenobarbital, oxidative stress |
| SF4724 | Induced | Muc68Ca | MF, unknown; BP, blood coagulation | N/A |
| SF2958 | Induced | Gie | MF, GTP binding; BP, sister chromatid segregation | N/A |
| SF1034 | Induced | Hml | MF, protein homodimerization; BP, hemolymph coagulation, hemostasis, wound healing | N/A |
| SF2502 | Induced | CG7715 | MF, chitin binding; BP, chitin metabolic process | N/A |
| SF8380 | Induced | CG5830 | MF, phosphatase; BP, unknown | N/A |
| SF8519 | Induced | DnaJ-1 | MF, heat shock protein binding, unfolded protein binding; BP, response to heat | N/A |
| SF693 | Induced | Hsc70-4 | MF, chaperone binding; BP:RNA interference, axon guidance, neurotransmitter secretion, embryonic development via the syncytial blastoderm, nervous system development, axonal fasciculation, vesicle-mediated transport | N/A |
| SF11108 | Induced | Rpp20 | MF, protein binding; BP, unknown | N/A |
| SF12295 | Induced | Mur89F | MF, chitin binding; BP, chitin metabolic process | N/A |
| SF10011 | Repressed | hoip | MF, mRNA binding; BP, nervous system development | N/A |
| SF2851 | Repressed | shep | MF, mRNA binding; BP, gravitaxis | N/A |
| SF12099 | Repressed | CoVa | MF, cytochrome-c oxidase, BP:positive regulation of cell cycle | Starvation, age |
| SF2493 | Repressed | eIF3-S10 | MF, translation initiation factor activity; BP, mitotic spindle elongation, mitotic spindle organization | N/A |
| SF3615 | Repressed | CG42336 | MF, unknown; BP, unknown | Starvation, age |
| SF271 | Repressed | dod | MF, peptidyl-prolyl cis–trans isomerase; BP, epidermal growth factor receptor signaling pathway | N/A |
| SF13113 | Repressed | CG7181 | MF, cytochrome-c oxidase; BP, mitochondrial electron transport, cytochrome c to oxygen | Starvation, age |
| SF3948 | Repressed | Cpr72Ec | MF, structural constituent of chitin-based cuticle; BP, unknown | N/A |
| SF1424 | Repressed | CG4169 | MF, ubiquinol-cytochrome-c reductase; BP, mitochondrial electron transport, ubiquinol to cytochrome c | Starvation, age |
| SF2735 | Repressed | DNaseII | MF, deoxyribonuclease II; BP, nurse cell apoptosis | Starvation |
The S. flava performance and growth rate experiments show that flies reared in WT plants are developmentally delayed relative to those reared on GKO plants (fig. 3). Thus, developmentally regulated genes that do not respond to dietary glucosinolates directly may confound gene expression differences between larvae reared on GKO or WT plants. To compensate for this potential confounding factor, we excluded S. flava transcripts from further analysis if developmental expression differences in the D. melanogaster homolog between second and early third instar larvae (Graveley et al. 2011), corresponding to possible stages at which larvae were harvested, explained >50% of the log2 fold expression difference in S. flava larvae reared on WT versus GKO plants. We found 11 transcripts induced by glucosinolate consumption and 10 transcripts repressed by glucosinolate consumption in S. flava that passed this extremely conservative test (table 3); for these 21 transcripts, developmental differences between larvae are unlikely to explain the transcriptional differences we observed. Of the 11 induced loci, two were among the four found to be induced by other stressors in D. melanogaster (table 3), and of the 10 repressed loci, five were found to be repressed by other stressors in D. melanogaster. This overlap suggests that the observed correlation between the transcriptional response to stress and the transcriptional response to dietary glucosinolates is not an artifact of developmental differences between larvae in the two samples.
Metabolic, tissue remodeling, and wound-associated genes were among the general classes of the 21 genes affected by glucosinolate uptake in S. flava (table 3). Thus, a set of stress-related genes appear to be induced by glucosinolates in the diet of even specialized insects that have adapted to grow in the presence of glucosinolates.
Evolutionary History of Genes Differentially Regulated by Glucosinolates
The assembled transcriptome allowed us to study the evolutionary history of glucosinolate-regulated S. flava transcripts for which we could find homologs in the other Drosophila species. We tested whether these transcripts exhibit more rapid rates of evolution than the bulk of the transcriptome, as found for immunity and stress-related genes in D. melanogaster (Sackton et al. 2007). We used alignments of all single copy orthologs in the S. flava transcriptome and D. grimshawi, D. virilis, and D. mojavensis genomes, to conduct a foreground–background analysis in PAML (Yang 1998, 2007) to estimate ω (dN/dS, the rate of nonsynonymous substitutions per nonsynonymous site/synonymous substitutions per synonymous site). Using our drosophilid species tree (fig. 1), we examined ω on the branch leading to S. flava (foreground) and across the rest of the subgenus Drosophila (background). With only three evolutionarily proximate taxa in the subgenus Drosophila, we lacked sufficient power to test for positive selection (a subset of codons with dN/dS>1) in the branch leading to S. flava.
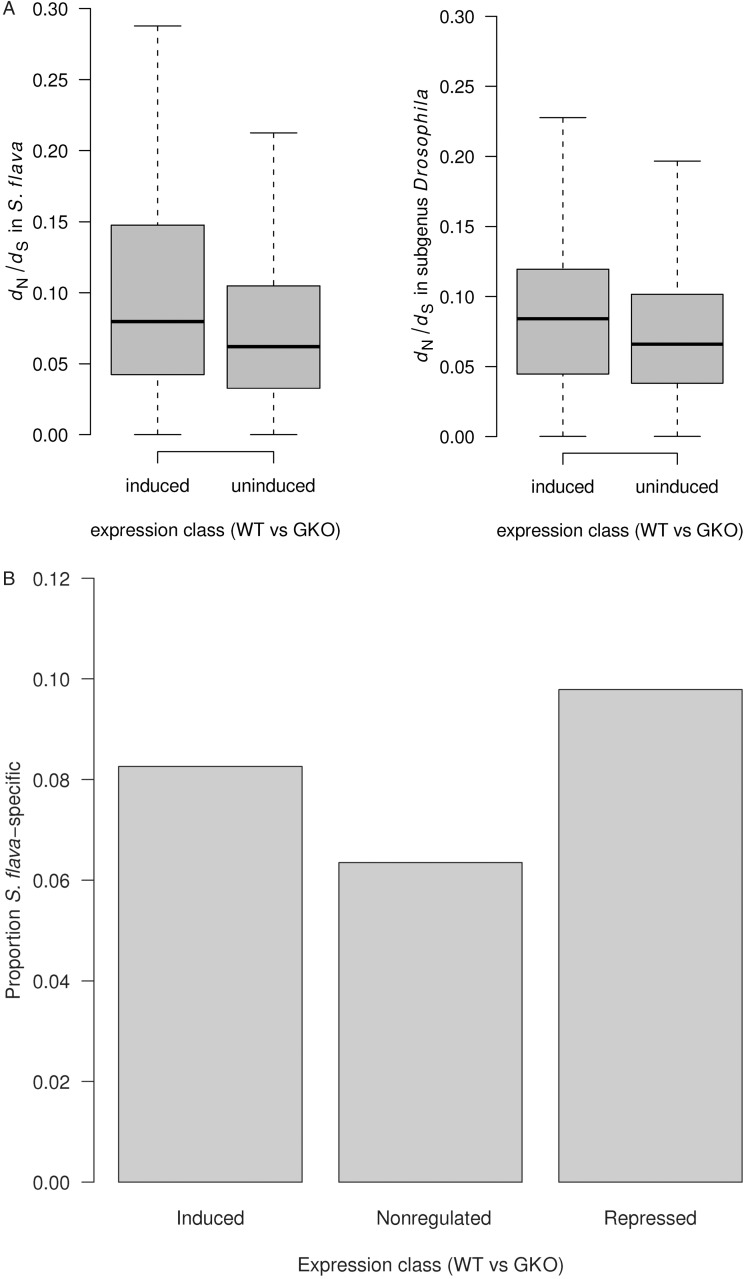
Transcripts that were more abundant in flies that fed on WT plants (those with glucosinolates; N = 62 transcripts) had a significantly higher ω value in the S. flava lineage than transcripts whose expression was unaffected by glucosinolates (N = 4,711) (P = 0.036, Mann–Whitney U test) (fig. 5A). Interestingly, this pattern is not specific to S. flava, as the induced transcripts also have significantly elevated ω values in the background lineages included from the subgenus Drosophila (D. grimshawi, D. mojavensis, and D. virilis) compared with the unregulated transcripts (P = 0.043, Mann–Whitney U test). Accelerated evolution of genes induced by glucosinolates in S. flava is consistent with other studies showing that genes involved in stress responses tend to evolve rapidly (Li et al. 2003, 2004; Clark et al. 2007; Low et al. 2007; Sackton et al. 2007; Thomas 2007; Matzkin 2008).
Fig. 5.—
(A) Results of dN/dS analysis in PAML for differentially regulated genes in S. flava with orthologs in other Drosophila species. Genes induced by dietary glucosinolates in S. flava evolve more rapidly than genes unaffected in expression by glucosinolates. Median dN/dS values are significantly higher in loci induced by dietary glucosinolates in S. flava compared with loci not induced by glucosinolates. (B) Results of comparison of the proportion of S. flava contigs lacking identifiable homologs in each expression class. The difference among classes is significant by a χ2 test (χ2 = 12.8346, df = 2, P = 0.00163). Candidate S. flava–specific transcripts are significantly more abundant among transcripts induced or repressed by glucosinolates in the diet of S. flava versus those that are uninduced by glucosinolates in the diet of S. flava.
Despite a concerted search for conserved domains on GenBank, we were able to identify D. melanogaster homologs of only 36% of genes (121 of 341) that were differentially regulated by glucosinolates in the diet of S. flava. More generally, a substantial number (5,967) of the 16,476 transcripts we identified in S. flava have no identifiable homologs in other species (supplementary table S6, Supplementary Material online). To provide reliable estimates of the extent of S. flava-specific transcripts, we therefore focused on an analysis of unassembled contigs, rather than scaffolds, because by definition scaffolds can only be assembled for transcripts with a D. grimshawi homolog. We used conservative criteria to define S. flava-specific contigs, requiring them to be at least 200 bp in length and to fail to produce a significant (E < 1e−10) blast hit against both the NCBI nonredundant protein database (nr) and Drosophila-specific databases. Using these criteria, 6,036 of 79,947 contigs were S. flava specific (7.55%). We then compared the proportion of contigs in the induced, repressed, and nonregulated expression categories that have no known homologs outside of S. flava (fig. 5B). The set of contigs that were induced or repressed in 5-day-old larvae reared on WT versus GKO plants had more candidate S. flava-specific contigs than those not regulated by the presence of glucosinolates (χ2 = 12.8346, df = 2, P < 0.01). This suggests that adaptation to glucosinolates in S. flava potentially involved novel genes. A preliminary blast against newly available genomic sequencing reads from S. flava confirms that they are of fly origin (see supplementary materials and methods, Supplementary Material online). A total of 99.3% (149/150) of the S. flava-specific transcripts that were differentially regulated by glucosinolate consumption were represented in the preliminary genome assembly (blastn, E-value cutoff: 1e−10).
Discussion
Herbivory (leaf mining) in the drosophilid fly genus Scaptomyza likely evolved relatively recently (6–16 MYBP) compared to the evolution of herbivory in the dominant orders of herbivorous insects. Because of this relatively recent acquisition of a phytophagous life history and the many genomic tools available for drosophilids and Arabidopsis, S. flava–Arabidopsis interactions represent an attractive and promising model system to study the evolutionary, ecological, and molecular aspects of genome adaptations that occur during the transition to herbivory in insects.
We primarily focused on the interactions between S. flava and glucosinolates, the major secondary compounds in their most typical host plants. Arabidopsis plants activate the indolic glucosinolate biosynthetic pathway in response to feeding by S. flava larvae, and S. flava, which is mostly restricted in host range to mustard plants (Martin 2004), is at least partially susceptible to glucosinolates synthesized by Arabidopsis as evidenced by reduced growth rate on WT versus GKO plants. Interestingly, S. flava larvae reared on WT plants were not significantly different in average mass from those reared on AGKO plants. AGKO plants still have indolic glucosinolates, and this suggests an important role for indolic glucosinolates or indole-derived camalexins in mediating quantitative resistance against S. flava. Rapid development time may be an advantage in leaf-mining species, which are heavily attacked by parasitoid wasps throughout their development (Connor and Taverner 1997; Whiteman et al. 2011). Because S. flava feeds mostly, but not exclusively, on plants in the order Brassicales, the finding of reduced performance of S. flava on WT versus GKO plants is not surprising. Oligophagous herbivores with broader host ranges, such as S. flava, are thought to be less efficient at coping with plant secondary compounds than are specialist species that feed on a more restricted range of hosts (Li et al. 2004). Future studies should dissect the independent role of camalexins and indolic glucosinolates in medating resistance against S. flava.
The assembly of an S. flava transcriptome allowed us to identify 341 S. flava genes that were differentially regulated by glucosinolates, most of which were induced rather than repressed. Only a limited number of transcriptional profiling studies have been carried out on herbivorous insects, including those in which larvae feed on plants with disabled defense pathways (Govind et al. 2010; Alon et al. 2011) or on different plant species with alternative toxins (Grbic et al. 2011). In each of these cases, hundreds of genes are differentially regulated by the presence of plant secondary compounds, a pattern similar to the one found here. However, in the case of S. flava, the benefit of being able to compare the transcriptome to the exceptionally well curated and annotated D. melanogaster genome greatly facilitated assignment of putative functions for differentially expressed genes, as well as the discovery of newly evolved loci.
The most prominent functional category identified was for stress-related genes. The fact that stress-related genes are regulated by glucosinolates in S. flava is intriguing because most specialist (monophagous) and oligophagous herbivores are thought to have evolved efficient and highly specialized detoxification systems to minimize physiological impacts of glucosinolate consumption. For example, Pieris spp. butterfly larvae do not encounter toxic glucosinolate breakdown products, such as isothiocyanates, and instead have evolved ways of preventing their formation (Wittstock et al. 2004).
We found that the S. flava homolog of the central D. melanogaster translational regulator Thor is particularly interesting because it is induced by bacterial infection, phenobarbital, oxidation stress, and starvation in D. melanogaster (Bernal and Kimbrell 2000; Landis et al. 2004; Harbison et al. 2005; Misra et al. 2011). Thor also serves as a “metabolic brake” during stressful conditions in D. melanogaster (Teleman et al. 2005). A homolog of Thor, 4E-BP1, is induced by phenethyl isothiocyanate in human cancer cell lines suggesting that there is conservation in this response to isothiocyanates across metazoans (Hu et al. 2007). Other differentially regulated S. flava genes with D. melanogaster homologs include the Tweedle genes, a large group of genes involved in formation of the insect cuticle (Cornman 2009). Isothiocyanates could potentially interact with the cuticle (and chitin) directly (McCormick et al. 1980), resulting in regulation of genes involved in cuticle and peritrophic membrane formation (Whiteman et al. 2011) and maintenance due to the presence of isothiocyanates in leaves that are encountered by endophagous feeders such as S. flava (van Ommen Kloeke et al. 2012). Genes involved in blood coagulation were also among those found to be differentially regulated by isothiocyantes and this finding is consistent with effects of dietary isothiocyanates on animals from other taxa. For example, dietary isothiocyanates in rats increase the rate of blood coagulation (Idris and Ahmad 1975; Shan et al. 2010).
Furthermore, we found that transcripts induced or repressed in larvae feeding on WT versus GKO plants are less evolutionarily conserved than transcripts that were not differentially regulated. We also uncovered a greater proportion of transcripts with no known homologs in the differentially regulated set, which could represent novel genes in this lineage or in the subgenus Drosophila. Taken together, our data indicate that the adaptation to mustard oil-bearing plants, and potentially herbivory in the lineage Scaptomyza, involved elaboration of pre-existing loci, potential exaptation of existing, rapidly evolving, stress-related genes, and the emergence of entirely new genes.
The functional basis of adaptation to glucosinolates in the diets of herbivores has been investigated in great detail in several lineages of insect, including the lepidopterans Pieris rapae (Wheat et al. 2007) and Plutella xylostella (Ratzka et al. 2002), the orthopteran Schistocerca gregaria (Falk and Gershenzon 2007), the sawfly Athalia rosae (Opitz et al. 2011), and the aphid Brevicoryne brassicae (Jones et al. 2001). In each of these species, single enzymes were identified as essential for detoxification or sequestration of glucosinolate breakdown products that appear to allow for minimal contact with isothiocyanates (Muller et al. 2010). In contrast, the identities of the genes actually involved in detoxifying glucosinolates in S. flava remain unknown. Our results suggest that S. flava may be directly encountering isothiocyanates given the significant growth rate reduction when cultured in WT versus GKO plants and the fact that hundreds of transcripts are differentially regulated by the presence or absence of glucosinolates in the diet of S. flava. Hundreds of genes are also differentially regulated by isothiocyanates in experiments where humans and rats are given glucosinolates or isothiocyanates (Hu et al. 2004; Bhamre et al. 2009). It is unknown whether other insects relatively specialized on Brassicales plants exhibit similar patterns to S. flava or whether their detoxification mechanisms are so efficient that gene expression differences between those that feed on plants with and without glucosinolates are minimal (Muller et al. 2010).
Transcriptomic approaches in other insect taxa such as D. melanogaster would likely yield valuable insights into common pathways influenced by glucosinolates in insects and how these molecules affect insect physiology more generally (Li et al. 2009). We did not detect transcriptional differences in S. flava homologs of most loci involved in induction of the Phase I or Phase II enzyme systems, which are candidate mustard oil detoxification proteins in humans and insects (Wadleigh and Yu 1988; Hu et al. 2004; Bhamre et al. 2009). This may be due to examination of one time point corresponding to 5 days posteclosion. For example, elevated levels of transcripts of some of these genes, such as glutathione S-transferases, which are involved in glucosinolate detoxification in some insects and in humans (Wadleigh and Yu 1988), are unlikely to remain elevated for such a long period of time (Tang and Tu 1995). Post-transcriptional regulation could explain increased GST levels in some insects treated with toxins rather than increases in mRNA levels per se (Tang and Tu 1995). Alternatively, structural changes could alter catalyitc efficiencies of genes encoding GSTs.
In addition to providing insight into the ecological pressures faced by Arabidopsis in nature, analysis of a leaf-mining parasite of Arabidopsis that is readily cultured in the laboratory and amenable to emerging genomic technologies may also have utility in agricultural research. Herbivorous insects cause major damage to crop plants around the world and are becoming increasingly resistant to pesticides. Understanding molecular mechanisms used in overcoming natural insecticidal toxins such as glucosinolates may provide direct or indirect routes to novel insecticides or breeding strategies. Because S. flava attacks and completes development in all Arabidopsis ecotypes (accessions) and mutants that have been screened (Whiteman et al. 2011), this system provides a platform for dissecting the genetic bases of interactions between plants and chewing herbivores.
Supplementary Material
Supplementary materials, figures S1–S7, and tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to the following for advice, or technical help, or both: Dr Amir Karger who facilitated the preliminary 454 transcriptome analyses; Dr Frans Tax and Dr Lisa Nagy who facilitated the GUS histochemical assays. Dr Doug Futuyma, Dr Jill Anderson, and anonymous reviewers provided helpful feedback on an earlier version of the manuscript. Fly specimens were collected at the Rocky Mountain Biological Laboratory and under permits from the U.S. Forest Service Rocky Mountain Region to N.K.W. N.K.W. and T.B.S. were each supported by Kirschstein National Research Service Awards (NRSA) from the National Institutes of Health (NIH) (F32AI069732 to N.K.W. and 1F32GM086950 to T.B.S.), N.K.W. by a University of Arizona Faculty Seed Grant and a set-up award from the University of Arizona, C.K. by NIH grant PO1 AI44220, F.M.A. by grants from the National Science Foundation (NSF) (grant MCB-0519898) and NIH (R37 GM48707; P30 DK040561), and N.E.P. by a grant from the NSF (grant SES-0750480). N.K.W., S.C.G., F.M.A., and N.E.P. were supported by a grant from the Harvard University’s Provost Fund for Interfaculty Collaboration and a grant from the Harvard University Science and Engineering Committee Seed Fund for Interdisciplinary Science. A.D.G. and P.T.H. were each supported by NSF IGERT Program in Comparative Genomics doctoral fellowships through the University of Arizona. R.T.L. was supported by an NIH Postdoctoral Excellence in Research and Teaching (PERT) fellowship through the Center for Insect Science at the University of Arizona (5K12GM000708-12).
Literature Cited
- Alon M, et al. Insights into the transcriptomics of polyphagy: Bemisia tabaci adaptability to phenylpropanoids involves coordinated expression of defense and metabolic genes. Insect Biochem Mol Biol. 2011;42:251–263. doi: 10.1016/j.ibmb.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, et al. EvoPipes.net: bioinformatic tools for ecological and evolutionary genomics. Evol Bioinform Online. 2010;6:143–149. doi: 10.4137/EBO.S5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci U S A. 2000;97:6019–6024. doi: 10.1073/pnas.100391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernays EA. Evolution of feeding behavior in insect herbivores. BioScience. 1998;48:35–44. [Google Scholar]
- Bernays EA, Chapman RF. Host plant selection by phytophagous insects. New York: Chapman and Hall; 1994. [Google Scholar]
- Bhamre S, Sahoo D, Tibshirani R, Dill DL, Brooks JD. Temporal changes in gene expression induced by sulforaphane in human prostate cancer cells. Prostate. 2009;69:181–190. doi: 10.1002/pros.20869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol I, Jackman SD, Nielsen CB, et al. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- Bock IR, Parsons PA. Adaptive radiation in the subgenus Scaptodrosophila of Australian Drosophila. Nature. 1975;258:602. doi: 10.1038/258602a0. [DOI] [PubMed] [Google Scholar]
- Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Ann Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chittenden FH. Some insects injurious to vegetable crops. 1902 Bulletin 33. Washington, DC: U.S. Department of Agriculture, Division of Entomology. [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Collinge SK, Louda SM. Scaptomyza nigrita Wheeler (Diptera: Drosophilidae), a leaf miner of the native crucifer, Cardamine cordifolia A. Gray (Bittercress) J Kans Entomol Soc. 1989;62:1–10. [Google Scholar]
- Connor EF, Taverner MP. The evolution and adaptive significance of the leaf-mining habitat. Oikos. 1997;79:6–25. [Google Scholar]
- Cornman RS. Molecular evolution of Drosophila cuticular protein genes. PLoS One. 2009;4:e8345. doi: 10.1371/journal.pone.0008345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JE, et al. De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS One. 2010;5:e14202. doi: 10.1371/journal.pone.0014202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I, Jones CD. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics. 2009;181:721–736. doi: 10.1534/genetics.108.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH. Butterflies and plants: a study in coevolution. Evolution. 1964;18:586–603. [Google Scholar]
- Falk KL, Gershenzon J. The desert locust, Schistocerca gregaria, detoxifies the glucosinolates of Schouwia purpurea by desulfation. J Chem Ecol. 2007;33:1542–1555. doi: 10.1007/s10886-007-9331-0. [DOI] [PubMed] [Google Scholar]
- Fan J, et al. Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science. 2011;331:1185–1188. doi: 10.1126/science.1199707. [DOI] [PubMed] [Google Scholar]
- Farrell BD. “Inordinate fondness” explained: why are there so many beetles? Science. 1998;281:555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- Frank MR, Fogleman JC. Involvement of cytochrome P450 in host-plant utilization by Sonoran Desert Drosophila. Proc Natl Acad Sci U S A. 1992;89:11998–12002. doi: 10.1073/pnas.89.24.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel G. The raison d'etre of secondary plant substances. Science. 1959;129:1466–1470. doi: 10.1126/science.129.3361.1466. [DOI] [PubMed] [Google Scholar]
- Gelbart WM, Emmert DB. FlyBase high throughput expression pattern data beta version. 2010 [cited 2012 Aug 1]. Available from: http://flybase.org/reports/FBrf0212041.html. [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind G, et al. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One. 2010;5:e8735. doi: 10.1371/journal.pone.0008735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic M, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackman W. On the genus Scaptomyza Hardy (Dipt., Drosophilidae) with descriptions of new species from various parts of the world. Acta Zool Fennica. 1959;97:3–73. [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, Mackay TF. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 2005;6:R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M. Ecological costs of induced resistance. Curr Opin Plant Biol. 2002;5:345–350. doi: 10.1016/s1369-5266(02)00267-4. [DOI] [PubMed] [Google Scholar]
- Hu R, et al. In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther. 2004;310:263–271. doi: 10.1124/jpet.103.064261. [DOI] [PubMed] [Google Scholar]
- Hu J, et al. Phenethyl isothiocyanate, a cancer chemopreventive constituent of cruciferous vegetables, inhibits cap-dependent translation by regulating the level and phosphorylation of 4E-BP1. Cancer Res. 2007;67:3569–3573. doi: 10.1158/0008-5472.CAN-07-0392. [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci U S A. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris R, Ahmad K. The effect of allylisothiocyanate and other antithyroid compounds on blood coagulation in rats. Biochem Pharmacol. 1975;24:2003–2005. doi: 10.1016/0006-2952(75)90393-7. [DOI] [PubMed] [Google Scholar]
- Jaenike J, Grimaldi DA, Sluder AE, Greenleaf AL. agr-Amanitin tolerance in mycophagous Drosophila. Science. 1983;221:165–167. doi: 10.1126/science.221.4606.165. [DOI] [PubMed] [Google Scholar]
- Jones AM, Bridges M, Bones AM, Cole R, Rossiter JT. Purification and characterisation of a non-plant myrosinase from the cabbage aphid Brevicoryne brassicae (L.) Insect Biochem Mol Biol. 2001;31:1–5. doi: 10.1016/s0965-1748(00)00157-0. [DOI] [PubMed] [Google Scholar]
- Jones CD. The genetic basis of larval resistance to a host plant toxin in Drosophila sechellia. Genet Res. 2001;78:225–233. doi: 10.1017/s0016672301005298. [DOI] [PubMed] [Google Scholar]
- Jones CD. The genetics of adaptation in Drosophila sechellia. Genetica. 2005;123:137–145. doi: 10.1007/s10709-004-2728-6. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kunitz M. Crystallization of a trypsin Inhibitor from soybean. Science. 1945;101:668–669. doi: 10.1126/science.101.2635.668. [DOI] [PubMed] [Google Scholar]
- Labandeira CC. Fossil history and evolutionary ecology of Diptera and their associations with plants. In: Weigmann B, Yeates D, editors. The evolutionary biology of flies. New York: Columbia University Press; 2003. pp. 217–273. [Google Scholar]
- Labandeira CC, Dilcher DL, Davis DR, Wagner DL. Ninety-seven million years of angiosperm-insect association: paleobiological insights into the meaning of coevolution. Proc Natl Acad Sci U S A. 1994;91:12278–12282. doi: 10.1073/pnas.91.25.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Li HM, et al. Transcriptional signatures in response to wheat germ agglutinin and starvation in Drosophila melanogaster larval midgut. Insect Mol Biol. 2009;18:21–31. doi: 10.1111/j.1365-2583.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- Li W, Schuler MA, Berenbaum MR. Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: specificity and substrate encounter rate. Proc Natl Acad Sci U S A. . 2003;100(2 Suppl):14593–14598. doi: 10.1073/pnas.1934643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Baudry J, Berenbaum MR, Schuler MA. Structural and functional divergence of insect CYP6B proteins: from specialist to generalist cytochrome P450. Proc Natl Acad Sci U S A. 2004;101:2939–2944. doi: 10.1073/pnas.0308691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WY, et al. Molecular evolution of glutathione S-transferases in the genus Drosophila. Genetics. 2007;177:1363–1375. doi: 10.1534/genetics.107.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnanou E, Malenke JR, Dearing MD. Expression of biotransformation genes in woodrat (Neotoma) herbivores on novel and ancestral diets: identification of candidate genes responsible for dietary shifts. Mol Ecol. 2009;18:2401–2414. doi: 10.1111/j.1365-294X.2009.04171.x. [DOI] [PubMed] [Google Scholar]
- Markow TA, O'Grady PM. Evolutionary genetics of reproductive behavior in Drosophila: connecting the dots. Annu Rev Genet. 2005;39:263–291. doi: 10.1146/annurev.genet.39.073003.112454. [DOI] [PubMed] [Google Scholar]
- Martin NA. History of an invader, Scaptomyza flava (Fallen, 1823) (Diptera: Drosophilidae) New Zealand J Zool. 2004;31:27–32. [Google Scholar]
- Matzkin LM. The molecular basis of host adaptation in cactophilic Drosophila: molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics. 2008;178:1073–1083. doi: 10.1534/genetics.107.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick CL, Lichatowich DK. Homogeneous solution reactions of cellulose, chitin, and other polysaccharides to produce controlled-activity pesticide systems. J Polym Sci. 1979;17:479–484. [Google Scholar]
- Millet YA, et al. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter C, Farrell BD, Wiegmann B. The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am Nat. 1988;132:107–128. [Google Scholar]
- Moran NA. A 48-million-year-old aphid–host plant association and complex life cycle: biogeographic evidence. Science. 1989;245:173–175. doi: 10.1126/science.245.4914.173. [DOI] [PubMed] [Google Scholar]
- Muller R, et al. Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. J Chem Ecol. 2010;36:905–913. doi: 10.1007/s10886-010-9825-z. [DOI] [PubMed] [Google Scholar]
- O'Grady P, Desalle R. Out of Hawaii: the origin and biogeography of the genus Scaptomyza (Diptera: Drosophilidae) Biol Lett. 2008;4:195–199. doi: 10.1098/rsbl.2007.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz SE, Mix A, Winde IB, Muller C. Desulfation followed by sulfation: metabolism of benzylglucosinolate in Athalia rosae (Hymenoptera: Tenthredinidae) Chembiochem. 2011;12:1252–1257. doi: 10.1002/cbic.201100053. [DOI] [PubMed] [Google Scholar]
- Ratzka A, Vogel H, Kliebenstein DJ, Mitchell-Olds T, Kroymann J. Disarming the mustard oil bomb. Proc Natl Acad Sci U S A. 2002;99:11223–11228. doi: 10.1073/pnas.172112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Glazebrook J, Ausubel FM. Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact. 1996;9:748–757. doi: 10.1094/mpmi-9-0748. [DOI] [PubMed] [Google Scholar]
- Sackton TB, et al. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Shan Y, et al. Protective effect of sulforaphane on human vascular endothelial cells against lipopolysaccharide-induced inflammatory damage. Cardiovasc Toxicol. 2010;10:139–145. doi: 10.1007/s12012-010-9072-0. [DOI] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen JG, Nielsen MM, Kruhoffer M, Justesen J, Loeschcke V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones. 2005;10:312–328. doi: 10.1379/CSC-128R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood TRE. The insect/plant relationship-an evolutionary perspective. Symp R Entomol Soc London. 1973;6:3–30. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tang AH, Tu C-PD. Pentobarbital-induced changes in Drosophila glutathione S-transferase D21 mRNA stability. J Biol Chem. 1995;270:13819–13825. doi: 10.1074/jbc.270.23.13819. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen Y-W, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor T, Gershenzon J. Herbivore induction of the glucsoinolate-myrosinase defense system: major trends, biochemical bases and ecological significance. Phytochem Rev. 2009;8:149–170. [Google Scholar]
- Thomas JH. Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet. 2007;3:e67. doi: 10.1371/journal.pgen.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AL, Varala K, Newman TC, et al. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science. 2007;318:441–444. doi: 10.1126/science.1146647. [DOI] [PubMed] [Google Scholar]
- van Ommen Kloeke AEE, van Gestel CAM, Styrishave B, et al. Molecular and life-history effects of a natural toxin on herbivorous and non-target soil arthropods. Ecotoxicology. 2012;21:1084–1093. doi: 10.1007/s10646-012-0861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadleigh RW, Yu SJ. Detoxification of isothiocyanate allelochemicals by glutathione transferase in three lepidopterous species. J Chem Ecol. 1988;14:1279–1288. doi: 10.1007/BF01019352. [DOI] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley P. Lower cretaceous lepidoptera. Nature. 1977;266:526–526. [Google Scholar]
- Wheat CW, et al. The genetic basis of a plant-insect coevolutionary key innovation. Proc Natl Acad Sci U S A. 2007;104:20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, et al. Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol Ecol. 2011;20:995–1014. doi: 10.1111/j.1365-294X.2010.04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, et al. Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc Natl Acad Sci U S A. 2004;101:4859–4864. doi: 10.1073/pnas.0308007101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zhao J, Last RL. Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell. 1996;8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]