Abstract
Both Chikungunya and Dengue virus belong to the acute arthropod-borne viruses. Because of the lack of specific symptoms, it is difficult to distinguish the two infections based on clinical manifestations. To identify and quantitatively detect Chikungunya and Dengue viruses, a real-time accelerated reverse-transcription-loop-mediated isothermal amplification (RT-LAMP) platform was developed, and 26-confirmed RNA samples, 42 suspects, and 18 healthy serum samples were evaluated by the method. The RT-polymerase chain reaction (PCR) and cDNA sequencing were used as references. The results showed that it could identify the Chikungunya and Dengue virus RNA correctly in all antibody-positive samples within 1 hour, without any cross-reactions. The virus load of the positive samples was quantitatively detected with a turbidimeter. The sensitivity was 100% and specificity was 95.25%. The findings indicate that the RT-LAMP is an effective method for rapid quantity detection of Chikungunya virus and Dengue virus in serum samples with convenient operation, high specificity, and high sensitivity.
Introduction
Chikungunya (CHIK) virus, the agent of Chikungunya fever, is an acute arthropod-borne alpha virus classified to the family Togaviridae, which was first isolated in Tanzania in 19531; it has become a new, unexpected public health problem in many tropical African and Asian countries within the past decade. Although it was historically thought of as a tropical disease, CHIK has spread to temperate regions by adapting to the Aedes albopictus mosquito caused by a genetic mutation that occurred during 2005–2006.2,3 The most serious outbreak swept over many parts of southern India in 2006 and affected more than 1,000,000 persons.4 Later, an event was reported in Italy in 2007 (Enserink, 2007; Rezza and others26) and an epidemic took place in Singapore in 2008,5 which brought about a serious public health problem and caused a significant economic loss. This arboviral disease is now a global concern.6,7
Dengue (DEN) virus is a member of genus Flavivirus of the family Flavivirus. Compared with other arboviral viruses, DEN causes a wide range of serious diseases, such as dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS). It was estimated that the annual worldwide occurrence of dengue fever is 100 million.8 Nearly 2.6 billion people live in areas at risk of an epidemic9; the latest outbreak of DEN took place in the Philippines in 2011, in which more than 38,000 people were affected, and 226 people have since died of the DEN virus.
The two kinds of infections have similar characteristics of geographical distribution and seasonal correlation,10,11 because they share the same vectors of transmission, Aedes albopictus and Aedes aegypti mosquitoes. Some factors such as population migration and transportation could also promote the spread of the diseases.12 Both CHIK and DEN viruses can cause similar symptoms such as fever, rash, and severe rheumatic. However, the prognoses of the CHIK and DEN infections are greatly different. For example, DEN has a higher mortality rate. It is extremely important to identify two viruses correctly in a short time frame with simple operations, especially in countries or areas where the CHIK virus is newly discovered.
Virus isolation from infected tissue and blood specimens of the viremic patients is the gold standard for both DEN and CHIK virus detection, but this process needs at least 7 days. As for CHIK isolation, a laboratory with BSL-3 is required in China. The enzyme-linked immunosorbent assay (ELISA) has been developed. This method, a cross-reaction that occurs between CHIK virus and O'nyong-nyong virus, can take place within 2–3 hours.13,14 Molecular tools such as the conventional or real-time reverse transcription-polymerase chain reaction (RT-PCR) are available for the detection of DEN and virus in culture supernatants or clinical samples,9,15 but precise and sophisticated equipment is needed.
In this study, a reliable, specific real-time method for quantity detection and differentiation of CHIK and DEN virus was developed based on loop-mediated isothermal amplification (LAMP). The LAMP technique is highly specific for target sequence and has been successfully used in the differential diagnosis of microorganisms such as Brettanomyces and Dekkera sp. yeasts16 and fungi identification.17 According to a previous study, the sensitivity of the LAMP method is below 10 copies/reaction.18 Because of the high amplification efficiency, the LAMP reaction can be ended within 1 hour under isothermal conditions (60–65°C) and sophisticated instruments are not required,18 which makes this method adaptive to field diagnosis. Furthermore, as a result of the invention of the turbidimeter, which enables the LAMP technique from a qualitative detection method, becomes a kind of quantitative technique.19,20 In this RT-LAMP platform, a reverse transcription and amplification was designed in one step with two tubes under the same reaction conditions for rapid identification and quantitative detection of RNA for CHIK and DEN viruses, respectively. There were 42 unknown serum samples detected by RT-LAMP compared with an antibodies test and conventional RT-PCR in the study.
Material And Methods
RNA of the viruses.
Ten RNA samples from CHIK and 12 DEN viruses were offered by the Guangdong Inspection and Quarantine Technology Center, which were collected over recent years from patient isolates. Four serotypes of DEN virus were involved, including 5 serotype I, 3 serotype II, 2 serotype III, and 2 serotype IV. All of these RNA samples have been confirmed by complementary DNA (cDNA) sequencing before being used as positive controls. West Nile virus, Japanese encephalitis virus, Yellow fever virus, and Avian influenza virus (H5N1) were selected as negative controls.
Serum samples and RNA extraction.
There were 42 unknown serum samples collected from a local hospital and imported persons, who presented the CHIK-like or DEN-like symptoms during June and October in 2010 from India and Nigeria. Meanwhile, 18 healthy serum samples were selected randomly as a negative control. Informed consent was obtained from all participants. The viral RNA was extracted from 100 μL of people's serum by using the QIAamp Viral RNA Mini kit (QIAGEN, Hilton Germany) according to the manufacturer's protocol. The RNA was eluted by 80 μL elution buffer and stored at − 80°C until use.
RT-LAMP for CHIK and DEN virus detection.
The structural E1 gene was selected as the target of RT-LAMP for CHIKV detection. To assure the specificity of the primers, the sequences of E1 was selected and downloaded from GenBank and aligned with multiple sequence alignment tools (DNAMAN software V6 Lynnon Corporation in Canada) to identify the conserved region. The potential target region of 570 bp was finally selected from the aligned sequence, in which the mutation site (A226V) of E1 gene was avoided in overlapping. The primers used in this study were designed using the PrimerExplorer V4 software (Eiken Chemical: http://primerexplorer.jp/elamp4.0.0/index.html). The sequence of RT-LAMP primers specified for CHIKV were as follows: CHIK-F3 (forward outer primer), 5′-CGCCCTCTTTAACGGACATG-3′; CHIK-B3 (the backward outer primer), 5′-AATTCGGCGCTGGCTAAG-3′ CHIK-FIP (the forward inner primer), 5′-TGCCTTTCTTGCTGGCTGCATATACCAGCCTGCACCCATT-3′; CHIK-BIP (the backward inner primer), 5′-AGTGTGCGGTGCATTCGATGATGCAGCTGAGAATTCCCTTC-3′.
As for the DEN detection, 850 bp of the potential target region of the C-prM gene was selected from the aligned sequence and the primers were designed based on the software mentioned previously. The final primer sets of the group, specific for all of the four serotypes of DEN virus were as follows: DEN-F3 (forward outer primer), 5′-GAGAAACCGCGTGTCAAC-3′; DEN-B3 (the backward outer primer), 5′-CCTTCCAATCTCTTTCCTGAA-3′; DEN-FIP (the forward inner primer), 5′-AGGGCCATGAACAGTTTTAATGGTCAGCTGACAAAGAGATTCTCA′; DEN-BIP (the backward inner primer,) 5′-CCTAACAATCCCACCAACAGCACCCTCTCAAAACATTGATAGC-3′. All primers were synthesized by Takara Biotechnology (Dalian) Co., Ltd., China.
RT-LAMP reaction.
The RT-LAMP assay was performed with a loopamp RNA amplification kit (Eiken Chemical). A total of 25 μL final reaction system composed of 2 μL of template RNA, 1 μL of Bst DNA polymerase mix, 1.6 μmol L-1 each of inner primers FIP and BIP, 0.2 μmol L-1 each of outer primers F3 and B3, and 1 × Reaction Mix (Eiken Chemical). The final volume was adjusted to 25 μL with distilled water. The reaction mixture was incubated at 63°C for 90 min in a loopamp real-time turbidimeter (LA-320; Teramecs, Kyoto, Japan). The reaction was terminated automatically by inactivating the polymerase at 80°C for 2 min. The reaction was considered to be positive when the turbidity reached 0.1 within 90 min in the loopamp real-time turbidimeter (LA-320; Teramecs, Kyoto, Japan). According to the program of LA-320, the data from 10 min after the start of reaction were taken as the baseline. In the preliminary experiment, turbidity of 0.05 that was more than 10 times the variance (SD) calculated was taken as the threshold. To facilitate comparison of the results in this study, the results from other two methods were simultaneously used as judgment. First, 1 μL SYBR Green I dye (Sigma-Aldrich, USA) was added into the tube to observe the color change after the amplification procedure. For LAMP assays, the lowest bands from amplicons were purified using the QIA quick PCR purification kit (QIAGEN, Germany) and cloned into the pMD18-T vector (Takara Biotechnology (Dalian) Co., Ltd., China).21 The sequencing was done by an ABI 3130XL genetic analyzer (Applied Biosystems, Japan).
Quantity detection of CHIK and DEN RNA.
To accurately test the sensitivity of the RT-LAMP assay for CHIK and DEN virus detection, the target region of 218 bp of E1 gene and 252 bp of C-prM gene were amplified by a RT-PCR method by using F3 and B3 primers described previously. The fragment was gel purified using the gel extraction kit (Ferment) and cloned in pMD 18-T Vector (Takara Biotechnology (Dalian) Co., Ltd., China) according to the manufacture's instruction. The plasmid with a correct insert was confirmed by the DNA sequencing. The concentration of the constructed plasmid was determined by measuring the optical density at 260 nm and determined by using the following formula: copies/μL = concentration of plasmid in g/μL (plasmid length × 660) × 6.022 × 1023. A further serial 10-fold dilution of the plasmid was used for limit detection. The standard curves were constructed using the Tt (time threshold) value against the concentration of serially diluted plasmids.
RT-PCR.
The RT-PCR method used in this study was used according to Reference 22 with a minor change of the following, verified by confirmed-RNA samples of CHIK and DEN viruses. The sequence of RT-PCR primers specified for CHIK virus was as follows (401 bp): CHIK-f: 5′-CGGTAAGAGCGGTGAACT-3′, CHIK-r: 5′- CCCGAGGGTGGTATGTGA-3′. The sequence of RT-PCR primers specified for DEN virus was as follow (1,013 bp): DEN-f: 5′-ACAGGTTCTTTTAGGGAG-3′, DEN-r 5′- TGCCATCGTCGTCAC -3′. The PCR amplification was carried out after reverse transcription was performed on the RNA samples. A total of 25 μL of reaction volume was used with 2 μL of the cDNA, 2.5 μL of the 10 × buffer, 1.5 μL (10 pmol/μL), a couple of appropriate primers, 4 μL of dNTPs mixture (2.5 mM of each dNTPs), and 0.5 μL (5 U/μL) of rTaq DNA polymerase. The reaction was performed on a BIO-RAD Peltier Thermal Cycler (California, USA) with the cycling conditions of 95°C for 5 min, followed by 35 cycles of 95°C for 30s, 55°C for 30 s, and 72°C for 60 s.
IgM and IgG detection in serum.
The serum samples from the patients and negative controls were tested for the presence of IgM- or IgG-specific antibodies against CHIKV or DENV according to Reference 23. An indirect immunofluorescence test (EUROIMMUN, Lübeck, Germany) was used for CHIK-specific antibody identification,24 and the Panbio Mac ELISA kit (Australia Panbio company in Melbourne) was used for the detection of DEN. In brief, the samples were diluted 1:10 –1:80, and 20 μL were applied to each reaction, and incubated for 1 hour following the manufacturer's instructions. For the detection of IgM and IgG, the rheumatic factor was pre-adsorbed with EUROSORB reagent (EUROIMMUN, Germany). For antibody detection, anti-human IgG or IgM antibodies labeled with fluorescein isothiocyanate were used, observed, and evaluated with fluorescence microscopy. Titers ≥ 1:10 were considered positive.
Results
Sensitivity of the RT-LAMP method for both CHIK and DEN viruses.
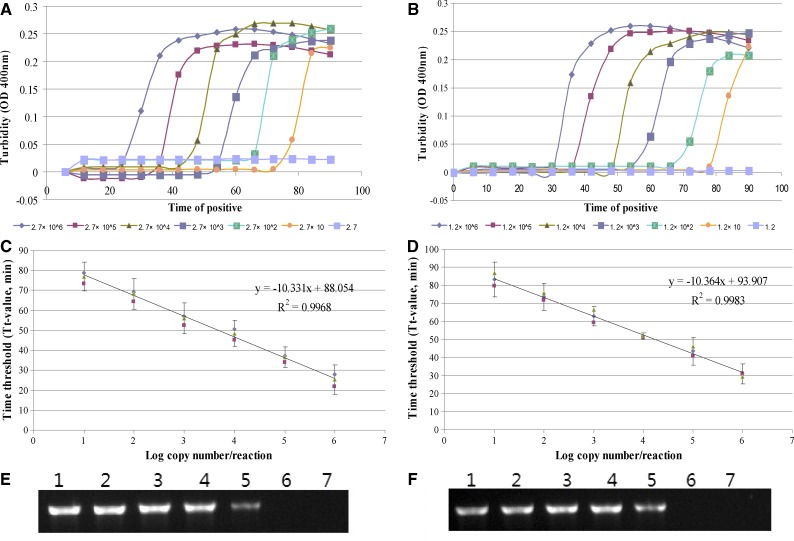
The sensitivity of the RT-LAMP method for the detection of CHIK and DEN viruses was determined by testing serial 10-fold dilutions of a cloned target that had previously been quantified and compared with that of the conventional RT-PCR. The correct cloned vector was constructed and verified by DNA sequencing. A representative sensitive result of the real-time RT-LAMP for CHIK or DEN virus detection is shown in Figure 1A and B, respectively. The Tt values of the RT-LAMP for CHIK detection were found to be between 25.13 and 77.73 min for templates ranging from 2.7 × 106 to 27 copies/reaction. The Tt values of RT-LAMP for DEN detection ranged from 31.62 to 82.54 min for the concentration of 1.2 × 106 to 12 copies/reaction. The linear relationship of each reaction was obtained and the standard curve was generated by plotting the graph between different concentrations of virus copy numbers to the Tt values (Figure 1C and D).The results of sensitivity from the conventional RT-PCR, which was as a comparison method in the study, were 270 and 120 copies/reaction for CHIK and DEN virus detection (Figure 2E and F) against that of the RT-LAMP 27 and 12 copies/reaction for CHIK and DEN virus, respectively. It was found to be 10-fold more sensitive than that of the RT-PCR.
Figure 1.
The sensitivity of the reverse transcription-loop-mediated isothermal amplification (RT-LAMP) and the RT-polymerase chain reaction (PCR) for detection of Chikungunya (CHIK) virus and Dengue (DEN) virus. (A) The sensitivity of the RT-LAMP for CHIK virus detection as monitored in a real-time turbidity assay. (B) The sensitivity of the RT-LAMP method for DEN virus detection. (C) The standard curves for CHIK-specific RT-LAMP assays generated from the amplification plots between concentration of templates and the time threshold (Tt-value). (D) The standard curves for DENe-specific RT-LAMP assays. (E) Electrophoretic analysis of RT-PCR products for detection of CHIK virus. Lanes 1–6, 2.7 × 106, 2.7 × 105, 2.7 × 104, 2.7 × 103, 2.7 × 102, and 27 copy/reaction, respectively; Lane 7, negative control. (F) Electrophoretic analysis of RT-PCR products for detection of DEN virus. Lanes 1–6, 1.2 × 106, 1.2 × 105, 1.2 × 104, 1.2 × 103, 1.2 × 102, and 12 copy/reaction, respectively; Lane 7, negative control.
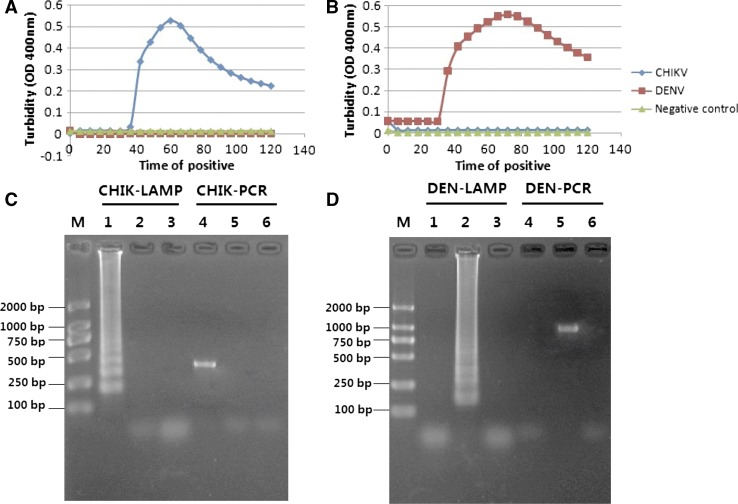
Figure 2.
The specificity of the reverse transcription-loop-mediated isothermal amplification (RT-LAMP) when identifying Chikungunya virus (B10H000991) and Dengue virus (DEN 1030 type II) with turbidimeter and agarose gel electrophoresis. (A) A turbidity judgment graph of the Chikungunya specified RT- LAMP. (B) A turbidity judgment graph of the Dengue specified RT- LAMP. (C) Agarose gel electrophoresis analysis of RT-LAMP and RT-polymerase chain reaction (PCR) method for Chikungunya detection. (D) Agarose gel electrophoresis analysis of RT-LAMP and RT-PCR method for Dengue virus detection. Lane M, DL2000 marker (TaKaRa, Japan). Lane 1, the products of the RT-LAMP method with RNA of CHIK. Lane 2, the RT-LAMP method with RNA of DEN. Lane 3, the RT-LAMP method with RNA from serum samples from healthy individuals as negative control. Lane 4, the products of the conventional RT-PCR method with RNA of CHIK. Lane 5, the conventional RT-PCR method with RNA of DEN. Lane 6, the conventional RT-PCR method with RNA from serum samples from healthy individuals.
Specificity of RT-LAMP for CHIK and DEN identification.
Except for positive RNA samples of CHIK virus and DEN viruses with four serotypes, West Nile virus, Japanese encephalitis virus, Yellow fever virus, and Avian influenza virus (H5N1) were used as negative controls. Each of the samples used in the study was tested twice and results were recorded. All 10 CHIK RNA samples received positive results from the RT-LAMP for CHIK and all 12 DEN RNA was positive for DEN. No cross-reactions and false-positive or false-negative results could be found (Table 1). Both CHIK- and DEN-specific RT-LAMP primers showed high degree of specificity by amplifying only the respective targets (Figure 2). For serotypes1–4 of the DEN viruses, the same high degree of specificity could be obtained by the RT-LAMP method.
Table 1.
Comparative analysis of CHIK- and DEN-specific RT-LAMP method with RT-PCR, and IgG and IgM antibodies test
| Type of cases and species | No. of samples | No. of samples positive by | |||||||
|---|---|---|---|---|---|---|---|---|---|
| RT-LAMP* | RT-PCR | IgG | IgM | ||||||
| CHIK | DEN | CHIK | DEN | CHIK | DEN | CHIK | DEN | ||
| Chikungunya virus | 10 | 10 | 0 | 10 | 0 | –† | – | – | – |
| Dengue virus | 12 | 0 | 12 | 0 | 12 | – | – | – | – |
| West Nile virus | 1 | 0 | 0 | 0 | 0 | – | – | – | – |
| Japanese encephalitis virus | 1 | 0 | 0 | 0 | 0 | – | – | – | – |
| Yellow fever virus | 1 | 0 | 0 | 0 | 0 | – | – | – | – |
| Avian influenza virus (H5N1) | 1 | 0 | 0 | 0 | 0 | – | – | – | – |
| Samples‡ | 42 | ||||||||
| CHIK-diagnosed | 3 | 3 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| DEN-diagnosed | 15 | 0 | 15 | 0 | 15 | 0 | 13 | 0 | 8 |
| Healthy | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
In comparison with the results of the reference method, the RT-LAMP assay had a sensitivity of 100%, and a specificity of 95.25%.
Not attempted.
Samples from import cases and clinical suspected patients for screening were referred as suspected samples.
CHIK = Chikungunya; DEN = Dengue; RT-LAMP = reverse-transcription-loop-mediated isothermal amplification; RT-PCR = reverse-transcription-polymerase chain reaction.
Evaluation of the samples for CHIK or DEN by RT-LAMP.
Forty-two unknown serum samples were collected from the local highly suspicious patients and imported persons from India, Congo, Nigeria, Malaysia, and Singapore who present the CHIK-like or DEN-like symptoms in entry quarantine during the period of June–October from 2008 to 2011. These samples were screened for the presence of CHIK or DEN RNA by the RT-LAMP method and compared with the real-time PCR and IgG or IgM antibody tests. There were three serum samples that obtained positive CHIK results, two of them RT-PCR positive, one can be diagnosed as CHIK based on the IgG antibody test. For DEN virus detection, 15 samples were positive by the real-time RT-LAMP, which were the same as the conventional RT-PCR. There were 13 positive IgG results and 8 positive IgM results among all of them. No cross-reactions, neither false-positive nor false-negative results, could be observed. All negative serum samples were not detected for CHIK or DEN virus by the RT-LAMP method or other reference assay (Table 1).
After being compared with the results from other tests, it was determined that the RT-LAMP method could detect the CHIK and DEN virus RNA in all 14 of the positive IgG samples with sensitivity of 100%. The CHIK-like or DEN-like cases can be distinguished as CHIK and DEN correctly by the newly developed RT-LAMP platform with no cross-reaction. After sequencing, all of the samples that obtained positive results by LAMP or PCR were verified. The specificity of RT-LAMP platform for CHIK and DEN viral RNA detection is 95.25% (40 of 42) and 95.25% (40 of 42).
Quantitative detection of CHIK and DEN virus load in the sample.
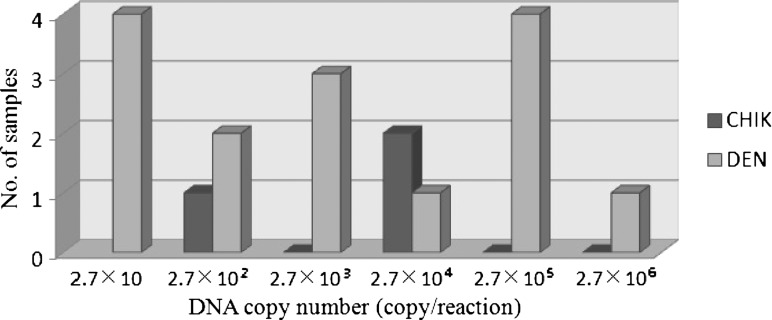
The standard curves of CHIK and DEN virus were generated by plotting the graph between various concentrations of virus genomic equivalents ranging from 2.7 × 106 to 27 copies/reaction and 1.2 × 106 to 12 copies/reaction to the time of positivity through real-time monitoring of the turbidity during amplification. The quantification of the virus load in the positive samples was extrapolated on the basis of their time of positivity (Tt-value) by employing the standard curve. The concentrations of the virus in the patient serum were found within the range of 10 to 106 copy/reaction. Among the three CHIK-positive samples, there were two samples of concentration that were 102 copies/reaction and one sample was 104 copies/reaction (Figure 3).
Figure 3.
Quantitative determination of virus concentrations in clinical samples by Chikungunya-loop-mediated isothermal amplification (CHIK-LAMP) and Dengue-loop-mediated isothermal amplification (DEN-LAMP) method developed in this study. The columns in the histogram showed the distribution of virus concentration in serum samples.
Discussion
Viral infections have become a serious public health problem in recent years.11,25 Emergence or reemergence of severe mosquito-borne viruses, such as CHIK virus and DEN virus, have been frequently reported worldwide and have caused a significant economic loss.6,11 There have been a few adaptive mutations of CHIK virus or a shift in viral genotypes; its epidemiologic characteristics or host-specific barriers have changed to distribute into new geographic ranges. It is now considered a real threat to temperate areas, such as Europe and the Americas, that are colonized by Aedes spp.12,26 In China, the first case infected with CHIK virus was reported in 2008.24 In other countries like India27 and South East Asia, numerous outbreaks have reemerged.28 The disease that emerges or reemerges in these countries or areas not only brings an anxiety about the expansion trend of the CHIK, but also causes new social and public health problems. Both CHIK and DEN infectious diseases share the same characteristic of geographical distribution and seasonal correlation. They also have similar symptoms such as fever, rash, and severe rheumatic disorder, which increased difficulties in the differential diagnose. Moreover, the report for discrimination of the CHIK and DEN virus infection within a platform was seldom described as a simple method for early identification and differentiation of the two viruses, which has great significance for field application.
In this study, one step with two tubes, a real-time RT-LAMP platform was developed successfully, and can be used for rapid CHIK and DEN virus identification with simple operational methods. All 10 CHIK and 12 DEN viruses had positive results. The negative control, West Nile virus, Japanese encephalitis virus, Yellow fever virus, and Avian influenza virus (H5N1) received negative results. No false-positive or false-negative results were found. For these reasons, this method is an effective means to diagnose the relative RNA viruses. For example, the CHIK virus could be identified and distinguished from the DEN virus correctly without cross-reaction by the RT-LAMP within 1 hour. The specificity of two RT- LAMP reactions was both 100%. In the sensitivity evaluation test, we found the detection limit of this RT-LAMP method to be 2.7 and 12 copies/reaction for CHIK virus and DEN virus, respectively. Furthermore, the new method can be used in quantitative detection of the virus copy number followed by the standard curves that were constructed. It is evident that the RT-LAMP can overcome the shortage of cross-reactions between CHIK virus and O'nyong-nyong virus by the ELISA method.13,29
Serum samples from 42 patients with DEN-like and CHIK-like symptoms, and 18 negative controls were tested by the RT-LAMP method compared with the real-time PCR and IgG or IgM antibody tests. We found three of them to be CHIK positive and 15 samples to be DEN positive. No cross-reactions, neither false-positive nor false-negative results, could be found. In the study, the specificity was calculated as follows: (the number of true positives and true negatives)/total number of samples. The specificity of the RT-LAMP method for CHIK and DEN detection in acute samples was 95.25% (40 of 42) and 95.25% (40 of 42).The sensitivity was 100% (1 of 1) and 100% (13 of 13), respectively. Because viral load is a useful marker of disease progression and a measure of the efficiency of antiviral compounds,30 the quantitative detection was constructed in the study. The virus load in the clinical samples was mainly distributed within the range of 10 to 106 copies/reaction. The results were satisfactory for both the clinical and the field detection, though a further confirmation test with more samples should be conducted. The fact that there were only three CHIK-positive cases identified in the study indicate that CHIK virus infection in China seems relatively rare compared with that of DEN virus. No large-scale local outbreak of CHIK infection has been reported in this country so far. However, according to the official data from China National Tourism Administration, more than 3.17 million people visit or return to China from epidemic countries each year, which would cause a potential public health problem. In addition, Ae. albopictus is widespread in southern China where the population density is high and many DEN virus infections have been reported.31,32 To determine the etiology of symptomatic visitors correctly are key measures for preventing CHIK epidemics.
As a most important mosquito-borne virus, Dengue virus is one of the major human pathogens and more than 2.5 billion people are at risk.8 It can cause DF and DHF/DSS. There were about 100 million cases of DF and 250,000 cases of DHF/DSS occurring annually. The first dengue case was reported as early as 1978 in China.33,34 One of the most serious outbreaks took place in Hainan Island, and a total of 437,468 cases were reported with a morbidity level of 8,097/100,000 populations in 1980.35 This shows that it is very important to establish a specific, sensitive, and rapid assay for the detection of DEN virus and to differentiate DEN from other viruses. Because it does not require a sophisticated instrument or a complex operation, the RT-LAMP assay can be applied easily at clinics, basic hospitals, field investigations, and especially in African and Asian countries or tropical regions where both CHIK and DEN virus diseases are major epidemics.6,36
The RT-LAMP method has emerged as a powerful gene amplification tool for microorganism identification, which is becoming increasingly accepted because of its advantages. The most attractive characteristic of the LAMP method is the visual determination of nucleic acid amplification,18 which cannot only be used in well-equipped hospitals or laboratories in developed countries, but also in small-scale hospitals or even in field detection.37 The LAMP method has been used in the differential diagnosis of the pathogens with close genetic relationships16,17,38,39 and even in the differentiation of DEN virus serotypes.40 The appearance and use of a turbidimeter allowed the LAMP method to achieve real-time detection. In fact, it is convenient to quantitatively detect CHIK and DEN viruses by monitoring the turbidity of the RT-LAMP reaction or by visually judging the reaction by using the SYBR Green I dye, based on different requirements.
ACKNOWLEDGMENTS
We sincerely thank Elizabeth Li for her great help with revising the article.
Footnotes
Financial support: This work was supported by State Key Laboratory of Respiratory Diseases (2007DA780154F0904), The Research Program General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China (2010IK222), The Research Project of Guangdong Entry-Exit Inspection and Quarantine Bureau (2009GDK11).
Authors' addresses: Xi Lu and Faguang Jin, Department of Respiratory, Tangdu Hospital, Fourth Military Medical University, Xi'an, China, E-mails: xi.lu@mail.scut.edu.cn and jinfag@fmmu.edu.cn. Xiaobo Li, Guangdong Entry-Exit Inspection and Quarantine Bureau, Guangzhou, China, E-mail: lxb_1980@yahoo.cn. Ziyao Mo and Hongbo Zhao, The State Key Laboratory of Respiratory Diseases, Guangzhou Medical University, Guangzhou, China, E-mails: ziyaomo@gmail.com and zhaohongbolt@163.com. Boliang Wang, Department of Emergency, Tangdu Hospital, Fourth Military Medical University, Xi'an China, E-mail: wang.fmmu@gmail.com. Xiaoxiao Shan, School of Bioscience & Bioengineering, South China University of Technology, Guangzhou, China. Lei Shi, College of Light Industry and Food Sciences, South China University of Technology, Guangzhou, China, E-mail: leishi@scut.edu.cn.
References
- 1.Ross R. The Newala epidemic: III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 1956;54:177–191. doi: 10.1017/s0022172400044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelvin AA. Outbreak of chikungunya in the Republic of Congo and the global picture. J Infect Developing Ctries. 2011;5:441–444. [PubMed] [Google Scholar]
- 3.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, Lavenir R, Pardigon N, Reynes JM, Pettinelli F. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravi V. Re-emergence of chikungunya virus in India. Indian J Med Microbiol. 2006;24:83–84. doi: 10.4103/0255-0857.25175. [DOI] [PubMed] [Google Scholar]
- 5.Leo YS, Chow AL, Tan LK, Lye DC, Lin L, Ng LC. Chikungunya outbreak, Singapore, 2008. Emerg Infect Dis. 2009;15:836–837. doi: 10.3201/eid1505.081390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon F, Javelle E, Oliver M, Leparc-Goffart I, Marimoutou C. Chikungunya virus infection. Curr Infect Dis Rep. 2011;13:218–228. doi: 10.1007/s11908-011-0180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 9.Saxena P, Dash PK, Santhosh S, Shrivastava A, Parida M, Rao PV. Development and evaluation of one step single tube multiplex RT-PCR for rapid detection and typing of dengue viruses. Virol J. 2008;5:20. doi: 10.1186/1743-422X-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charrel RN, de Lamballerie X, Raoult D. Chikungunya outbreaks—the globalization of vector-borne diseases. N Engl J Med. 2007;356:769–771. doi: 10.1056/NEJMp078013. [DOI] [PubMed] [Google Scholar]
- 11.Lakshmi V, Neeraja M, Subbalaxmi MV, Parida MM, Dash PK, Santhosh SR, Rao PV. Clinical features and molecular diagnosis of chikungunya fever from South India. Clin Infect Dis. 2008;46:1436–1442. doi: 10.1086/529444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parola P, de Lamballerie X, Jourdan J, Rovery C, Vaillant V, Minodier P, Brouqui P, Flahault A, Raoult D, Charrel RN. Novel chikungunya virus variant in travelers returning from Indian Ocean islands. Emerg Infect Dis. 2006;12:1493–1498. doi: 10.3201/eid1210.060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karabatsos N. Antigenic relationships of group A arboviruses by plaque reduction neutralization testing. Am J Trop Med Hyg. 1975;24:527–532. doi: 10.4269/ajtmh.1975.24.527. [DOI] [PubMed] [Google Scholar]
- 14.Powers AM, Brault AC, Tesh RB, Weaver SC. Re-emergence of chikungunya and o'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 15.Telles J-N, Le Roux K, Grivard P, Vernet G, Michault A. Evaluation of real-time nucleic acid sequence-based amplification for detection of chikungunya virus in clinical samples. J Med Microbiol. 2009;58:1168–1172. doi: 10.1099/jmm.0.010736-0. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi N, Arai R, Tada S, Taguchi H, Ogawa Y. Detection and identification of Brettanomyces/Dekkera sp. yeasts with a loop-mediated isothermal amplification method. Food Microbiol. 2007;24:778–785. doi: 10.1016/j.fm.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Ohori A, Endo S, Sano A, Yokoyama K, Yarita K, Yamaguchi M, Kamei K, Miyaji M, Nishimura K. Rapid identification of Ochroconis gallopava by a loop-mediated isothermal amplification (LAMP) method. Vet Microbiol. 2006;114:359–365. doi: 10.1016/j.vetmic.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 18.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 20.Siyi C, Beilei G. Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus. BMC Microbiol. 2010;10:41. doi: 10.1186/1471-2180-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, Wang L, Chu J, Li Y, Xu Z, Li L, Shirtliff ME, He X, Liu Y. Rapid detection of Vibrio parahaemolyticus strains and virulent factors by loop-mediated isothermal amplification assays. Food Sci Biotechnol. 2010;19:1191–1197. [Google Scholar]
- 22.Bonilauri P, Bellini R, Calzolari M, Angelini R, Venturi L, Fallacara F, Cordioli P, Angelini P, Venturelli C, Merialdi G. Chikungunya virus in Aedes albopictus, Italy. Emerg Infect Dis. 2008;14:852. doi: 10.3201/eid1405.071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai Z, Liu L, Tu Z, Yao L, Liu J, Xu B, Tang B, Wan Y, Fang M. Real-time PCR for detecting circulating dengue virus in the Guangdong Province of China in 2006. J Med Microbiol. 2008;57:1547–1552. doi: 10.1099/jmm.0.2008/003418-0. [DOI] [PubMed] [Google Scholar]
- 24.Zheng K, Li J, Zhang Q, Liang M, Li C, Lin M, Huang J, Li H, Xiang D, Wang N. Genetic analysis of chikungunya viruses imported to mainland China in 2008. Virol J. 2008;7:8. doi: 10.1186/1743-422X-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coker RJ, Hunter BM, Rudge JW, Liverani M, Hanvoravongchai P. Emerging infectious diseases in southeast Asia: regional challenges to control. Lancet. 2011;377:599–609. doi: 10.1016/S0140-6736(10)62004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli A, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 27.Ravi V. Re-emergence of chikungunya virus in India. Indian J Med Microbiol. 2006;24:83–84. doi: 10.4103/0255-0857.25175. [DOI] [PubMed] [Google Scholar]
- 28.Laras K, Sukri NC, Larasati RP, Bangs MJ, Kosim R. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans R Soc Trop Med Hyg. 2005;99:128–141. doi: 10.1016/j.trstmh.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Powers AM, Brault AC, Tesh RB, Weaver SC. Re-emergence of chikungunya and o'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 30.Parida MM, Santhosh SR, Dash PK, Tripathi NK, Lakshmi V, Mamidi N, Shrivastva A, Gupta N, Saxena P, Babu JP, Rao PV, Morita K. Rapid and real-time detection of chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2007;45:351–357. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G, Dong H, Shi N, Liu S, Zhou A, Cheng Z, Chen G, Liu J, Fang T, Zhang H. An outbreak of dengue virus serotype 1 infection in Cixi, Ningbo, People's Republic of China, 2004, associated with a traveler from Thailand and high density of Aedes albopictus. Am J Trop Med Hyg. 2007;76:1182–1188. [PubMed] [Google Scholar]
- 32.Lu L, Lin H, Tian L, Yang W, Sun J, Liu Q. Time series analysis of dengue fever and weather in Guangzhou, China. BMC Public Health. 2009;9:395. doi: 10.1186/1471-2458-9-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan W, Yu S, Cosgriff TM. The reemergence of dengue in China. Rev Infect Dis. 1989;11:S847–S853. doi: 10.1093/clinids/11.supplement_4.s847. [DOI] [PubMed] [Google Scholar]
- 34.Qiu FX, Chen QQ, Ho QY, Chen WZ, Zhao ZG, Zhao BW. The first epidemic of dengue hemorrhagic fever in the People's Republic of China. Am J Trop Med Hyg. 1991;44:364–370. doi: 10.4269/ajtmh.1991.44.364. [DOI] [PubMed] [Google Scholar]
- 35.Qiu FX, Gubler DJ, Liu JC, Chen QQ. Dengue in China: a clinical review. Bull World Health Organ. 1993;71:349–359. [PMC free article] [PubMed] [Google Scholar]
- 36.Gould E, Gallian P, De Lamballerie X, Charrel R. First cases of autochthonous dengue fever and chikungunya fever in France: from bad dream to reality! Clin Microbiol Infect. 2010;16:1702–1704. doi: 10.1111/j.1469-0691.2010.03386.x. [DOI] [PubMed] [Google Scholar]
- 37.Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonizzoni M, Afrane Y, Yan G. Loop-mediated isothermal amplification (LAMP) for rapid identification of Anopheles gambiae and Anopheles arabiensis mosquitoes. Am J Trop Med Hyg. 2009;81:1030–1034. doi: 10.4269/ajtmh.2009.09-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inacio J, Flores O, Spencer-Martins I. Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J Clin Microbiol. 2008;46:713–720. doi: 10.1128/JCM.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parida M, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, Islam MA, Inoue S, Hosaka N, Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]