Abstract
Intracellular transport is an essential biological process that is highly conserved throughout the eukaryotic organisms. In fungi, adaptor proteins implicated in the endocytic cycle of endocytosis and exocytosis were found to be important for growth, differentiation, and/or virulence. For example, Saccharomyces cerevisiae Pan1 is an endocytic protein that regulates membrane trafficking, the actin cytoskeleton, and signaling. In Cryptococcus neoformans, a multi-modular endocytic protein, Cin1, was recently found to have pleiotropic functions in morphogenesis, endocytosis, exocytosis, and virulence. Interestingly, Cin1 is homologous to human intersectin ITSN1, but homologs of Cin1/ITSN1 were not found in ascomycetous S. cerevisiae and Candida albicans, or zygomycetous fungi. Moreover, an Eps15 protein homologous to S. cerevisiae Pan1/Ede1 and additional relevant protein homologs were identified in C. neoformans, suggesting the existence of either a distinct endocytic pathway mediated by Cin1 or pathways by either Cin1 or/and Pan1/Ede1 homologs. Whether and how the Cin1-mediated endocytic pathway represents a unique role in pathogenesis or reflects a redundancy of a transport apparatus remains an open and challenging question. This review discusses recent findings of endocytic adaptor proteins from pathogenic fungi and provides a perspective for novel endocytic machinery operating in C. neoformans. An understanding of intracellular trafficking mechanisms as they relate to pathogenesis will likely reveal the identity of novel antifungal targets.
Keywords: intersectin, endocytosis, exocytosis, fungal pathogenesis
Introduction to membrane trafficking
Membrane trafficking or intracellular transport is a cellular process by which membrane materials containing proteins, lipids, and other macromolecules are shuttled between endo-membrane compartments and the plasma membrane [1]. It fosters cellular functions such as the maintenance of cellular homeostasis and compartmental communications during nutrient uptake and signal perception, which is important for the survival of eukaryotic organisms. There are two major intracellular trafficking routes: endocytosis and exocytosis. Endocytosis consists of an inward flux of endocytic vesicles from the plasma membrane, whereas exocytosis is an outward flux of exocytic vesicles to the plasma membrane [2 – 6].
During endocytosis, plasma membrane proteins and lipids are internalized into vesicles and then delivered to early endosomes. They are subsequently sent to late endosomes and vacuoles for degradation, or, as in the case of certain protein receptors, may be recycled back to the plasma membrane [5]. Endocytosis is a complex molecular process involving clathrin and/or adaptor proteins, various protein and lipid kinases, phosphatases, and the actin cytoskeleton. In higher eukaryotic cells, clathrin-dependent endocytosis involves the formation of vesicles coated with clathrin, while clathrin-independent endocytosis involves several internalization pathways that use an alternative protein coat or do not require a protein coat for the formation of endocytic vesicles [5,7].
In exocytosis, the vesicles of early exocytic pathways can be formed similarly with participation of coat proteins complex I (COPI) and II (COPII), which determine vesicle specificity and target destinations [3]. COPI is involved in the trafficking of proteins within the Golgi compartment and retrograde trafficking back to the endoplasmic reticulum (ER), while COPII is involved in the trafficking of newly synthesized proteins from the ER to the Golgi apparatus [8]. The final stage of vesicle trafficking is the fusion of secretory vesicles to target plasma membrane, which is controlled by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) [3,9].
Altered membrane trafficking is thought to be the basis of several human diseases, such as Down syndrome and Alzheimer's disease, which provides the impetus for gaining an understanding of normal and abnormal cellular trafficking events. Fungi are fundamentally comparable to, though less complex than, multicellular higher eukaryotes with respect to the endomembrane system, which comprises the ER, the Golgi, endosomes, and lytic compartments [10]. Not surprisingly, many genetically identified endocytic proteins in S. cerevisiae have homologues in mammalian cells. Indeed, the study of membrane trafficking in S. cerevisiae is not only providing mechanistic insights into mammalian cellular trafficking events, but in other fungi as well. In filamentous and dimorphic fungi, either pathogenic to host plants or saprophytic, intracellular transport is required for polarity establishment, hyphal growth, and/or virulence. A wealth of information is available detailing the roles of specific endocytic components in these fungi, such as Aspergillus oryzae End4/Sla2, Fusarium graminearum End1 (a S. cerevisiae End3 homolog), and Ustilago maydis Yup1[11 – 13]. In addition, there are also excellent reviews discussing roles of endocytic transport in growth and pathogenicity of U. maydis [14,15]. Intracellular transport, particularly the exocytotic route, has an established role in virulence attributes in human fungal pathogens such as C. albicans and C. neoformans [16 – 26]. Despite this importance, we are just beginning to decipher the constituents of the endocytic machinery responsible for membrane trafficking in these pathogenic organisms.
In this review, we discuss recent discoveries in membrane trafficking of pathogenic fungi, specifically the endocytic adaptor proteins involved in both endocytosis and exocytosis. We emphasize recent findings from C. neoformans and compare them with a background of data from model yeast S. cerevisiae and pathogenic yeast C. albicans. One significant feature of the endocytic machinery in C. neoformans is that it differs from that of S. cerevisiae and C. albicans. The significance of this distinction in functions and whether it plays a role in virulence mechanisms of the fungus remains to be the subject of future investigations.
Intracellular transport and endocytic proteins in Saccharomyces cerevisiae
The budding yeast S. cerevisiae remains one of few models in fungi in which membrane trafficking, particularly endocytosis, has been examined. The availability of the lypophilic styryl dye N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (FM4-64) and seven transmembrane domain pheromone receptors, Ste2 and Ste3, have allowed identification of various mutants whose characterization provided a scheme for the endocytic pathway [27,28]. Many features of the pathway are surprisingly conserved in those of mammalian systems, including clathrin and Eps15 protein homologs that function in endocytosis and membrane trafficking [10,29,30]. In this pathway, clathrin assembly and adaptor proteins are required. Clathrin and Eps15 homolog Ede1 are recruited to the endocytic site along with additional proteins such as Ent1/Ent2, Yap1801/1802, and AP2 [31,32]. In addition, Las17/Bee1, an ortholog of the human Wiskott-Aldrich syndrome protein (WASP) that activates the Arp2/3 complex to promote actin assembly, Sla2, an actin-binding protein, and Pan1, a homolog of human Eps15 protein are also recruited. Following complex formation with Vrp1, a Las17-interacting WIP homolog, and Bzz1, a Las17-interacting synapsin homolog, actin assembly is initiated and class I myosin is recruited. Finally, the coat module is internalized and, following coat/actin disassembly, vesicles are released and fusion with early endosomes occurs [32,33].
Among proteins involved in coat protein assembly and vesicle formation and fusion, the adaptor protein Pan1 appears to play a prominent role. Pan1 is an essential protein forming the core of an endocytic complex, which is involved in both the internalization step of endocytosis and the organization of the actin cytoskeleton [34,35]. Pan1 contains two N-terminal Eps15 homology (EH) domains for binding with an Asn-Pro-Phe (NPF) motif containing Ent1/2 and Yap1801/2 proteins [35]. It also binds to End3, an EH domain-containing protein involved in the internalization step of endocytosis and is required for actin cytoskeleton organization [36]. It has been suggested that the central coiled-coil region mediates the formation of dimers or oligomers with either Pan1 itself or other coiled-coil domain-containing proteins [35,37]. The carboxyl-terminal region is proline rich and can bind with Src homolog 3 (SH3) and WW domain-containing proteins, such as Rsp5, an ubiquitin protein ligase required for endocytosis [38,39]. Also involved in endocytosis is Ede1, a protein containing three EH domains and genetically interacting with Pan1, End3, and Rsp5 [40]. Based on multiple proteins that bind to Pan1 and their function, it was surmised that Pan1 serves as an adaptor to regulate and coordinate the activities of the endocytic and actin cytoskeleton machinery, which include clathrin coat assembly (Yap1801/2 and Ede1 proteins), ubiquination (Rsp5), and actin regulation (End3) [35]. A graphic illustration of the Pan1-mediated endocytic pathway involved in intracellular trafficking of S. cerevisiae is shown in Fig. 1.
Fig. 1.

In Saccharomyces cerevisiae, Pan1 associates through its N-terminus with Sla1, which binds to End3. Pan1 also interacts with End3 through its EH domains to be recruited to cortical actin cytoskeleton. Pan1 interacts through EH domains with Ent1/2 and Yap1801/2, which bind to Clathrin, to regulate endocytosis. Additionally, Pan1 activates actin patch polymerization through binding of its C-terminal A domain to the Arp2/3 complex. Moreover, Pan1 associates through its proline-rich (PR) domain with type I Myosin Myo3/5 and SH3 domains, contributing to the late stage of endocytosis. Sla1 inhibits the function of Las17/Bee1, a Wiskott-Aldrich Syndrome Protein (WASP) homologue, which activates the Arp2/3 complex to initiate the branch filament actin assembly [32,34,35,104,105]. EH, Eps15 homology; α-helix, α-helical coiled-coil domain; A, acidic domain; PR, proline rich domain.
Secretory transport is linked to virulence of Candida albicans
Studies in S. cerevisiae have demonstrated the importance of intracellular transport in the growth and differentiation of lower eukaryotic organisms. Additionally, characterizations of Pan1 and Pan1-interacting proteins have revealed the complexity of the intracellular transport event in unicellular yeasts. As secretory properties in particular have the propensity to be linked to virulence, intracellular transport in medically important pathogenic fungi, such as C. albicans and C. neoformans, has been the focus for several recent studies.
C. albicans is a commensal and the most frequent fungal pathogen of humans causing infections of mucosal membranes and serious systemic diseases. The ability to cause the disease is attributable to both the ability to undergo a reversible morphogenic transition from yeast to hyphae and the ability to secrete lytic proteinases such as aspartic proteinases (see reviews in [41 – 44]). Both events are largely dependent on intracellular transport of macromolecules, but knowledge about the details of intracellular trafficking remains fragmented, particularly for the endocytic machinery that is involved in intracellular transport.
Nevertheless, a number of studies primarily focusing on the secretory pathway showed that vesicular proteins, constituents of the endosomal-sorting complexes required for transport (ESCRT) pathway, and proteins involved in actin cytoskeleton organization, which are largely conserved in those of S. cerevisiae, are important in the morphogenic transition, intracellular trafficking, and virulence of C. albicans ([45] and reviewed by [46]).
Endocytosis was first described for C. albicans in 1990, when Basrai and colleagues demonstrated the uptake of lucifer yellow (LY) by the fungus [47]. As with many other molecular systems, S. cerevisiae has continued to serve as the model from which analogous conclusions are derived. In a study of C. albicans ORFs whose homologs in S. cerevisiae are involved in endocytosis, several genes encoding endocytic coat components, such as Ede1 and the adaptor protein Pan1, were found [48]. An actin-binding protein, Abp1, and a marker for actin polymerization in cortical patches, Bzz1, that are involved in endocytosis of C. albicans were also found [48 – 50]. C. albicans Ede1 is similar to yeast Ede1 by containing three EH domains [48] and End3 is also similar to the yeast homolog that binds to Pan1 [51]. Interestingly, whether Abp1 encodes an essential function or not remains uncertain, as an ede1 mutant was viable [48,50]. In accordance with studies in yeast, mutations of both alleles of Pan1 were not possible for C. albicans [48].
Also worth noting are the Sla2 and Sla1 proteins of C. albicans. S. cerevisiae Sla2 is a Pan1-interacting protein that negatively regulates Pan1 activity [52]. The coiled-coil region of Sla2 binds to Pan1 and inhibits its ability to activate Arp2/3-mediated actin nucleation [52]. It is not known whether the role of Sla2 on Pan1 also exists in C. albicans, but a recent study showed a similar function for Sla2 in the regulation of cell cycle in both S. cerevisiae and C. albicans [53]. The SH3 domain containing Sla1 protein is a cytoskeleton-binding protein of S. cerevisiae and the yeast sla1 sla2 mutant is synthetically lethal [54]. A Sla1 homolog was among three proteins recently identified from C. albicans that contain the SH3 domain [55]. Consistently, a sla1 mutant was found to be defective in hyphal growth, cytokinesis, and vacuolar formation [55].
Endocytosis was found to have a role in the acquisition of hemoglobin-iron in C. albicans [16]. Various vacuolar sorting proteins, such as Vps28 and Vps32 that are part of the ESCRT complexes, small GTPases, such as Vps21 and Ypt72 are required for trafficking through the prevacuolar compartment (PVC) and vacuole biogenesis, vacuolar ATPase subunit Vma7, and bin-amphiphysin-rvs (BAR) domain proteins Rvs161 and Rvs167 are all involved in endocytosis, hyphal transition, and virulence [17,19,22,56]. Deletion of C. albicans Sur7, an S. cerevisiae Sur7 homolog that is found on eisosomes, affected plasma membrane organization, endocytosis, and normal growth of yeast and hyphae [57].
Based on these findings, it is reasonable to hypothesize that the endocytic machinery operating in C. albicans closely follows that of S. cerevisiae. The C. albicans endocytic protein components and actin-related proteins involved in intracellular traffic, morphogenic transition, and/or virulence, as well as their orthologs in S. cerevisiae are summarized in Table 1.
Table 1.
Endocytosis-related proteins in Saccharomyces cerevisiae, Candida albicans, and Cryptococcus neoformans.
| S. cerevisiae protein [reference] | Proposed function | C. albicans ortholog [reference] | C. neoformans ortholog [reference]* |
|---|---|---|---|
| Endocytic coat component | |||
| Chc1 [82] | Stabilization of membrane curvature | XP_710836 | XP_567407 |
| Clc1 [82] | Binding clathrin and recruiting other coat proteins | XP_722811 | XP_572797 |
| Ede1 [40] | Binding Ent proteins and ubiquitin | Cta3 [48] | XP_567624 |
| Ent1 [31, 32] | Binding clathrin, part of coat complex | XP_722494 | XP_570888 |
| Ent2 [31, 32] | Binding clathrin, part of coat complex | no homolog | |
| End3 [36] | Binding Pan1 and recruiting Sla1 | End3 [51] | no homolog |
| Pan1 [34, 35] | Binding Clc1, Yap180 and activating Arp2/3 | Pan1 [48] | Cin1 [limited similarity, 26] |
| Sla1 [32, 33] | Binding cargo and inhibiting Las17 | Sla1 [55] | XP_571337 |
| Sla2 [32, 33] | Coupling actin to vesicle coat | Sla2 [52] | XP_570853 |
| YAP1801 [31, 32] | Clathrin coat assembly, binding clathrin and Pan1 | XP_712638 | XP_572163 |
| YAP1802 [31, 32] | Clathrin coat assembly, binding clathrin and Pan1 | XP_774414 | |
| Actin related protein | |||
| Abp1 [83] | Binding actln to coat and recruiting regulatory kinases Ark1 and Prk1 | Abp [48] | same as XP_570077 |
| Arf3 [84] | Regulating PIP2 levels and actin | Arf3 [85] | XP_568186 |
| Arp2 [86] | Part of the machinery of branching filament actin | Arp2 [87] | XP_572870 |
| Arp3 [86] | Part of the machinery of branching filament actin | Arp3 [87] | XP_570501 |
| Bbc1 [88] | Inhibiting Las17 | Bbc1 [55] | XP_570166 |
| Bzz1 [89] | Binding to Las17 and myosin | Bzz1 [48] | XP_570077 |
| Cap1, Cap2 [90] | Funnelling monomer to growing actin filament ends | XP_717926, XP_722814 | XP_569038, XP_569217 |
| Cof1 [91] | Depolymerizing actin filaments | EEQ46087 | XP_569980 |
| Las17 [92] | Activating Arp2/3 complex to iniate branch actin filament | Wal1 [93] | Wsp1 [Shen et al., unpublished] |
| Myo3, Myo5 [94] | Arp2/3 activation, actin motor for invagination, and vesicle scission | Myo5 [95] | XP 568008 |
| Rvs161/Rvs167 [96] | Vesicle scission | Rvs161/Rvs167 [22] | XP_568976 |
| Sac6 [97] | Bundling and cross-linking actin | Sac6 | XP_567345 |
| Scp1 [98] | Bundling and cross-linking actin | Scp1 | no homolog |
| Vps1 [99] | Sla1 binding and actin organization | Vps1 [20] | XP_566870 |
| Vrp1 [100] | Regulation of type 1 myosins | Vrp1 [33] | no homolog |
| Ysc84 [101] | Binds to Las17 and increases rate of actin polymerization | EEQ45902 | XP_571248 |
| Other protein | |||
| Ark1 [102] | Kinase regulating disassembly of the endocytic coat | Ark1 [103] | XP_569322 |
| Prk1 [102] | Kinase regulating disassembly of the endocytic coat | no homolog |
All are putative, with the exception of Cin1 and Wsp1.
Exosomal transport is linked to virulence of Cryptococcus neoformans
C. neoformans is an encapsulated yeast belonging to a class of fungi named basidiomycetes, which is distinct from ascomycetous S. cerevisiae and C. albicans. Basidiomycetous fungi develop distinct dikaryotic mycelia or filaments with clamp connections and produce basidia and basidiospores as a result of genetic cross [58]. C. neoformans infects primarily immunocompromised individuals causing meningoencephalitis, which is fatal if left untreated (see the review in [59]). This fungus employs a multifaceted virulence mechanism involving the ability to produce an antiphagocytic polysaccharide capsule, the antioxidant melanin pigment, and to secrete phospholipase B and urease (see the reviews in [59,60]). Because of this, intracellular trafficking, particularly of those involving the secretory pathways, was the recent focus for a number of studies, leading to identification of protein components that suggests the existence of distinct, as well as conserved, intracellular transport mechanisms.
The production of the polysaccharide capsule is one of the primary virulence factors and a great array of references has illustrated its importance and likely mechanisms in the virulence of the fungus [61,62]. Components of the capsule materials such as glucuronoxylomannan (GXM) are synthesized in the endoplasmic reticulum system, similar to proteins, and are then transported through the post trans Golgi network (TGN) to the extracellular environment in the form of exosomes and vesicular bodies [63]. Lipids such as glucosylceramide and enzymes such as laccases are also found in the extracellular vesicles [64,65]. Additional studies have validated the presence of those vesicular bodies [66]. Moreover, a recent study provided genetic evidence indicating that the secretory transport of GXM-containing vesicles follows the conserved secretory pathways demonstrated in S. cerevisiae. In this study, the authors created a mutant strain of Sec4/Sav1 that resulted in failed secretion and accumulation of GXM-containing exosomal vesicles [67]. The finding of a conserved secretory pathway in C. neoformans was corroborated by studies of C. neoformans Sec6, a homolog for the component of the exocyst complex in S. cerevisiae, and vacuolar protein Vps41, which were also involved in regulation of exosomal transport of the vesicular bodies containing laccase, phospholipases and other proteins [24,25]. These studies demonstrated that secretory transport in C. neoformans is likely achieved through a conserved mechanism, which also contributes to the virulence traits of the fungus.
Cin1 is a novel endocytic adaptor protein
Studies in S. cerevisiae have illustrated that intracellular transport, being endocytic or exocytic, is comprised of highly organized events involving orchestrated functions of many proteins, including Pan1 and others that link the endocytic cycle to actin cytoskeleton and signaling [2,68]. A recent study of a novel multi-modular protein, Cin1, which is more homologous to human intersectin ITSN1 than Pan1, has revealed a distinct key component of the endocytic machinery in C. neoformans [26].
Intersectins are cytoplasmic membrane associated human proteins involved in membrane trafficking, actin cytoskeleton maintenance, and signaling. The long-splicing variant, ITSN1, is neuronal-specific and required for normal transport of synaptic vesicles. ITSN1 overexpression, as in the case of chromosome 21 trisomy, was linked to Alzheimer's disease and Down syndrome [69]. ITSN1 contain two N-terminal EH domains, a central coiled-coil region, five SH3 domains, a tandem RhoGEF (DH)-Plectrin homology (PH) domain motif, and a C-terminal C2 domain that binds calcium [70,71]. The N-terminal EH and central coiled-coil configurations are similar to Pan1 (E-value: 6.1e-26). The SH3 domains are found in signaling proteins including phospholipases, tyrosine kinases, and Ras GTPase-activating protein (GAP), Cdc24, and Cdc25 [72]. The DH domain is the structural component of guanine nucleotide exchange factor (GEF) that activates Rho/Rac/Cdc42-like small GTPases to control the actin dynamics, membrane trafficking and signaling [73 – 78], whereas the PH domain occurs in proteins involved in intracellular signaling or as constituents of the cytoskeleton [79 – 81].
Cryptococcal intersectin homolog, Cin1, was identified in a search for protein partners of Gib2, a Gβ-like/RACK protein homolog [26]. Similar to ITSN1, Cin1 contains an N-terminal EH domain, a coiled-coil region, two SH3 domains, and a C-terminal DH-PH domain. In contrast to ITSN1, Cin1 also has a unique WH2 domain, which is known in other systems to bind to actin monomers and to be involved in the regulation of the actin cytoskeleton [74,82 – 84], but lacks a C2 binding domain [26].
Cin1 encodes important functions as disruption of the CIN1 gene resulting in a wide array of phenotypic defects. The cin1 mutant exhibited reduced growth rate at ambient temperature and was nonviable at body temperature (37 °C). In addition, the mutant failed to respond to the presence of pheromones by the opposite mating type and to undergo genetic cross [26]. Cytological examination revealed that the cin1 mutant was unable to undergo normal cytokinesis separation thus appearing as clusters of multicellular structures, and the mutant was unable to endocytose FM4-64, suggesting a defect in endocytic transport. The defect in exocytosis was demonstrated by in vivo examination of a fluorescence-labeled laccase fusion protein, DsRed-Lac1 [26]. Importantly, the cin1 mutant failed to display common virulence traits such as capsule, melanin, and secretory proteins phospholipase B, urease, and laccase [26]. In a murine model of cryptococcosis, freshly prepared cin1 cells, which exhibited less severe multicellular structural defects, were infected via the nasal cavity of mice. The cin1 mutant was verified to be present inside the lung following inoculation, however, they were completely cleared by the host within 34 days (Whittington and Wang, unpublished result), indicating that the cin1 mutant is avirulent.
Cin1 defines novel endocytic machinery of Cryptococcus neoformans
Characterization of Cin1 reveals its functions in both endocytosis (FM4-64 uptake) and exocytosis (DsRed-Lac1 localization). This is highly analogous to functions of human ITSN1 proteins. Moreover, Cin1 exists in two ITSN1-like isoforms due to the similar event of alternate mRNA splicing: a 2004 amino acid long isoform (Cin1) and a 1282 amino acid short isoform (Cin1s). Although the SH3 and DH-PH domains unique to Cin1 were found to exhibit in vitro and in vivo functions, distinctions between Cin1 and Cin1s remain largely unknown [26].
As S. cerevisiae Pan1 epitomizes fungal endocytic adaptor proteins, a comparison in domain structures was made between Cin1 and Pan1. Cin1 and Pan1 both contain the EH domain at the N-terminal (Cin1 has a single EH domain), but Cin1 has the WH2, SH3, and DH-PH domains that are not present in Pan1. Proteins with domain structures similar to Cin1 were not found in the S. cerevisiae genome (www.yeastgenome.org), indicating divergence in the evolution of endocytic adaptor proteins between the two. However, it is intriguing as to how and why C. neoformans retains a protein that is more homologous to humans than yeast. To better understand such a conundrum, a search of available fungal genome databases for homologs of either Cin1 or Pan1 was made. Unexpectedly, proteins homologous to Cin1 can only be found in other members of basidiomycetous fungi, such as Ustilago maydis (GenBank accession: XP_762160 or UM06013), Coprinopsis cinerea (GenBank accession: XP_001839934), and Phanerochaete chrysosporium (GenBank accession: XP_001878432), but not in ascomycetous including S. cerevisiae and C. albicans and deuteromycetous fungi. Additionally, a EH domain-containing protein (GenBank accession: XP_567624) was identified in C. neoformans to bear some similarities to either Pan1 or S. cerevisiae Ede1. XP_567624 contains two N-terminal EH domains, a coiled-coil domain, and a proline-rich (PR) domain similar to Pan1 (E-value: 2.5e-12). Conversely, its C-terminal ubiquitin-associated (UBA) domain and the centrally located PR domain mimic S. cerevisiae Ede1 (E-value: 7.1e-29) [40]. Moreover, proteins homologous to S. cerevisiae Arp2/Arp3 (GenBank accessions: XP_572870/XP_570501), Rsp5 (GenBank accession: XP_572216), and Sla2/Sla2 (GenBank accession: XP_570853/XP_572163), which are components of the S. cerevisiae Pan1 endocytic pathway, and homologs of actin-related proteins were also present in the C. neoformans genome (Table 1). Thus, it is likely that a pathway similar to Pan1 operates to regulate membrane trafficking and the actin cytoskeleton in this fungus. If functional characterization of XP_567624 reveals that it is analogous to yeast Ede1, not Pan1, Cin1 may provide a `Pan1' function through its EH, coiled-coil, and other domains. Cin1 and XP_567624 form a large protein complex regulating the endocytic cycle of endocytosis and exocytosis, as well as coupling both events to the actin cytoskeleton organization and signaling though its multi-domains. Further functional dissections of XP_567624, along with continual characterization of Cin1, are therefore necessary. Regardless, current findings provide compelling evidence supporting that C. neoformans has evolved a major endocytic adaptor protein that not only shares functions of S. cerevisiae Pan1 but also functions of human ITSN1. The putative endocytic pathway mediated by the adaptor protein Cin1 is depicted in Fig. 2.
Fig. 2.
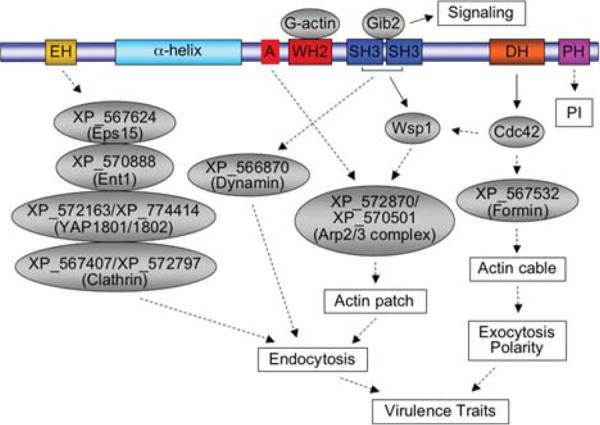
In Cryptococcus neoformans, Cin1 has a pleiotropic function through associations with multiple proteins. Cin1 is hypothesized to interact with Eps15, epsin, and clathrin homologs through the EH domain to mediate endocytosis. The putative A domain binds to the Arp2/3 complex, promoting the extension of the branched actin filaments, and the monomeric actin binds to WH2 to supply the globular actin. These two processes contribute to endocytosis. Through SH3 domains, Cin1 interacts with and activates Wsp1, leading to the assembly of actin patches. Wsp1 is also activated at the same time by binding of Cdc42 to its GBD domain. Additionally, Cin1 functions as a GEF for Cdc42 and activated Cdc42 binds formin to regulate the assembly of actin cables. Moreover, Cin1 could bind to dynamin proteins through the SH3 domain to regulate the actin cytoskeleton. The signifi cance of Cin1 interacting with Gib2 remains to be determined. Solid arrows indicate experimentally demonstrated interactions, while dotted arrows suggest potential interactions based on studies of other model organisms ([26] and Shen et al., manuscript in preparation). EH, Eps15 homology; α-helix, α-helical coiled-coil domain; A, acidic domain; WH2, WASP homology 2; SH3, Src homology 3; DH, Rho-GEF domain; PH, Plectrin homology; PI, phosphoinositides.
Concluding remarks
Intracellular trafficking is a conserved cellular process essential for biological functions of all eukaryotic cells. In pathogenic fungi, it also introduces new functions associated with the organism's ability to cause diseases. Whether the endocytic process, such as signal perception, nutrient uptake, and ion homeostasis, or the secretory transport of vesicular bodies containing virulence constituents (virulence cargo), or both play a role in virulence remains to be further studied and identified. The multi-functional role of Cin1 underscores the importance of endocytic proteins in physiology and virulence of fungi such as C. neoformans. Studies to further dissect additional proteins that interact with Cin1 and to delineate the significance of these interactions will further reveal the importance of Cin1. Thus, the identification of Cin1 marks a key step in our understanding of novel endocytic machinery required for membrane trafficking, which will facilitate the understanding of virulence mechanisms in C. neoformans.
Acknowledgements
We thank J. Cutler, A. Whittington, and anonymous reviewers for helpful suggestions and comments regarding this review. Research in the Wang Laboratory is supported in part by NIH grants AI054958 and AI074001, and a fund from the Research Institute for Children, New Orleans, Louisiana, USA.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 2.Wendland B, Emr SD, Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- 3.Battey NH, James NC, Greenland AJ, Brownlee C. Exocytosis and endocytosis. Plant Cell. 1999;11:643–660. doi: 10.1105/tpc.11.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samaj J, Read ND, Volkmann D, Menzel D, Baluska F. The endocytic network in plants. Trends Cell Biol. 2005;15:425–433. doi: 10.1016/j.tcb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Miaczynska M, Stenmark H. Mechanisms and functions of endocytosis. J Cell Biol. 2008;180:7–11. doi: 10.1083/jcb.200711073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung AY, de Vries SC. Membrane trafficking: intracellular highways and country roads. Plant Physiol. 2008;147:1451–1453. doi: 10.1104/pp.104.900266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson MS, Watts C, Marino Z. Membrane dynamics in endocytosis. Cell. 1996;84:13–21. doi: 10.1016/s0092-8674(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 8.Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 9.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munn AL. The yeast endocytic membrane transport system. Microsc Res Tech. 2000;51:547–562. doi: 10.1002/1097-0029(20001215)51:6<547::AID-JEMT5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi Y, Shoji JY, Arioka M, et al. Endocytosis is crucial for cell polarity and apical membrane recycling in the fi lamentous fungus Aspergillus oryzae. Eukaryot Cell. 2009;8:37–46. doi: 10.1128/EC.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JH, Kim HW, Heo DH, et al. FgEnd1 is a putative component of the endocytic machinery and mediates ferrichrome uptake in F. graminearum. Curr Genet. 2009;55:593–600. doi: 10.1007/s00294-009-0272-8. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs U, Hause G, Schuchardt I, et al. Endocytosis is essential for pathogenic development in the corn smut fungus Ustilago maydis. Plant Cell. 2006;18:2066–2081. doi: 10.1105/tpc.105.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs U, Steinberg G. Endocytosis in the plant-pathogenic fungus Ustilago maydis. Protoplasma. 2005;226:75–80. doi: 10.1007/s00709-005-0109-3. [DOI] [PubMed] [Google Scholar]
- 15.Steinberg G. On the move: endosomes in fungal growth and pathogenicity. Nat Rev Microbiol. 2007;5:309–316. doi: 10.1038/nrmicro1618. [DOI] [PubMed] [Google Scholar]
- 16.Weissman Z, Shemer R, Conibear E, et al. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- 17.Johnston DA, Eberle KE, Sturtevant JE, et al. Role for endosomal and vacuolar GTPases in Candida albicans pathogenesis. Infect Immun. 2009;77:2343–2355. doi: 10.1128/IAI.01458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palanisamy SK, Ramirez MA, Lorenz M, et al. Candida albicans PEP12 is required for biofilm integrity and in vivo virulence. Eukaryot Cell. 2010;9:266–277. doi: 10.1128/EC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornet M, Bidard F, Schwarz P, et al. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect Immun. 2005;73:7977–7987. doi: 10.1128/IAI.73.12.7977-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernardo SM, Khalique Z, Kot J, et al. Candida albicans VPS1 contributes to protease secretion, filamentation, and biofilm formation. Fungal Genet Biol. 2008;45:861–877. doi: 10.1016/j.fgb.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf JM, Johnson DJ, Chmielewski D, et al. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot Cell. 2010;9:1203–1215. doi: 10.1128/EC.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas LM, Martin SW, Konopka JB. BAR domain proteins Rvs161 and Rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence. Infect Immun. 2009;77:4150–4160. doi: 10.1128/IAI.00683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornet M, Gaillardin C, Richard ML. Deletions of the endocytic components VPS28 and VPS32 in Candida albicans lead to echinocandin and azole hypersensitivity. Antimicrob Agents Chemother. 2006;50:3492–3495. doi: 10.1128/AAC.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Hu G, Panepinto J, et al. Role of a VPS41 homologue in starvation response, intracellular survival and virulence of Cryptococcus neoformans. Mol Microbiol. 2006;61:1132–1146. doi: 10.1111/j.1365-2958.2006.05299.x. [DOI] [PubMed] [Google Scholar]
- 25.Panepinto J, Komperda K, Frases S, et al. Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol Microbiol. 2009;71:1165–1176. doi: 10.1111/j.1365-2958.2008.06588.x. [DOI] [PubMed] [Google Scholar]
- 26.Shen G, Whittington A, Song K, et al. Pleiotropic function of intersectin homologue Cin1 in Cryptococcus neoformans. Mol Microbiol. 2010;76:662–676. doi: 10.1111/j.1365-2958.2010.07121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis NG, Horecka JL, Sprague GF. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raths S, Rohrer J, Crausaz F, et al. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benmerah A, Begue B, Dautry-Varsat A, et al. The ear of alpha-adaptin interacts with the COOH-terminal domain of the Eps 15 protein. J Biol Chem. 1996;271:12111–12116. doi: 10.1074/jbc.271.20.12111. [DOI] [PubMed] [Google Scholar]
- 30.van Delft S, Govers R, Strous GJ, et al. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272:14013–14016. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- 31.Kaksonen M. Taking apart the endocytic machinery. J Cell Biol. 2008;180:1059–1060. doi: 10.1083/jcb.200802174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toret CP, Drubin DG. The budding yeast endocytic pathway. J Cell Sci. 2006;119:4585–4587. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- 33.Borth N, Walther A, Reijnst P, et al. Candida albicans Vrp1 is required for polarized morphogenesis and interacts with Wal1 and Myo5. Micro-biology. 2010;156:2962–2969. doi: 10.1099/mic.0.041707-0. [DOI] [PubMed] [Google Scholar]
- 34.Tang HY, Munn A, Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benedetti H, Raths S, Crausaz F, et al. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tebar F, Confalonieri S, Carter RE, et al. Eps15 is constitutively oligomerized due to homophilic interaction of its coiled-coil region. J Biol Chem. 1997;272:15413–15418. doi: 10.1074/jbc.272.24.15413. [DOI] [PubMed] [Google Scholar]
- 38.Galan JM, Moreau V, Andre B, et al. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 39.Zoladek T, Tobiasz A, Vaduva G, et al. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. Genetics. 1997;145:595–603. doi: 10.1093/genetics/145.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagny B, Wiederkehr A, Dumoulin P, et al. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- 41.Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9:327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- 42.Cutler JE. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 43.Haynes K. Virulence in Candida species. Trends Microbiol. 2001;9:591–596. doi: 10.1016/s0966-842x(01)02237-5. [DOI] [PubMed] [Google Scholar]
- 44.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veses V, Gow NA. Vacuolar dynamics during the morphogenetic transition in Candida albicans. FEMS Yeast Res. 2008;8:1339–1348. doi: 10.1111/j.1567-1364.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 46.Palmer GE, Askew DS, Williamson PR. The diverse roles of autophagy in medically important fungi. Autophagy. 2008;4:982–988. doi: 10.4161/auto.7075. [DOI] [PubMed] [Google Scholar]
- 47.Basrai MA, Naider F, Becker JM. Internalization of lucifer yellow in Candida albicans by fluid phase endocytosis. J Gen Microbiol. 1990;136:1059–1065. doi: 10.1099/00221287-136-6-1059. [DOI] [PubMed] [Google Scholar]
- 48.Martin R, Hellwig D, Schaub Y, et al. Functional analysis of Candida albicans genes whose Saccharomyces cerevisiae homologues are involved in endocytosis. Yeast. 2007;24:511–522. doi: 10.1002/yea.1489. [DOI] [PubMed] [Google Scholar]
- 49.Veses V, Casanova M, Murgui A, et al. ABG1, a novel and essential Candida albicans gene encoding a vacuolar protein involved in cytokinesis and hyphal branching. Eukaryot Cell. 2005;4:1088–1101. doi: 10.1128/EC.4.6.1088-1101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veses V, Casanova M, Murgui A, et al. Candida albicans ABG1gene is involved in endocytosis. FEMS Yeast Res. 2009;9:293–300. doi: 10.1111/j.1567-1364.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- 51.Yin Z, Stead D, Selway L, et al. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]
- 52.Toshima J, Toshima JY, Duncan MC, et al. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gale CA, Leonard MD, Finley KR, et al. SLA2 mutations cause SWE1-mediated cell cycle phenotypes in Candida albicans and Saccharomyces cerevisiae. Microbiology. 2009;155:3847–3859. doi: 10.1099/mic.0.033233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holtzman DA, Yang S, Drubin DG. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993;122:635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reijnst P, Jorde S, Wendland J. Candida albicans SH3-domain proteins involved in hyphal growth, cytokinesis, and vacuolar morphology. Curr Genet. 2010;56:309–319. doi: 10.1007/s00294-010-0301-7. [DOI] [PubMed] [Google Scholar]
- 56.Poltermann S, Nguyen M, Gunther J, et al. The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology. 2005;151:1645–1655. doi: 10.1099/mic.0.27505-0. [DOI] [PubMed] [Google Scholar]
- 57.Alvarez FJ, Douglas LM, Rosebrock A, et al. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol Biol Cell. 2008;19:5214–5225. doi: 10.1091/mbc.E08-05-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hibbett DS, Binder M, Bischoff JF, et al. A higher-level phylogenetic classification of the fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS - 100 years after the discovery of Cryptococcus neoformans. Clinical Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchanan KL, Murphy JW. What makes Cryptococcus neoformans a pathogen? Emerg Infect Dis. 1998;4:71–83. doi: 10.3201/eid0401.980109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozel TR, Gotschlich EC. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J Immunol. 1982;129:1675–1680. [PubMed] [Google Scholar]
- 62.Kozel TR, Pfrommer GS, Guerlain AS, et al. Role of the capsule in phagocytosis of Cryptococcus neoformans. Rev Infect Dis. 1988;10(Suppl.2):S436–S439. doi: 10.1093/cid/10.supplement_2.s436. [DOI] [PubMed] [Google Scholar]
- 63.De Jesus M, Nicola AM, Rodrigues ML, et al. Capsular localization of the Cryptococcus neoformans polysaccharide component galactoxylomannan. Eukaryot Cell. 2009;8:96–103. doi: 10.1128/EC.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rittershaus PC, Kechichian TB, Allegood JC, et al. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Investig. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salas SD, Bennett JE, Kwon-Chung KJ, et al. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodrigues ML, Nakayasu ES, Oliveira DL, et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang HY, Xu J, Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pucharcós C, Fuentes JJ, Casas C, et al. Alu-splice cloning of human Intersectin (ITSN), a putative multivalent binding protein expressed in proliferating and differentiating neurons and overexpressed in Down syndrome. Eur J Hum Genet. 1999;7:704–712. doi: 10.1038/sj.ejhg.5200356. [DOI] [PubMed] [Google Scholar]
- 70.Guipponi M, Scott HS, Chen H, et al. Two isoforms of a human intersectin (ITSN) protein are produced by brain-specific alternative splicing in a stop codon. Genomics. 1998;53:369–376. doi: 10.1006/geno.1998.5521. [DOI] [PubMed] [Google Scholar]
- 71.Yamabhai M, Hoffman NG, Hardison NL, et al. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 72.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 73.Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 74.Rohatgi R, Ma L, Miki H, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 75.Hussain NK, Jenna S, Glogauer M, et al. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 76.Zamanian JL, Kelly RB. Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent src homology 3 domains that are also involved in endocytosis. Mol Biol Cell. 2003;14:1624–1637. doi: 10.1091/mbc.E02-08-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang JB, Wu WJ, Cerione RA. Cdc42 and Ras cooperate to mediate cellular transformation by intersectin-L. J Biol Chem. 2005;280:22883–22891. doi: 10.1074/jbc.M414375200. [DOI] [PubMed] [Google Scholar]
- 78.Malacombe M, Ceridono M, Calco V, et al. Intersectin-1L nucleotide exchange factor regulates secretory granule exocytosis by activating Cdc42. EMBO J. 2006;25:3494–3503. doi: 10.1038/sj.emboj.7601247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haslam RJ, Koide HB, Hemmings BA. Pleckstrin domain homology. Nature. 1993;363:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 80.Shaw G. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 81.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 82.Chu DS, Pishvaee B, Payne GS. The light chain subunit is required for clathrin function in Saccharomyces cerevisiae. J Biol Chem. 1996;271:33123–33130. doi: 10.1074/jbc.271.51.33123. [DOI] [PubMed] [Google Scholar]
- 83.Drubin DG, Mulholland J, Zhu ZM, et al. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990;343:288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- 84.Huang CF, Liu YW, Tung L, et al. Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:3834–3847. doi: 10.1091/mbc.E03-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goyard S, Knechtle P, Chauvel M, et al. The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albi-cans. Mol Biol Cell. 2008;19:2251–2266. doi: 10.1091/mbc.E07-09-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Epp E, Walther A, Lepine G, et al. Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol Microbiol. 2010;75:1182–1198. doi: 10.1111/j.1365-2958.2009.07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 89.Soulard A, Lechler T, Spiridonov V, et al. Saccharomyces cerevisiae Bzz1p is implicated with type I myosins in actin patch polarization and is able to recruit actin-polymerizing machinery in vitro. Mol Cell Biol. 2002;22:7889–7906. doi: 10.1128/MCB.22.22.7889-7906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amatruda JF, Gattermeir DJ, Karpova TS, et al. Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J Cell Biol. 1992;119:1151–1162. doi: 10.1083/jcb.119.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moon AL, Janmey PA, Louie KA, et al. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walther A, Wendland J. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich syndrome protein homolog Wal1p. Eukaryot Cell. 2004;3:471–482. doi: 10.1128/EC.3.2.471-482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goodson HV, Anderson BL, Warrick HM, et al. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J Cell Biol. 1996;133:1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oberholzer U, Marcil A, Leberer E, et al. Myosin I is required for hypha formation in Candida albicans. Eukaryot Cell. 2002;1:213–228. doi: 10.1128/EC.1.2.213-228.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sivadon P, Crouzet M, Aigle M. Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett. 1997;417:21–27. doi: 10.1016/s0014-5793(97)01248-9. [DOI] [PubMed] [Google Scholar]
- 97.Adams AE, Botstein D, Drubin DG. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Science. 1989;243:231–233. doi: 10.1126/science.2643162. [DOI] [PubMed] [Google Scholar]
- 98.Goodman A, Goode BL, Matsudaira P, et al. The Saccharomyces cerevisiae calponin/transgelin homolog Scp1 functions with fimbrin to regulate stability and organization of the actin cytoskeleton. Mol Biol Cell. 2003;14:2617–2629. doi: 10.1091/mbc.E03-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vater CA, Raymond CK, Ekena K, et al. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mochida J, Yamamoto T, Fujimura-Kamada K, et al. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics. 2002;160:923–934. doi: 10.1093/genetics/160.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dewar H, Warren DT, Gardiner FC, et al. Novel proteins linking the actin cytoskeleton to the endocytic machinery in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:3646–3661. doi: 10.1091/mbc.E02-05-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cope MJ, Yang S, Shang C, et al. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blankenship JR, Fanning S, Hamaker JJ, et al. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miliaras NB, Park JH, Wendland B. The function of the endocytic scaffold protein Pan1p depends on multiple domains. Traffic. 2004;5:963–978. doi: 10.1111/j.1600-0854.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- 105.Duncan MC, Cope MJ, Goode BL, et al. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]


