Abstract
ATP-binding cassette (ABC) transporters harvest the energy present in cellular ATP to drive the translocation of a structurally diverse set of solutes across the membrane barriers of eubacteria, archaebacteria, and eukaryotes. The positively cooperative ATPase activity (Hill coefficient, 1.7) of a model soluble cassette of known structure, MJ0796, from Methanococcus jannaschii indicates that at least two binding sites participate in the catalytic reaction. Mutation of the catalytic base in MJ0796, E171Q, produced a cassette that can bind but not efficiently hydrolyze ATP. The equivalent mutation (E179Q) in a homologous cassette, MJ1267, had an identical effect. Both mutant cassettes formed dimers in the presence of ATP but not ADP, indicating that the energy of ATP binding is first coupled to the transport cycle through a domain association reaction. The non-hydrolyzable nucleotides adenosine 5′-(β,γ-imino)triphosphate and adenosine 5′-3-O-(thio)triphosphate were poor analogues of ATP in terms of their ability to promote dimerization. Moreover, inclusion of MgCl2, substitution of KCl for NaCl, or alterations in the polarity of the side chain at the catalytic base all weakened the ATP-dependent dimer, suggesting that electrostatic interactions are critical for the association reaction. Thus, upon hydrolysis of bound ATP and the release of product, both electrostatic and conformational changes drive the cassettes apart, providing a second opportunity to couple free energy changes to the transport reaction.
ATP-binding cassette (ABC)1 transporters are ubiquitous membrane proteins that couple ATP hydrolysis to the energy-dependent transport of a wide variety of molecules across lipid bilayers. They comprise the single largest gene family in several sequenced prokaryotic genomes (1). Mutations in human ABC transporters underlie diseases such as cystic fibrosis, hypercholesterolemia, adrenoleukodystrophy, and Stargardt’s disease, while multidrug resistance in cancer cells and infectious microorganisms often arises from the overexpression of ABC transporters that serve as drug efflux pumps (2, 3).
The ABC transporters share an invariant domain organization of two conserved cytoplasmic nucleotide binding cassettes associated with two transmembrane (TM) domains (2). The cassettes contain three highly conserved motifs required for nucleotide binding and hydrolysis: the Walker A site (GX4GK(S/T), where X = any residue) and the Walker B site (RX6 – 8Φ4D, where Φ = hydrophobic residue) (4), which reside in the αβ core of the cassette (5–11), and the LSGGQ signature sequence (1, 2), which lies more toward the periphery of the cassette in an α-helical subdomain (5–11). The TM domains that mediate the movement of the structurally diverse solutes exhibit less sequence conservation (1, 2). The organization of prokaryotic transporter operons and of single polypeptide chain transporters suggests that the minimal functional unit consists of at least two cassettes and two TM domains (5).
The crystal structures of a homodimeric half-ABC transporter (one TM domain and one cassette in each monomer), MsbA (6), and isolated cassettes (7–11) have not resolved the oligomeric organization of these domains. The structures reveal a wide variety of potential, but mutually exclusive, dimeric organizations (6–13). Moreover these structures possess surprisingly open nucleotide binding sites and largely lack physical interactions between the LSGGQ signature sequence and the bound nucleotide, although mutations in this motif affect ATP hydrolysis (14, 15). The crystal structure of the distantly related DNA repair protein Rad50 (12) suggests a resolution of this conundrum and provides a potential mechanism for the power stroke of ABC transporters.
In Rad50, two opposing nucleotide binding domains bind the non-hydrolyzable ATP analogue AMP-PNP with the Walker A and B sites of one monomer and an LSGGQ-like sequence of the other monomer completing the two binding pockets in a dimer that sandwiches the two nucleotides at the interface (12). This arrangement (13) forms a much more occluded active site and is consistent with the effect of LSGGQ sequence mutations on ATP hydrolysis (14, 15). In the absence of AMP-PNP Rad50 is monomeric, suggesting a model for the mechanism of ABC-type ATPases wherein an ATP-driven dimerization of the cassettes couples ATP binding and hydrolysis to useful thermodynamic output (12). In the transporters, the formation of such an ATP-dependent dimer and/or dissociation of the dimer driven by ATP hydrolysis could mediate rearrangements of the TM domains that support solute transport across the membrane. This model suggests that a cassette mutant that binds but is unable to hydrolyze ATP might form a stable ATP sandwich dimer. To test this hypothesis and gain insight into the mechanism of ABC transporters, we mutated the catalytic base of two model archaeal nucleotide binding cassettes and characterized their ability to form nucleotide-dependent dimers.
MATERIALS AND METHODS
Protein Expression and Purification
MJ0796 and MJ1267 expression plasmids (9, 10) were mutated (E171Q and E179Q, respectively) using the QuikChange mutagenesis kit (Stratagene). Both wild type and mutant proteins were purified using previously applied methods (9–10). Wild type and mutant proteins were purified to greater than 99% as assessed by densitometry of Coomassie-stained protein, which migrated near the expected 26,500 Da upon SDS-PAGE (see Fig. 1B).
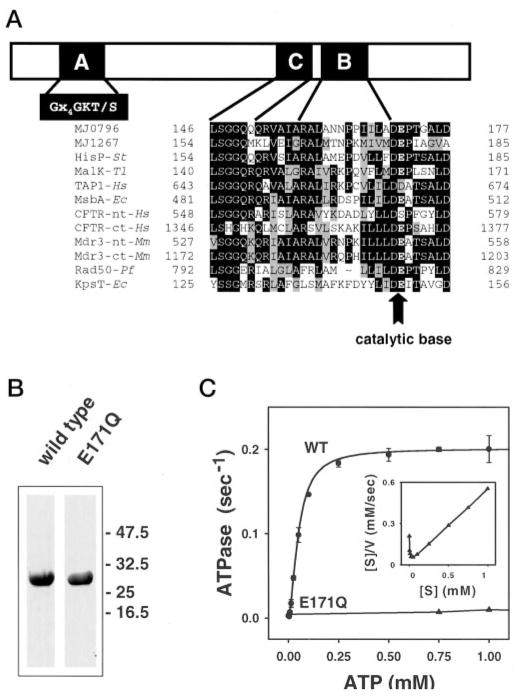
Fig. 1. Mutation of the catalytic base in the ABC ATPase cassettes.
A, consensus sequences in the ATP-binding cassettes of ABC transporters. An alignment of the Walker B and transporter consensus (LSGGQ) regions of several transporters including those of known three-dimensional structure is shown. The Glu (E) (arrow) in the DEP sequence at the end of the Walker B sequence is in position to serve as the catalytic base. B, Coomassie Blue-stained 10% SDS-polyacrylamide gel of wild type MJ0796 and the E171Q mutant. Positions of protein marker bands are illustrated by arrows. C, wild type MJ0796 exhibited cooperative ATPase activity, whereas the mutant E171Q exhibited no measurable activity at ATP levels as high as 5 mM (data not shown). A Vmax of 0.2 s−1, a Km of 50 μM, and a Hill constant of 1.7 were used to produce the fit (solid line). A Hanes-Woolf plot of the data (inset) shows positive cooperativity. WT, wild type.
Dimerization Assays
Cassettes were analyzed on a GFC-200 gel filtration column (TOSOH Bioseparations) at room temperature. Protein samples in 10% glycerol, 10 mM Tris, pH 7.6 (30 μM) were injected and resolved at 1 ml/min flow rate in a mobile phase of 200 mM NaCl, 50 mM Tris-Cl, pH 7.6. Alternatively, analysis of the monomer-dimer transition (16) was carried out in a Beckman XLI centrifuge in an AN60Ti rotor using 56 μM (1.48 mg/ml) protein in the same buffer used for the gel filtration assays except when KCl replaced NaCl or MgCl2 was added as noted in Table I. Data were collected at 17 krpm and 4 °C using interference optics at 675 nm and analyzed using the Beckman Optima software. The calculated molecular weight for the monomer was 26,565, and the partial specific volume was 0.7365. Solvent densities were calculated to be 1.00968 g/ml for the NaCl solutions and 1.01088 for the KCl solutions using Sednterp.
Table I. Analytical ultracentrifugation results.
Data were analyzed using the Beckman Optima software (see “Materials and Methods”).
| [ATP] | [ADP] | [NaCl] | [KCl] | KD | Single species fit |
|---|---|---|---|---|---|
|
| |||||
| mM | μM | Da | |||
| 0 | 0 | 200 | 0 | 208 | 37,286 |
| 0.1 | 0 | 200 | 0 | 134 | 41,612 |
| 0.4 | 0 | 200 | 0 | 147 | 41,345 |
| 2 | 0 | 200 | 0 | 0.071 | 52,884 |
| 10 | 0 | 200 | 0 | 0.019 | 53,079 |
| 0 | 10 | 200 | 0 | 208 | 39,263 |
| 10 | 0 | 0 | 200 | 0.601 | 52,211 |
ATPase Activity
ADP production was assessed by monitoring NADH production via a coupled pyruvate kinase/lactate dehydrogenase assay system as described previously (17).
RESULTS AND DISCUSSION
Mutational analyses of the bacterial cassettes HisP (14) and KpsT (18) and of mouse Mdr3 (19) indicate that a highly conserved glutamic acid residue, found directly adjacent to the Walker B aspartic acid residue in the sequence RX6 – 8Φ4DEP (Fig. 1A), is critical for ATPase and transport activity. This glutamic acid residue is in position to activate a nearby water in the HisP structure and was thus predicted to serve as the catalytic carboxylate (7, 20, 21). To further investigate the role of this glutamic acid residue, we generated either glutamate to glutamine or alanine mutations in MJ0796 and MJ1267, two well characterized ABC transporter nucleotide binding cassettes (9, 10).
ATPase activity was assayed using a coupled ATPase assay (17). Wild type MJ0796 (1 μM) exhibited a Vmax of 0.2 s−1 with a Km of 50 μM (Fig. 1C). The Hanes-Woolf plot (Fig. 1C, inset) presents a shape diagnostic of positively cooperative ATP binding/hydrolysis consistent with the determined Hill coefficient of 1.7. Positive, two-site cooperativity was previously observed for wild type bacterial maltose (22) and histidine (23) transporters. By contrast, the E171Q mutant of MJ0796 had undetectable ATPase activity (Fig. 1C). The equivalent mutation in the MJ1267 cassette (E179Q) also eliminated its ATPase activity (data not shown). These results show that the carboxylate functional group of the highly conserved glutamate residue at the C terminus of the Walker B sequence (Fig. 1A) is required for ATP hydrolysis.
Based on the Rad50 model for the transport cycle, the hydrolysis-deficient glutamate to glutamine mutants of ABC transporter nucleotide binding cassettes would be expected to form a similar ATP-bound homodimer provided their nucleotide binding ability is unaltered by the mutation. To assay the ability of mutant cassettes to form stable, nucleotide-dependent dimers, we performed experiments wherein protein samples were mixed with ADP or ATP and then resolved on a size exclusion column. The data (Fig. 2, A and B) show that both wild type and E171Q mutant MJ0796 migrated as monomers in the absence of ATP. However, the E171Q mutant migrated largely as a homodimer in the presence of ATP (Fig. 2B). Wild type MJ0796 migrated as a monomer regardless of the presence of ATP (Fig. 2A). ADP did not support the formation of a stable dimer of either wild type (Fig. 2A) or mutant (Fig. 2B) MJ0796. Moreover the addition of ADP inhibited ATP-dependent dimer formation in the E171Q mutant (data not shown). The fact that the mutant ATP-containing dimer was stable during elution in the absence of nucleotide in the mobile phase indicates that the nucleotide sandwich dimers only slowly dissociate in the absence of ATP hydrolysis. Consistent with our findings, previous studies of wild type HisP (24) and MalK (25) exhibit no more than a small degree of dimerization in the presence of nucleotide.
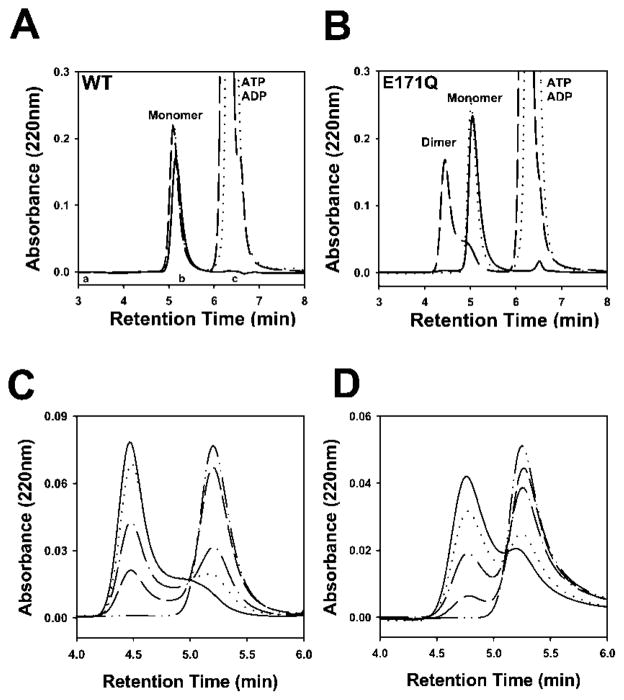
Fig. 2. Analytical gel filtration assays of nucleotide-dependent cassette dimerization.
Wild type (A) or E171Q (B) MJ0796 were analyzed at a 30 μM concentration without nucleotide (——), with 10 mM ATP (— —), or with 10 mM ADP (· · · ·). Column calibration standards are labeled: a, bovine serum albumin (66 kDa); b, carbonic anhydrase (34 kDa); c, lysozyme (14 kDa). Assays were conducted on both the E171Q mutant of MJ0796 (5 μM) in the presence of 0 μM (— · · —), 5 μM (— —), 10 μM (— · —), 20 μM (· · · ·), or 200 μM (——) ATP (C) and the equivalent E179Q mutant of MJ1267 (30 μM) in the presence of 0 μM (— · · —), 25 μM (— —), 50 μM (— · —), 500 μM (· · · ·), or 1 mM (——) ATP (D). WT, wild type.
The dependence of the dimerization of the E171Q mutant of MJ0796 on ATP concentration is shown in the analytical gel filtration data in Fig. 2C. The midpoint of the titration was between 50 and 100 μM, consistent with the Km for ATP hydrolysis observed for wild type MJ0796 (Fig. 2C). Analogous results demonstrating the ATP-dependent dimerization of the E179Q mutant of the MJ1267 nucleotide binding cassette are shown in Fig. 2D. The midpoint of the titration occurred at a slightly higher ATP concentration for this protein. Like MJ0796, wild type MJ1267 did not form stable dimers in the presence of ATP (data not shown).
To determine the energetics of the ATP-dependent homodimerization of MJ0796-E171Q observed by analytical gel filtration, samples were analyzed at a protein concentration of 56 μM by equilibrium analytical ultracentrifugation (data summarized in Table I). In the absence of ATP, a KD of 208 μM was calculated for monomer-homodimer equilibrium, indicating that the protein was present primarily as a monomer under these conditions, consistent with the gel filtration results (Fig. 1B). Again ADP did not support dimerization in the equilibrium analytical ultracentrifugation analysis. By contrast, the addition of 2 mM ATP resulted in a reduction of the KD to 70 nM, indicating that the protein was present primarily as a dimer under these conditions. The steep dependence of the KD on ATP concentration is expected if dimerization is coupled to positively cooperative ATP binding in the symmetric sandwich dimer (27).
The non-hydrolyzable ATP analogues ATPγS and AMP-PNP failed to promote cassette dimerization in gel filtration experiments on wild type MJ0796 and MJ1267 (data not shown) and only poorly promoted dimerization of the mutant cassettes (Fig. 3A). These results suggest that these analogues, while useful in numerous other applications, are not always accurate mimetics of their natural counterparts. The subtle electrostatic and steric differences between conventional nucleotides and the non-hydrolyzable analogs apparently prevent stable cassette dimerization. This observation likely explains the difficulty experienced in isolating wild type cassette dimers in the presence of non-hydrolyzable ATP analogues.
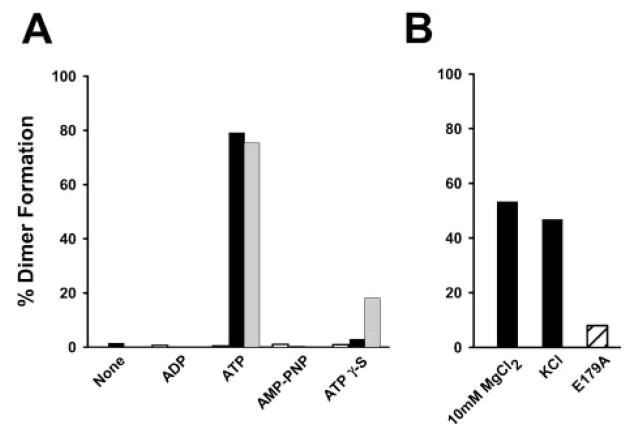
Fig. 3. Effect of non-hydrolyzable nucleotide analogues and cations on dimerization of MJ0796-E171Q and MJ1267-E179Q.
The percentage of dimer in gel filtration assays was calculated based on measurement of peak heights. Protein concentration was 30 μM. A, ADP, ATP, AMP-PNP, and ATPγS were assayed at 10 mM for their ability to form MJ0796-WT (open bars), MJ0796-E171Q (black bars), or MJ1267-E179Q (gray bars) dimers in a buffer containing 200 mM NaCl, 50 mM Tris-Cl, pH 7.6. B, the effects of changes in the salt environment during MJ0796-E179Q dimer formation and column chromatography were evaluated in the presence of 10 mM ATP. Either 10 mM MgCl2 was added to the buffer or 200 mM KCl was substituted for the 200 mM NaCl. The E179A mutant of MJ1267 was also evaluated in the standard NaCl buffer (hatched bar).
The addition of Mg2+ or the substitution of K+ for Na+ during analytical gel filtration and ultracentrifugation experiments inhibited the formation of the MJ0796-E171Q dimer (Fig. 3B and Table I). Addition of 10 mM MgCl2 lowered the dimer level by ~25% in the presence of 10 mM ATP. Likewise a decrease in dimerization was seen when KCl was substituted for NaCl (Fig. 3B and Table I). In KCl the KD for dimerization increased from 20 to 600 nM. Interestingly mutating the catalytic glutamate in MJ1267 to alanine (E179A) diminished ATP-dependent dimerization to 10% of the level achieved with the E179Q mutant (Fig. 3B). The ability of these modifications of the ionic environment to alter the energetics of dimer formation reinforces the idea that carefully balanced electrostatic effects play a critical role in mediating the ATP-dependent dimerization of ABC transporter nucleotide binding cassettes. Upon ATP hydrolysis, additional alterations in the electrostatics of the interface due to deprotonations and/or product release could effectively destabilize the sandwich dimer (Fig. 4). The structure of the stable ATP sandwich dimer of MJ0796-E171Q has been solved by x-ray crystallography and details the specific nature of these interactions.2
Fig. 4. A model for the reaction cycle of ABC transporters based on ATP-dependent dimerization of ATP-binding cassettes.
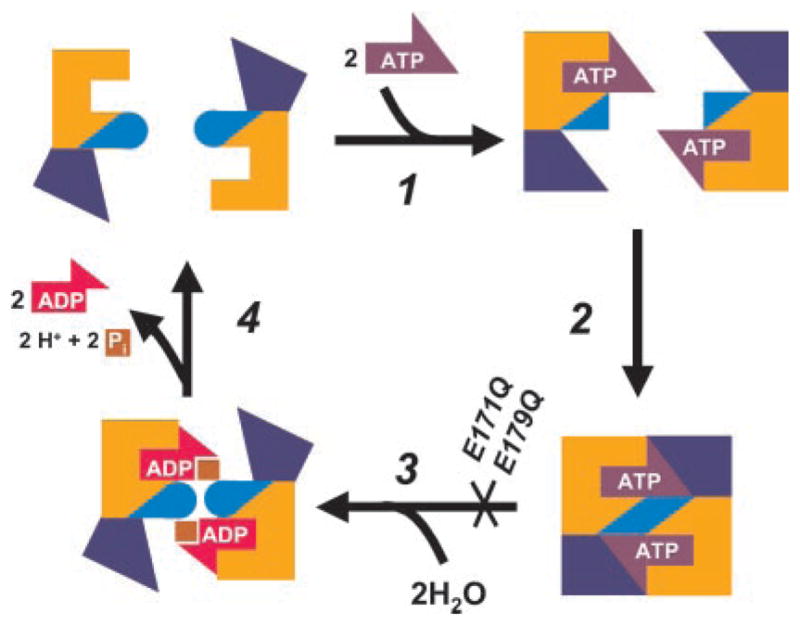
The ATP-binding core and antiparallel β-subdomain are shown in yellow. The initial step (step 1) combines binding of ATP to protein monomers with the α-helical subdomain (dark blue) rotation and γ-phosphate linker (light blue) shift. This step is rapidly followed by cassette dimerization (step 2). Hydrolysis of ATP (step 3) results in the reversal of the previous conformational changes coupled to dissociation of the dimer and release of hydrolysis products (step 4). The E171Q mutant of MJ0796 and the equivalent E179Q mutant of MJ1267 become locked in the dimeric state because hydrolysis cannot occur, and the protein remains in the thermodynamically favorable ATP-bound conformation. The cooperativity of ATP hydrolysis exhibited by wild type MJ0796 (Hill constant of 1.7) supports this model as the dimer formation occurs concomitantly with the cooperative binding of two ATP molecules.
Interestingly in the cystic fibrosis transmembrane conductance regulator (CFTR), a chloride channel (28, 29), only the C-terminal cassette contains the catalytic glutamate residue. A recent study of the non-equivalency of CFTR cassettes demonstrated that nucleotide binds stably and dissociates slowly from the N-terminal cassette, while the C-terminal cassette rapidly hydrolyzes nucleotide (26). Thus, based on the data presented here, it is possible that the N-terminal cassette of CFTR, which contains a serine rather than glutamate at the end of the Walker B sequence, forms associations that stabilize the protein in a substate that has channel activity.
Conformational changes coupled to hydrolysis of the bound ATP also likely play a role in driving the cassettes apart (Fig. 4). Comparison of ADP-bound conformations of the MJ0796 and MJ1276 cassettes with the ATP-bound form of the HisP cassette suggests that ATP binding produces a significant rotation of an α-helical subdomain in the cassettes accompanied by rearrangement of the γ-phosphate linker segment and rotation of a conserved histidine residue out of the active site (7, 9, 10). However, this subdomain rotation is unlikely to be the power stroke of the transport process since the mutation of the phylogenetically invariant glutamine residue (the equivalent of Gln-90 in MJ0796 and Gln-89 in MJ1267) mediating the rotation slows but does not abolish transport.3 In this regard, while these subdomain rearrangements are most likely critical for determining the rate of product release, the best candidate for the power stroke of ABC transporters is the ATP-driven formation of the cassette dimer as proposed by Hopfner et al. (12) and verified in this study. In this model, in the intact transporter the cassette dimer-monomer transition would be coupled to conformational changes in the TM domains that modulate the affinity and differential exposure of a transport substrate binding site on alternating sides of the membrane barrier.
Acknowledgments
We thank Elizabeth Goldsmith and the members of the Thomas laboratory for helpful discussions.
Footnotes
This work was supported by Robert Welch Foundation Grant I-1284 and National Institutes of Health Grant DK49835 (to P. J. T.).
The abbreviations used are: ABC, ATP-binding cassette; AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate; ATPγS, adenosine 5′-3-O-(thio)-triphosphate; TM, transmembrane; CFTR, cystic fibrosis transmembrane conductance regulator.
P. C. Smith, N. Karpowich, L. Millen, J. Moody, J. Rosen, P. J. Thomas, and J. F. Hunt, submitted.
L. Millen, J. E. Moody, and P. J. Thomas, unpublished results.
References
- 1.Young J, Holland IB. Biochim Biophys Acta. 1999;1461:177–200. doi: 10.1016/s0005-2736(99)00158-3. [DOI] [PubMed] [Google Scholar]
- 2.Higgins CF. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Hamon Y, Chimini G. J Lipid Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- 4.Walker JE, Saraste M, Runswick MJ, Gay NJ. EMBO J. 1981;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas PJ, Hunt JF. Nat Struct Biol. 2001;8:920–923. doi: 10.1038/nsb1101-920. [DOI] [PubMed] [Google Scholar]
- 6.Chang G, Roth CB. Science. 2001;293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 7.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 8.Diederichs K, Diez J, Greller G, Muller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF. Structure (Lond) 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 10.Yuan YR, Blecker S, Martsinkevich O, Millen L, Thomas PJ, Hunt JF. J Biol Chem. 2001;276:32313–32321. doi: 10.1074/jbc.M100758200. [DOI] [PubMed] [Google Scholar]
- 11.Gaudet R, Wiley DC. EMBO J. 2001;20:4964– 4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Cell. 2000;101:789– 800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 13.Jones PM, George AM. FEMS Microbiol Lett. 1999;179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 14.Shyamala V, Baichwal V, Beall E, Ames GF. J Biol Chem. 1991;266:18714–18719. [PubMed] [Google Scholar]
- 15.Schmees G, Stein A, Hunke S, Landmesser H, Schneider E. Eur J Biochem. 1999;266:420– 430. doi: 10.1046/j.1432-1327.1999.00871.x. [DOI] [PubMed] [Google Scholar]
- 16.Minton AP. Anal Biochem. 1990;190:1–6. doi: 10.1016/0003-2697(90)90125-s. [DOI] [PubMed] [Google Scholar]
- 17.Rossing J, Harris DA, Kemp A, Slater EC. Biochim Biophys Acta. 1975;376:13–26. doi: 10.1016/0005-2728(75)90201-7. [DOI] [PubMed] [Google Scholar]
- 18.Bliss JM, Garon CF, Silver RP. Glycobiology. 1996;6:445–452. doi: 10.1093/glycob/6.4.445. [DOI] [PubMed] [Google Scholar]
- 19.Urbatsch IL, Julien M, Carrier I, Rousseau ME, Cayrol R, Gros P. Biochemistry. 2000;39:14138–14149. doi: 10.1021/bi001128w. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida M, Amano T. FEBS Lett. 1995;359:1–5. doi: 10.1016/0014-5793(94)01438-7. [DOI] [PubMed] [Google Scholar]
- 21.Geourjon C, Orelle C, Steinfels E, Blanchet C, Deleage G, Di Pietro A, Jault JM. Trends Biochem Sci. 2001;26:539–544. doi: 10.1016/s0968-0004(01)01907-7. [DOI] [PubMed] [Google Scholar]
- 22.Davidson AL, Laghaeian SS, Mannering DE. J Biol Chem. 1996;271:4858– 4863. [PubMed] [Google Scholar]
- 23.Liu CE, Liu PQ, Ames GF. J Biol Chem. 1997;272:21883–21891. doi: 10.1074/jbc.272.35.21883. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido K, Liu PQ, Ames GF. J Biol Chem. 1997;272:27745–27752. doi: 10.1074/jbc.272.44.27745. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy KA, Traxler B. J Biol Chem. 1999;274:6259– 6264. doi: 10.1074/jbc.274.10.6259. [DOI] [PubMed] [Google Scholar]
- 26.Aleksandrov L, Aleksandrov AA, Chang X-b, Riordan JR. J Biol Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- 27.Goldsmith EJ. FASEB J. 1996;10:702–708. doi: 10.1096/fasebj.10.7.8635687. [DOI] [PubMed] [Google Scholar]
- 28.Rich DP, Anderson MP, Gregory RJ, Cheng SH, Paul S, Jefferson DM, McCann JD, Klinger KW, Smith AE, Welsh MJ. Nature. 1990;347:358–363. doi: 10.1038/347358a0. [DOI] [PubMed] [Google Scholar]
- 29.Bear CE, Li CH, Kartner N, Bridges RJ, Jensen TJ, Ramjeesingh M, Riordan JR. Cell. 1992;68:809– 818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]