Abstract
Microbes colonizing and/or infecting chronic wounds undoubtedly play a major and interactive role in impaired healing, especially in amplifying and perpetuating the host innate immune response. The development of molecular techniques to identify and quantify microbial organisms has revolutionized our view of the microbial world. These less-biased, high throughput methods greatly enable investigations regarding host-microbe interactions in the chronic wound environment. This review focuses on the mounting evidence implicating microbes and excessive inflammation in chronic wounds, as well as the challenges associated with understanding how microbes modulate wound healing and the innate immune response.
1 Introduction
Our bodies are colonized inside and out by microbes, estimated to exceed the number of eukaryotic cells of our bodies 10-fold. In most cases, these microbes are harmless and provide functions that we as humans have not had to evolve on our own (Gill et al., 2006). Despite constant confrontation with the immune system, commensal microbial populations peacefully coexist with the host for the most part. The immune system strikes a careful balance between tolerance and activation. Commensal microbes occupy a niche, competing with potentially pathogenic organisms for nutrients and space.
The skin is a critical interface between the human body and its external environment, preventing the loss of moisture and barring entry of pathogenic organisms and foreign substances (Segre, 2006). The skin is also colonized by myriad microbes (the “microbiome”), which in most cases are commensal and cause little harm to the host. However, when the skin barrier is wounded, subcutaneous tissue that is moist, warm, and nutritive is dangerously exposed to the microbial populations colonizing the skin. The presence of microbes rapidly induce the innate immune response, followed by the adaptive immune response, in an effort to combat potential invasion and infection. The immune response must be carefully calibrated to destroy invasive, pathogenic organisms yet avoid an overly exuberant response that harms the host.
This review will describe the mounting evidence suggesting that microbes inhibit wound healing, focusing on genomic approaches of characterizing microbial populations. We will also describe the innate immune response in wound healing, the aberrant innate immune response in chronic wound healing, and how the microbiota may modulate these responses. To develop a complete understanding of wound healing, the interactive nature of the host and colonizing microbiota must be considered.
2 The Burden of Chronic Wounds
Chronic non-healing wounds represent a major health care burden, cause disability, and decrease quality of life. An estimated 15–25% of persons with diabetes will develop foot ulcers (ADA, 1999). One to two percent of the elderly suffer from venous stasis ulcers per year (Margolis et al., 2002). Current direct and indirect costs of chronic wounds are estimated to exceed $12 billion annually in the United States and this number is expected to rise as the population ages (Bickers et al., 2006). Chronic wounds result in considerable morbidity with prolonged hospitalizations, antibiotic exposure, pain, and restricted mobility. A significant number of patients may require amputation, further worsening morbidity (ADA, 1999).
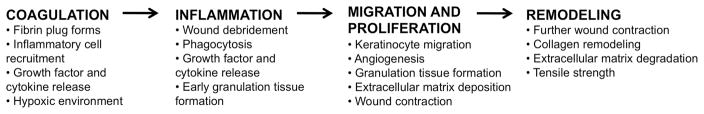
Normal wound repair follows a precisely orchestrated sequence of events of coagulation and hemostasis, inflammation, cell proliferation, cell migration and tissue remodeling, resulting in rapid closure of the wound within 3–14 days (Fig. 1) (Gurtner et al., 2008; Singer and Clark, 1999). Chronic wounds fail to proceed through this series of events and are characterized by persistent inflammation (Loots et al., 1998). Most chronic wounds occur in patients with underlying pathology or systemic disease, which includes an aberrant and/or impaired immune response (i.e. diabetes) (Fonder et al., 2008). Ischemic, necrotic, and/or hypoxic tissue provides an ideal environment for the colonization and proliferation of large microbial populations. Microbial colonization and proliferation induces neutrophil and macrophage infiltration of the, resulting in the wound with subsequent release of damaging free oxygen species, cytotoxic enzymes, and proteases in the wound environment. These molecules destroy cellular components and structural proteins of the extracellular matrix while amplifying persistent inflammation in chronic wounds (Eming et al., 2007).
Fig. 1.
The phases of wound healing and repair. Though depicted as linear, the stages of wound healing are overlapping. Chronic wounds are often characterized by a persistent inflammatory phase.
We postulate that modulating the cycle of inflammation followed by microbial invasion and/or infection is a critical step toward better treatment strategies for chronic wounds. Although there is a general acknowledgment that microbes inhibit the normal wound healing process, their specific role remains unclear. Chronic wounds that fail to progress in healing after 2–4 weeks are commonly treated with systemic and/or topical antibiotics, yet the efficacy of these treatments is uncertain. Unwarranted use of antimicrobials is a major concern as the emergence of antibiotic-resistance bacteria poses a significant threat to public health. Development of better treatment strategies of chronic wounds is dependent on understanding the relationship between the microbiome, the innate immune system, and impaired healing.
3 Surveying the microbial diversity of chronic wounds
The microflora associated with chronic wounds, as examined by traditional culture-based approaches has been reviewed extensively (Bowler and Davies, 1999; Davies et al., 2001; Gardner and Frantz, 2008; Martin et al., 2010). The unifying feature among these studies is that a wide variety of bacterial species can be isolated from chronic wounds, but no convincing link between clinical phenotype and microbial colonization has been established. Staphylococci, streptococci, enterococci, and Pseudomonas spp., are frequently isolated from chronic wounds. However, many of those species isolated in chronic wounds are also resident commensals living on the skin of healthy individuals (Grice et al., 2009).
A major problem with traditional culture-based approaches is that only a small minority of bacteria are able to thrive in isolation (Dunbar et al., 2002). Culture-based techniques essentially select for lab “weeds”, species that flourish under the typical nutritional and physiological conditions employed by diagnostic microbiology laboratories. These are not necessarily the most abundant or influential organisms in the community. Isolation of anaerobes is particularly problematic using routine culture-based approaches (Bowler et al., 2001; Davies et al., 2001). Anaerobic organisms are often slow growing and require special conditions not only for growth but also during sample transport and processing. In the case of chronic wounds, anaerobes are postulated to be particularly pathogenic, both on their own and in synergy with aerobic bacteria (Bowler and Davies, 1999).
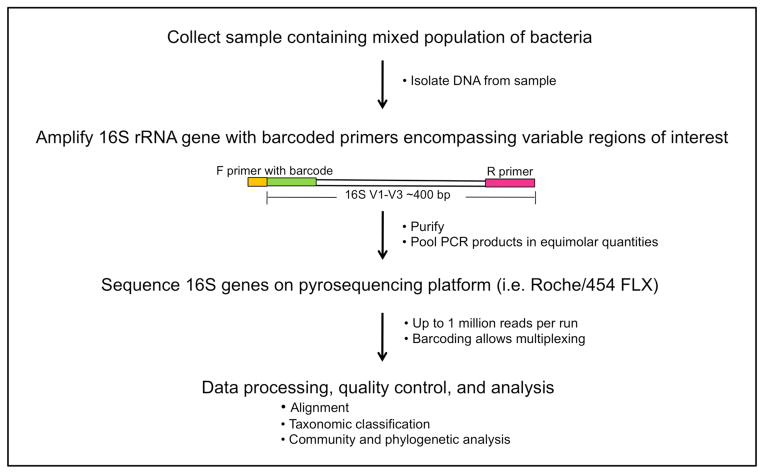
As such, the development of molecular techniques to identify and quantify microbial organisms has revolutionized our view of the microbial world. Characterization of the bacterial microbiome takes advantage of the 16S ribosomal RNA (rRNA) gene, present in all prokaryotes but not eukaryotes (Fig. 2). The 16S rRNA gene encodes a structurally and functionally essential component of the ribosome. The 16S rRNA gene contains species-specific hypervariable regions, allowing taxonomic classification, and highly conserved regions that act as a molecular clock and a substrate for PCR priming (Hugenholtz and Pace, 1996). Following PCR amplification, 16S rRNA genes are sequenced and analyzed (Fig. 2). Classification is enabled by the enormous databases of rRNA gene sequences that have been compiled in an effort to reconstruct the Tree of Life. For example, the Ribosomal Database Project, an online database of rRNA sequences, now contains >600,000 annotated 16S rRNA sequences (Cole et al., 2007). The advent of new sequencing technologies (i.e. 454/Roche pyrosequencing, Illumina, and ABI SOLiD platforms) has massively increased throughput while decreasing the cost of sequencing per base. Importantly, an organism does not need to be cultured to determine its type by 16S rRNA sequencing. Figure 2 outlines the experimental workflow of a typical bacterial microbiome sequencing project. Approaches for analyzing fungal and viral diversity of the human microbiome are currently under development. Fungal diversity of chronic wounds is relatively unexplored but may be a contributing component to wound complications and outcome.
Fig. 2.
Example of an experimental and analytical workflow to survey bacterial diversity of chronic wounds using a genomic approach of sequencing 16S ribosomal RNA genes.
A handful of chronic wound analyses by 16S rRNA sequencing have been performed (Table 1), yet no single organism has been identified in wounds of the same etiology (i.e. diabetic foot ulcer or venous leg ulcer). In a 16S gene survey of debridement material from 40 diabetic foot ulcers, the most prevalent bacterial genus, Corynebacteria, was found in 75% of samples (Dowd et al., 2008). Other common genera (present in at least 15 of the 40 ulcer samples) were Bacteroides, Peptoniphilus, Fingoldia, Anaerococcus, Streptococcus, and Serratia spp. In a study of 24 chronic wounds of mixed etiology, Price et al. report Clostridiales Family XI, which includes many anaerobes, as being most prevalent and abundant (Price et al., 2009). Streptococcaceae spp. were also more abundant among the diabetic ulcers as compared to the venous leg ulcers. In a study of 49 decubitus (pressure) ulcers, the microbial community was found to be highly variable with no clear significance attributed to a single bacterium (Smith et al., 2010).
Table 1.
Organisms associated with chronic wounds by 16S rRNA gene sequencing.
| Organism | Wound type | Respiration mode | Gram stain |
|---|---|---|---|
| Phylum Firmicutes | |||
| Staphylococcus | D,V | FA | + |
| Enterococcus | D | FA | + |
| Clostridium | D,V | OA | + |
| Veillonella | D | OA | − |
| Peptoniphilus | D,V | OA | + |
| Streptococcus | D,P | FA | + |
| Anaerococcus | D,P | OA | + |
| Dialister | P | OA | − |
| Finegoldia | D,P | OA | + |
| Peptostreptococcus | D | OA | + |
| Phylum Proteobacteria | |||
| Proteus | V | FA | − |
| Pseudomonas | D,V | A | − |
| Stenotrophomonas | D,V | A | − |
| Serratia | V,P | FA | − |
| Haemophilus | D | A,FA | − |
| Rhodopseudomonas | D | A | − |
| Citrobacter | D | FA | − |
| Sphingomonas | V | A | − |
| Acinetobacter | V | A | − |
| Phylum Bacteroidetes | |||
| Bacteroides | D,V | OA | − |
| Phylum Actinobacteria | |||
| Corynebacteria | D,V | A,FA | + |
D=diabetic ulcer, V=venous leg ulcer, P=pressure ulcer, FA=facultative anaerobe, OA=obligate anaerobe, A=aerobe, +=gram positive, −=gram negative.
The above-described studies focused on characterizing the bacterial diversity of the wound (determining the relative abundance of organisms) but do not give any indication of total bacterial load. One theory put forth is that wound bioburden and wound outcome are related (Gardner and Frantz, 2008). Wound bioburden refers to three dimensions of wound microbiology: total microbial load, microbial diversity, and presence of pathogenic organisms. While some studies assert that a microbial load >105 is related to poor outcome (Robson et al., 1999), others have challenged this, citing interactions between different species of microbes as more important (Bowler, 2003; Bowler et al., 2001). Therefore, it may be valuable to develop molecular techniques to better quantify microbial load and analyze this data in conjunction with genomic microbial diversity datasets to better predict wound outcomes.
A persistent problem in wound microbiota studies, both cultivation-based and molecular-based, is the lack of uniformity in sampling method. Some studies utilize debridement or curettage material, which is essentially nonviable tissue, to analyze microbial diversity. This is problematic because nonviable tissue likely supports the growth of a greater number and different diversity of organisms. While punch biopsies of viable wound tissue would likely provide the best representation of wound microbiota, obtaining a punch biopsy is not always feasible. Gardner et al. have demonstrated that swab samples obtained by Levine’s technique (Levine et al., 1976) provide comparable measures of wound bioburden when compared to punch biopsies of viable wound tissue (Gardner et al., 2006). Levine’s technique only samples wound microbiota from viable tissue, obtained by expressing tissue fluid from deep tissue layers.
Another challenge with existing genomic surveys of wound microbiota is the lack of meticulous clinical phenotyping and metadata associated with patient samples. Therefore, rigorous analysis of microbiome datasets stratified according to precise clinical criteria is not possible. For example, blood glucose control in diabetic wounds, wound location and topography, and oxygenation of surrounding tissue are several key factors that likely modulate the microbiota and host response. Duration of wound before sampling, presence of infection, and history of antibiotic treatment also need to be considered. Since the etiologies of chronic wounds (diabetes, pressure, venous disease) are likely different and environmental and genetic factors are difficult to control, analyzing host-microbe interactions of specific wound types will likely prove to be critical to draw meaningful conclusions.
Standardizing genetics and environment can be achieved in animal models of impaired wound healing. While animal models may not fully recapitulate human phenotypes of impaired wound healing, they can provide valuable insights that inform our understanding of human diseases. Furthermore, mechanistic analyses that dissect cause and effect of microbiota on impaired wound healing are only possible in animal models. For example, our group has demonstrated that impaired wound healing in a type 2 diabetic mouse model (Leprdb/db, deficient for the leptin receptor) is associated with a quantitative and qualitative shift in microbial colonization (Grice et al., 2010). In this model, diabetic mouse skin is characterized by a much greater bacterial load and greater representation by Staphylococcus spp. than healthy mouse skin. This shift in bacterial colonization is sustained throughout the entire wound healing time course. Other organisms with greater representation in the non-healing wounds included Aerococcus, Klebsiella, and Weissella spp. Diabetic mouse wounds are also colonized by a much lower diversity of organisms as compared to wounds in healthy mice. We postulate that studies in animal models will be critical toward uncovering the mechanistic features that drive the destructive relationship between microbes and the innate immune response in chronic wounds.
4 The skin is the first line of defense
The structural and functional integrity of the epidermis is the critical first line of defense against invasion by foreign and pathogenic substances. As such, the skin is a key component of the innate immune response even before injury occurs. A major challenge of maintaining the integrity of the skin barrier is modulating the immune response upon physical, chemical, or microbial insult. Response to the barrier breach must be carefully balanced between tolerance and activation, to rapidly control microbial invasion and infection without eliciting a potentially harmful, excessive inflammatory response.
The epidermal surface is both a structural and functional antimicrobial shield. Terminally differentiated, enucleated keratinocytes encased in lipid bilayers form the “bricks and mortar” of the stratum corneum, a formidable physical barrier to the exterior environment (Segre, 2006). The lipid component of the epidermis, along with proton pumps, and free amino acids, render the skin surface slightly acidic (pH of approximately 5.0) (Marples, 1965). The acidic and desiccated nature of the skin surface creates a hostile environment for most microorganisms (i.e. Staphylococcus aureus), yet allow colonization by skin commensals that are adapted to these conditions (i.e. Staphylococcus epidermidis). Antimicrobial substances (i.e. antimicrobial peptides, lysozymes, RNases) are secreted in the sweat and sebum, moisturizing the skin surface while dispersing an antimicrobial shield (Elias, 2007).
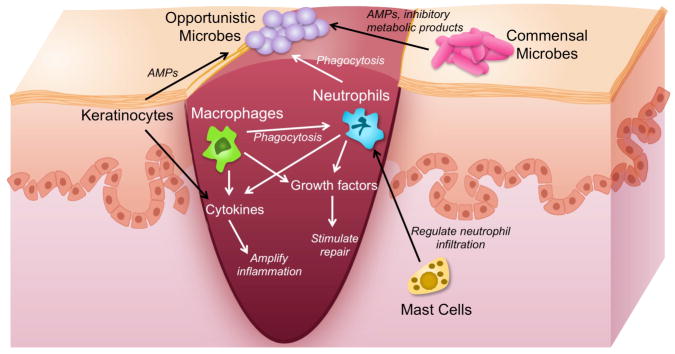
Keratinocytes are a rich source of innate immune sentinels and mediators (Fig. 3)(Nestle et al., 2009). Keratinocytes express pattern recognition receptors (PRRs) including Toll-like receptors (TLRs), mannose receptors, and Nod-like receptors (NLRs). PRRs recognize pathogen-associated molecular patterns (PAMPs; i.e. lipopolysaccharide, peptidoglycan, flagellin, nucleic acids) that are relatively conserved among microorganisms. Activation of TLR receptors induces production of cytokines, chemokines, adhesion molecules, and antimicrobial peptides, ultimately resulting in killing of microorganisms and initiation of the adaptive immune response.
Fig. 3.
Interaction of microbes with the innate immune system during wound healing. Interactions include microbe-microbe and host-microbe. AMP=antimicrobial peptide. Drawing not to scale.
5 The innate immune response during normal wound healing
The innate immune response plays a central role in wound repair, but is especially vital during the coagulative and inflammatory phases (Fig. 3). Immediately following wounding, platelet aggregation and fibrin matrix formation triggers inflammatory and immune cell recruitment (Singer and Clark, 1999). Neutrophils are the first inflammatory cells on the scene, responding to platelet degranulation, complement activation, and bacterial degradation (Grose 2004). Neutrophils clear the wound bed of foreign debris, nonviable tissue, and microorganisms while secreting proteolytic enzymes (i.e. matrix metalloproteases, elastase, cathelpsin G), antimicrobial peptides, reactive oxygen species, and cytokines. At this stage, mast cells may help regulate neutrophil infiltration, and some evidence suggests that they are required for normal cutaneous wound healing in mice (Egozi et al., 2003; Weller et al., 2006).
Neutrophils are gradually replaced by monocytes around day 2–3 post-wounding. Monocytes differentiate and become activated macrophages. In addition to phagocytosing remaining neutrophils, activated macrophages continue to phagocytose microorganisms and clear away debris in the wound bed. Macrophages sense and respond to their environment via Toll-like receptors, complement receptors, and Fc receptors (Gordon, 2003; Karin et al., 2006). A plethora of cytokines and growth factors are secreted by macrophages including TNFα, IL-1β, IL-6, IL-10, ILGF-1, PDGF, VEGF, TGFα, TGFβ, and CSF1.
Upon wounding, keratinocytes, neutrophils, and macrophages are induced to produce antimicrobial peptides (AMPs) (Gallo et al., 1994). AMPs are multi-functional cationic peptides capable of directly killing pathogens, recruiting immune cells, and inducing cytokine production (Lai and Gallo, 2009). Microbes stimulate expression of AMPs primarily through TLR signaling (Abtin et al., 2008; Buchau et al., 2007; Lai et al., 2010; Sumikawa et al., 2006). Importantly, AMPs promote wound healing. The human β-defensins hBD-2, -3, and -4 stimulate keratinocyte migration and proliferation and chemokine/cytokine production (Niyonsaba et al., 2007). Cathelicidin is expressed at high levels upon cutaneous wounding (Dorschner et al., 2001) and inhibition of cathelicidin in wounded organ-cultured human skin resulted in inhibition of re-epithelialization (Heilborn et al., 2003). Cathelicidin-deficient mice (targeted deletion of the Cnlp gene) display impaired wound healing and increased susceptibility to bacterial colonization and infection as compared to their wild-type counterparts (Braff et al., 2005; Nizet et al., 2001). Furthermore, cathelicidin can promote angiogenesis and neovascularization (Koczulla et al., 2003).
The inflammatory phase of wound repair overlaps the migrative/proliferative phase where re-epithelialization and angiogenesis occur. This is followed by the remodeling phase, which can persist for up to a year. Collagen is remodeled and tissue is strengthened, though it is only restored to a fraction of its original strength before wounding (Gurtner et al., 2008). Successful wound repair also requires resolution of inflammation. This can be achieved by downregulation of inflammatory molecules by anti-inflammatory cytokines (i.e. IL-10, TGFβ1) or upregulation of anti-inflammatory molecules (i.e. IL-1 receptor antagonist). Little is known about the mechanisms regulating resolution of the inflammatory phase. As we will see in the next section, unresolved inflammation is a hallmark of chronic wounds.
5 An aberrant innate immune response in chronic wounds
Most chronic wounds, including diabetic, venous, and pressure wounds, are stalled in a chronic inflammatory state (Loots et al., 1998). Microbes are a critical component in amplifying and perpetuating inflammation in the chronic wound environment. Bacteria and their components can directly stimulate the influx of neutrophils and macrophages (Singer and Clark, 1999). Leukocytes can be extraordinarily harmful to the wound environment. Non-viable tissue propagates the cycle of bacterial colonization/infection followed by leukocyte infiltration.
Since invading neutrophils and macrophages are a potent source of proteases, the chronic wound microenvironment is highly proteolytic. Of note, matrix metalloprotease (MMPs) activity is upregulated and MMP-inhibitor activity is downregulated (Eming et al.; Moor et al., 2009; Mwaura et al., 2006; Norgauer et al., 2002; Saarialho-Kere, 1998). As a result, mediators of repair, provisional wound matrix components, and growth factors are targeted and inactivated by proteolytic cleavage (Lauer et al., 2000; Moor et al., 2009; Roth et al., 2006; Wlaschek et al., 1997).
Oxidative stress amplifies chronic inflammation in non-healing wounds. Leukocytes are a major source of reactive oxygen species (ROS; superoxide anion, hydroxyl radicals, hydrogen peroxide, singlet oxygen), rendering the wound microenvironment highly pro-oxidant (James et al., 2003; Mendez et al., 1998; Wenk et al., 2001; Wlaschek and Scharffetter-Kochanek, 2005). ROS can directly damage structural proteins of the extracellular matrix and alter signaling pathways and transcriptional regulation of proinflammatory cytokines and chemokines (Wenk et al., 2001).
While leukocytes are plentiful in the chronic wound environment, their phagocytosis, chemotaxis, and bactericidal activity appears to be diminished, at least in chronic diabetic wounds (Calhoun et al., 2002; Naghibi et al., 1987; Nolan et al., 1978; Zykova et al., 2000). This may in part be due to bacteria interference with cell-matrix interactions (Athanasopoulos et al., 2006; Chavakis et al., 2002). As a result, the wound becomes even more susceptible to increased bacterial burden and infection. Further complicating treatment decisions, many chronic wounds do not outwardly display clinical signs and symptoms of infection despite high bacterial burden and the presence of pathogenic organisms (Gardner et al., 2001). This is likely due to population-specific factors, including tissue perfusion and oxygenation, hyperglycemia, and other aspects of immunocompetence.
Recent data indicates that deficiency of either neutrophils or macrophages is not deleterious to wound healing. In some cases, depletion of either or both cell types can enhance rate of wound repair and decrease scarring (Martin and Leibovich, 2005). This would suggest that there is redundancy present in the inflammatory response and modulating recruitment and activity of different leukocyte lineages may prove to be therapeutically beneficial in chronic wounds.
Activation of TLR pathways by microbial components may be a factor in the chronic inflammation associated with non-healing wounds. Our group recently demonstrated persistent expression of several TLRs (TLR1, 2, 4, 6, 7, 8 and 13) coinciding with impaired healing and a shift in microbes colonizing the wounds of db/db diabetic mice as compared to healthy mice (Grice et al., 2010). Diabetic (streptozotocin-induced) Tlr2−/− mice demonstrated improved wound healing and a decreased inflammatory response as compared to Tlr2+/+ diabetic mice (Dasu et al., 2010). Wound fluids from non-healing chronic venous leg ulcers demonstrated persistent levels of TLR-2 and TLR-4 activity over time, while healing wounds showed diminishing levels as they healed (Pukstad et al., 2010). Activation of TLR receptors ultimately leads to NF-κB-mediated transcription and production of inflammatory cytokines, amplifying the inflammatory state of the chronic wound microenvironment.
AMP production and secretion has a direct effect on microbial killing, and an indirect effect on cytokine/chemokine secretion, angiogenesis, and wound repair. Cathelicidin, an AMP normally upregulated during cutanous wound healing, has been demonstrated to be absent in chronic venous leg ulcers (Dressel et al., 2010; Heilborn et al., 2003). RNase 7, a potent antimicrobial ribonuclease, was also found to be absent, while psoriasin and hBD-2 were upregulated in chronic venous leg ulcers (Butmarc et al., 2004) (Dressel et al., 2010). Proteomic analysis of wound exudates identified increased amounts of the AMPs lactotransferrin, azurocidin-1, lipocalin, and bacterial/permeability-increasing protein in non-healing wounds as compared to healing wounds (Eming et al. 2010).
In a mouse model of type 2 diabetes (db/db), our group has globally analyzed the inflammatory and host defense response associated with impaired wound healing and how it correlates with colonization of microbiota (Grice et al., 2010). Non-healing wounds in db/db mice were characterized by persistent upregulation of inflammatory and host defense genes, including TLR pathway genes, complement pathway genes, and inflammatory cytokine genes. This pattern of gene expression is closely correlated with the relative abundance of Staphylococcus colonizing db/db wounds. While it is clear from this work and others that microbial community structure is closely associated with the host innate immune system, the mechanisms by which microbes interact and contribute to chronic inflammation in non-healing wounds remains unclear.
6 Conclusions and Perspectives
Wound healing is a complex process, further complicated by underlying pathology and systemic disease such as diabetes. The microbiome of the chronic wound undoubtedly plays a major and interactive role in impaired healing, especially in amplifying and perpetuating the host innate immune response. Technological and conceptual advances now provide unprecedented opportunities to fully delineate the role of host-microbe interactions in chronic wound healing. Recent advances in high-throughput sequencing technology allow unparalleled sampling depth for surveying microbial diversity. Generating resources to characterize the human microbiota and its role in health and disease is a major mission of the NIH Roadmap Human Microbiome Project (HMP) (Peterson et al., 2009). Yet interpreting the huge datasets generated by these studies will only provide meaningful results when the study is carefully designed and the experimental procedures meticulously validated. Teasing apart the molecular mechanisms governing host-microbe interactions in chronic wounds will also likely require studies in animal models. Tools for these studies (i.e. germ-free mice, selective colonization) have been developed for gut microbiome analysis but will need to be adapted and optimized for analyzing host-microbe interactions in a wound environment.
Gaining a better understanding of the interaction between wound microbiota and the innate immune system may provide insight into effective non-antimicrobial treatment strategies. Potential therapies could involve manipulating and/or normalizing microbiota, through inhibition of pathogenic bacteria or promotion of symbiotic bacteria, as a low-cost noninvasive target for the management of chronic wounds. Wound microbiome diversity profiling could also be utilized as a biomarker to predict wound outcomes or to identify clinical populations at risk for impaired wound healing or wound complications. On the host side of the relationship, inflammatory and/or innate immune factors may be targeted, for both control of persistent inflammation and to modulate closely associated, potentially pathogenic microbial populations. As we have shown, the cycle of inflammation triggered and amplified by microbial colonization and/or infection is highly deleterious for wound healing. Future studies aimed at dissecting the mechanisms of this relationship, though challenging, will provide a valuable foundation for clinical translation into improved diagnostics and therapeutics.
Acknowledgments
The authors thank Heidi Kong and Sue Gardner for their underlying contributions, thoughtful discussions and critical reading of the manuscript.
Footnotes
The authors have no conflict of interest.
Contributor Information
Elizabeth A. Grice, Email: gricee@mail.nih.gov.
Julia A. Segre, Email: jsegre@nhgri.nih.gov.
References
- Abtin A, Eckhart L, Mildner M, Gruber F, Schroder JM, Tschachler E. Flagellin is the principal inducer of the antimicrobial peptide S100A7c (psoriasin) in human epidermal keratinocytes exposed to Escherichia coli. FASEB J. 2008;22:2168–76. doi: 10.1096/fj.07-104117. [DOI] [PubMed] [Google Scholar]
- ADA . Consensus Development Conference on Diabetic Foot Wound Care: 7–8 April 1999, Boston, Massachusetts. American Diabetes Association. Diabetes Care. 1999;22:1354–60. doi: 10.2337/diacare.22.8.1354. [DOI] [PubMed] [Google Scholar]
- Athanasopoulos AN, Economopoulou M, Orlova VV, Sobke A, Schneider D, Weber H, et al. The extracellular adherence protein (Eap) of Staphylococcus aureus inhibits wound healing by interfering with host defense and repair mechanisms. Blood. 2006;107:2720–7. doi: 10.1182/blood-2005-08-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickers DR, Lim HW, Margolis D, Weinstock MA, Goodman C, Faulkner E, et al. The burden of skin diseases: 2004 a joint project of the American Academy of Dermatology Association and the Society for Investigative Dermatology. J Am Acad Dermatol. 2006;55:490–500. doi: 10.1016/j.jaad.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Bowler PG. The 10(5) bacterial growth guideline: reassessing its clinical relevance in wound healing. Ostomy Wound Manage. 2003;49:44–53. [PubMed] [Google Scholar]
- Bowler PG, Davies BJ. The microbiology of infected and noninfected leg ulcers. Int J Dermatol. 1999;38:573–8. doi: 10.1046/j.1365-4362.1999.00738.x. [DOI] [PubMed] [Google Scholar]
- Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14:244–69. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braff MH, Zaiou M, Fierer J, Nizet V, Gallo RL. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect Immun. 2005;73:6771–81. doi: 10.1128/IAI.73.10.6771-6781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchau AS, Hassan M, Kukova G, Lewerenz V, Kellermann S, Wurthner JU, et al. S100A15, an antimicrobial protein of the skin: regulation by E. coli through Toll-like receptor 4. J Invest Dermatol. 2007;127:2596–604. doi: 10.1038/sj.jid.5700946. [DOI] [PubMed] [Google Scholar]
- Butmarc J, Yufit T, Carson P, Falanga V. Human beta-defensin-2 expression is increased in chronic wounds. Wound Repair Regen. 2004;12:439–43. doi: 10.1111/j.1067-1927.2004.12405.x. [DOI] [PubMed] [Google Scholar]
- Calhoun JH, Overgaard KA, Stevens CM, Dowling JP, Mader JT. Diabetic foot ulcers and infections: current concepts. Adv Skin Wound Care. 2002;15:31–42. doi: 10.1097/00129334-200201000-00011. quiz 4–5. [DOI] [PubMed] [Google Scholar]
- Chavakis T, Hussain M, Kanse SM, Peters G, Bretzel RG, Flock JI, et al. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med. 2002;8:687–93. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–72. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasu MR, Thangappan RK, Bourgette A, DiPietro LA, Isseroff R, Jialal I. TLR2 expression and signaling-dependent inflammation impair wound healing in diabetic mice. Lab Invest. 2010;90:1628–36. doi: 10.1038/labinvest.2010.158. [DOI] [PubMed] [Google Scholar]
- Davies CE, Wilson MJ, Hill KE, Stephens P, Hill CM, Harding KG, et al. Use of molecular techniques to study microbial diversity in the skin: chronic wounds reevaluated. Wound Repair Regen. 2001;9:332–40. doi: 10.1046/j.1524-475x.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol. 2001;117:91–7. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressel S, Harder J, Cordes J, Wittersheim M, Meyer-Hoffert U, Sunderkotter C, et al. Differential expression of antimicrobial peptides in margins of chronic wounds. Exp Dermatol. 2010;19:628–32. doi: 10.1111/j.1600-0625.2009.01030.x. [DOI] [PubMed] [Google Scholar]
- Dunbar J, Barns SM, Ticknor LO, Kuske CR. Empirical and theoretical bacterial diversity in four Arizona soils. Appl Environ Microbiol. 2002;68:3035–45. doi: 10.1128/AEM.68.6.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egozi EI, Ferreira AM, Burns AL, Gamelli RL, Dipietro LA. Mast cells modulate the inflammatory but not the proliferative response in healing wounds. Wound Repair Regen. 2003;11:46–54. doi: 10.1046/j.1524-475x.2003.11108.x. [DOI] [PubMed] [Google Scholar]
- Elias PM. The skin barrier as an innate immune element. Semin Immunopathol. 2007;29:3–14. doi: 10.1007/s00281-007-0060-9. [DOI] [PubMed] [Google Scholar]
- Eming SA, Koch M, Krieger A, Brachvogel B, Kreft S, Bruckner-Tuderman L, et al. Differential proteomic analysis distinguishes tissue repair biomarker signatures in wound exudates obtained from normal healing and chronic wounds. J Proteome Res. 9:4758–66. doi: 10.1021/pr100456d. [DOI] [PubMed] [Google Scholar]
- Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127:514–25. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. Treating the chronic wound: A practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol. 2008;58:185–206. doi: 10.1016/j.jaad.2007.08.048. [DOI] [PubMed] [Google Scholar]
- Gallo RL, Ono M, Povsic T, Page C, Eriksson E, Klagsbrun M, et al. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc Natl Acad Sci U S A. 1994;91:11035–9. doi: 10.1073/pnas.91.23.11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SE, Frantz RA. Wound bioburden and infection-related complications in diabetic foot ulcers. Biol Res Nurs. 2008;10:44–53. doi: 10.1177/1099800408319056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SE, Frantz RA, Doebbeling BN. The validity of the clinical signs and symptoms used to identify localized chronic wound infection. Wound Repair Regen. 2001;9:178–86. doi: 10.1046/j.1524-475x.2001.00178.x. [DOI] [PubMed] [Google Scholar]
- Gardner SE, Frantz RA, Saltzman CL, Hillis SL, Park H, Scherubel M. Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen. 2006;14:548–57. doi: 10.1111/j.1743-6109.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HK, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Snitkin ES, Yockey LJ, Bermudez DM, Liechty KW, Segre JA. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 2010;107:14799–804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–21. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Heilborn JD, Nilsson MF, Kratz G, Weber G, Sorensen O, Borregaard N, et al. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J Invest Dermatol. 2003;120:379–89. doi: 10.1046/j.1523-1747.2003.12069.x. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–7. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- James TJ, Hughes MA, Cherry GW, Taylor RP. Evidence of oxidative stress in chronic venous ulcers. Wound Repair Regen. 2003;11:172–6. doi: 10.1046/j.1524-475x.2003.11304.x. [DOI] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–35. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Koczulla R, von Degenfeld G, Kupatt C, Krotz F, Zahler S, Gloe T, et al. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J Clin Invest. 2003;111:1665–72. doi: 10.1172/JCI17545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–21. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–41. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer G, Sollberg S, Cole M, Flamme I, Sturzebecher J, Mann K, et al. Expression and proteolysis of vascular endothelial growth factor is increased in chronic wounds. J Invest Dermatol. 2000;115:12–8. doi: 10.1046/j.1523-1747.2000.00036.x. [DOI] [PubMed] [Google Scholar]
- Levine NS, Lindberg RB, Mason AD, Jr, Pruitt BA., Jr The quantitative swab culture and smear: A quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma. 1976;16:89–94. [PubMed] [Google Scholar]
- Loots MA, Lamme EN, Zeegelaar J, Mekkes JR, Bos JD, Middelkoop E. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. J Invest Dermatol. 1998;111:850–7. doi: 10.1046/j.1523-1747.1998.00381.x. [DOI] [PubMed] [Google Scholar]
- Margolis DJ, Bilker W, Santanna J, Baumgarten M. Venous leg ulcer: incidence and prevalence in the elderly. J Am Acad Dermatol. 2002;46:381–6. doi: 10.1067/mjd.2002.121739. [DOI] [PubMed] [Google Scholar]
- Marples M. The Ecology of the Human Skin. Charles C Thomas, Bannerstone House; Springfield, Ill: 1965. [Google Scholar]
- Martin JM, Zenilman JM, Lazarus GS. Molecular microbiology: new dimensions for cutaneous biology and wound healing. J Invest Dermatol. 2010;130:38–48. doi: 10.1038/jid.2009.221. [DOI] [PubMed] [Google Scholar]
- Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 2005;15:599–607. doi: 10.1016/j.tcb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Mendez MV, Stanley A, Park HY, Shon K, Phillips T, Menzoian JO. Fibroblasts cultured from venous ulcers display cellular characteristics of senescence. J Vasc Surg. 1998;28:876–83. doi: 10.1016/s0741-5214(98)70064-3. [DOI] [PubMed] [Google Scholar]
- Moor AN, Vachon DJ, Gould LJ. Proteolytic activity in wound fluids and tissues derived from chronic venous leg ulcers. Wound Repair Regen. 2009;17:832–9. doi: 10.1111/j.1524-475X.2009.00547.x. [DOI] [PubMed] [Google Scholar]
- Mwaura B, Mahendran B, Hynes N, Defreitas D, Avalos G, Adegbola T, et al. The impact of differential expression of extracellular matrix metalloproteinase inducer, matrix metalloproteinase-2, tissue inhibitor of matrix metalloproteinase-2 and PDGF-AA on the chronicity of venous leg ulcers. Eur J Vasc Endovasc Surg. 2006;31:306–10. doi: 10.1016/j.ejvs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Naghibi M, Smith RP, Baltch AL, Gates SA, Wu DH, Hammer MC, et al. The effect of diabetes mellitus on chemotactic and bactericidal activity of human polymorphonuclear leukocytes. Diabetes Res Clin Pract. 1987;4:27–35. doi: 10.1016/s0168-8227(87)80030-x. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–91. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyonsaba F, Ushio H, Nakano N, Ng W, Sayama K, Hashimoto K, et al. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J Invest Dermatol. 2007;127:594–604. doi: 10.1038/sj.jid.5700599. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–7. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Nolan CM, Beaty HN, Bagdade JD. Further characterization of the impaired bactericidal function of granulocytes in patients with poorly controlled diabetes. Diabetes. 1978;27:889–94. doi: 10.2337/diab.27.9.889. [DOI] [PubMed] [Google Scholar]
- Norgauer J, Hildenbrand T, Idzko M, Panther E, Bandemir E, Hartmann M, et al. Elevated expression of extracellular matrix metalloproteinase inducer (CD147) and membrane-type matrix metalloproteinases in venous leg ulcers. Br J Dermatol. 2002;147:1180–6. doi: 10.1046/j.1365-2133.2002.05025.x. [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–23. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, et al. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4:e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukstad BS, Ryan L, Flo TH, Stenvik J, Moseley R, Harding K, et al. Non-healing is associated with persistent stimulation of the innate immune response in chronic venous leg ulcers. J Dermatol Sci. 2010;59:115–22. doi: 10.1016/j.jdermsci.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Robson MC, Mannari RJ, Smith PD, Payne WG. Maintenance of wound bacterial balance. Am J Surg. 1999;178:399–402. doi: 10.1016/s0002-9610(99)00208-1. [DOI] [PubMed] [Google Scholar]
- Roth D, Piekarek M, Paulsson M, Christ H, Bloch W, Krieg T, et al. Plasmin modulates vascular endothelial growth factor-A-mediated angiogenesis during wound repair. Am J Pathol. 2006;168:670–84. doi: 10.2353/ajpath.2006.050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarialho-Kere UK. Patterns of matrix metalloproteinase and TIMP expression in chronic ulcers. Arch Dermatol Res. 1998;290(Suppl):S47–54. doi: 10.1007/pl00007453. [DOI] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–8. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AJ, Clark RA. Cutaneous wound healing. N Engl J Med. 1999;341:738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- Smith DM, Snow DE, Rees E, Zischkau AM, Hanson JD, Wolcott RD, et al. Evaluation of the bacterial diversity of Pressure ulcers using bTEFAP pyrosequencing. BMC Med Genomics. 2010;3:41. doi: 10.1186/1755-8794-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumikawa Y, Asada H, Hoshino K, Azukizawa H, Katayama I, Akira S, et al. Induction of beta-defensin 3 in keratinocytes stimulated by bacterial lipopeptides through toll-like receptor 2. Microbes Infect. 2006;8:1513–21. doi: 10.1016/j.micinf.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Weller K, Foitzik K, Paus R, Syska W, Maurer M. Mast cells are required for normal healing of skin wounds in mice. FASEB J. 2006;20:2366–8. doi: 10.1096/fj.06-5837fje. [DOI] [PubMed] [Google Scholar]
- Wenk J, Foitzik A, Achterberg V, Sabiwalsky A, Dissemond J, Meewes C, et al. Selective pick-up of increased iron by deferoxamine-coupled cellulose abrogates the iron-driven induction of matrix-degrading metalloproteinase 1 and lipid peroxidation in human dermal fibroblasts in vitro: a new dressing concept. J Invest Dermatol. 2001;116:833–9. doi: 10.1046/j.1523-1747.2001.01345.x. [DOI] [PubMed] [Google Scholar]
- Wlaschek M, Peus D, Achterberg V, Meyer-Ingold W, Scharffetter-Kochanek K. Protease inhibitors protect growth factor activity in chronic wounds. Br J Dermatol. 1997;137:646. doi: 10.1111/j.1365-2133.1997.tb03804.x. [DOI] [PubMed] [Google Scholar]
- Wlaschek M, Scharffetter-Kochanek K. Oxidative stress in chronic venous leg ulcers. Wound Repair Regen. 2005;13:452–61. doi: 10.1111/j.1067-1927.2005.00065.x. [DOI] [PubMed] [Google Scholar]
- Zykova SN, Jenssen TG, Berdal M, Olsen R, Myklebust R, Seljelid R. Altered cytokine and nitric oxide secretion in vitro by macrophages from diabetic type II-like db/db mice. Diabetes. 2000;49:1451–8. doi: 10.2337/diabetes.49.9.1451. [DOI] [PubMed] [Google Scholar]