Abstract
The proteasome plays a central role in the degradation of regulatory and misfolded proteins. Current models suggest that substrates access the internal catalytic sites by processively threading their termini through the gated substrate channel. Here, we found that latent (closed) and activated (open) proteasomes degraded two natively disordered substrates at internal peptide bonds even when they lacked accessible termini, suggesting that these substrates themselves promoted gating of the proteasome. This endoproteolysis provides a molecular mechanism for regulated release of transcription factors from inactive precursors as well as a means of accessing internal folding defects of misfolded multidomain proteins.
The proteasome degrades the majority of cellular proteins in eukaryotes. Its activity allows for surveillance by the immune system, controls the levels of various regulatory proteins, and prevents the accumulation of misfolded mutant and damaged proteins (1). The architecture of the proteasome occludes its active sites within the lumen of a cylinder formed by four heptameric rings (2, 3). This arrangement prevents well-folded proteins from entering the constricted annulus, protecting them from degradation, and has led to a model by which unfolded substrates are fed from their termini into the central catalytic cavity as extended chains and are degraded processively (4, 5). However, this model cannot account for the proteasome-dependent release of active transcription factor domains from inactive precursors by partial degradation (6 – 8). For example, the p50 subunit of nuclear factor κ B transcription factor is produced by proteasome-dependent cotranslational processing of p105 (6). Because p50 constitutes the NH2-terminus of p105 and the COOH-terminus is presumably blocked by the ribosome, the proteasome must either wait for release of nascent p105 from the ribosome or activate another protease that catalyzes the initial endoproteolytic step, as in the proteasome-dependent activation of Esp1 protease during sister chromatid exchange (9). Alternatively, the proteasome itself could enter ribosomally bound p105 at an internal site and initiate processing with an endoproteolytic cleavage.
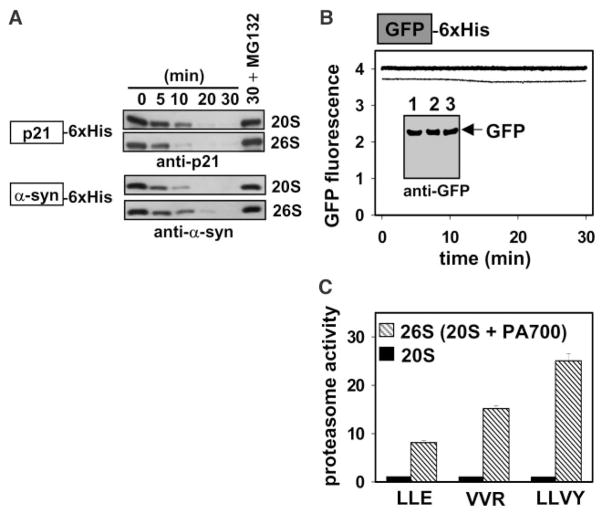
To determine whether proteasome-mediated degradation is initiated from an internal site of a protein, we used two physiological substrates: the cyclin-dependent kinase (CDK) inhibitor p21cip1 (p21) and α-synuclein (α-syn). Both proteins are “natively disordered” (10) and are efficiently degraded by the proteasome in the absence of polyubiquitin modification (11, 12). We compared the degradation of these unstable substrates by the latent 20S proteasome (13) with that of the activated, assembled 26S particle (14). The 26S particle, the accepted physiological form of the proteasome, is composed of the 20S core particle and the PA700 (19S) regulatory cap (14). PA700 contains polyubiquitin (15, 16) and misfolded protein (17, 18) binding sites, isopeptidase activity (19, 20), and adenosine triphosphatase subunits presumably involved in unfolding substrates (21). Both the 20S and 26S proteasomes efficiently degraded p21 and α-syn (Fig. 1A) (22). In contrast, the hyperstable green fluorescent protein (GFP) (melting temperature, Tm, > 65°C) (23) was not degraded by either 20S or 26S (Fig. 1B). The disordered substrates were efficiently degraded by the latent 20S proteasome despite its inability to hydrolyze short peptide substrates (Fig. 1C). Thus, unfolded proteins could open the “gate” that controls access to the otherwise occluded catalytic sites, thereby initiating a process similar to that employed by the proteasome regulators PA700 (24) and PA28 (25).
Fig. 1.
Proteasomal degradation of natively disordered substrates. (A) For p21 degradation, 400 nM p21 was incubated with 5 nM latent 20S or active 26S proteasome at 37°C in buffer A [20 mM Tris-HCl, pH = 7.1; 200 mM NaCl; 10 mM MgCl2; 0.25 mM adenosine triphosphate (ATP); and 1 mM dithiothreitol (DTT)]. For α-syn degradation, 500 nM α-syn was incubated with 10 nM latent 20S or active 26S proteasome at 37°C in buffer B (20 mM Tris-HCl, pH = 7.1; 20 mM NaCl; 10 mM MgCl2; 0.25 mM ATP; and 1 mM DTT). The degradation time course was monitored by immunoblotting with monoclonal antibodies to p21 or α-syn. 6XHis indicates the hexameric histidine metal affinity tag. 30+ MG132 represents the thirty minute time point in the presence of the proteasome inhibitor. (B) 100 nM GFP was incubated with 20 nM 20S or 26S proteasome at 37°C in buffer B. The stability of GFP was assessed by continuously monitoring fluorescence (λex/em = 395/508 nm; +20S, thin trace; +26S, thick trace) and by immunoblotting (inset) with an antibody to GFP at the beginning (lane 1) and on completion of fluorescence monitoring (+20S, lane 2; +26S, lane 3). (C) The latency of the 20S proteasome (black bars) was assessed by comparing its multiple catalytic activities to the activated 26S proteasome (hatched bars) with the use of fluorogenic peptide substrates Z-Leu-Leu-Glu-βNA (LLE, capase-like activity), Z-Val-Val-Arg-AMC (VVR, trypsin-like activity), and suc-Leu-Leu-Val-Tyr-AMC (LLVY, chymotrypsin-like activity) (14), where Z is benzyloxycarbonyl, βNA is 2-napthylamine, AMC is 7-amino-4-methylcoumarin, and suc is succinyl.
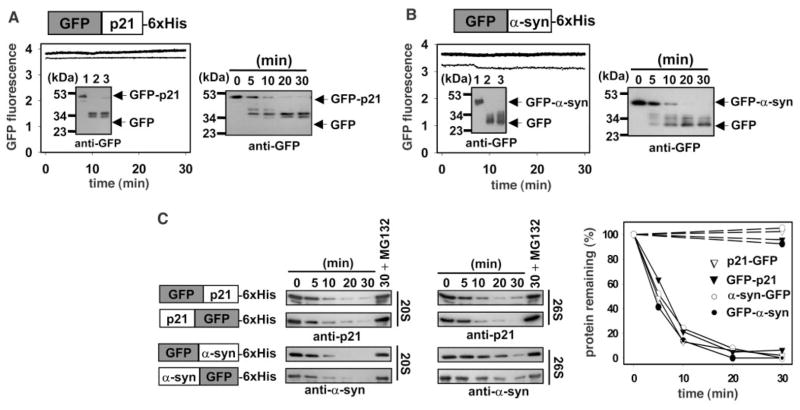
When fusions between the unfolded substrates and the stable GFP were assessed, both the latent 20S and the active 26S proteasomes efficiently degraded the p21 and α-syn domains but not the GFP domain of each fusion protein (Fig. 2, A and B), suggesting that additional factors were required for efficient unfolding of this stable domain. We also examined whether either the 20S or the 26S proteasome exhibited a directional preference by comparing the degradation of fusion proteins that contained GFP at either the NH2- or COOH-terminus (Fig. 2C). The relative activities of the 20S and 26S proteasomes were equivalent toward the p21 fusions. For the α-syn fusions, the 20S proteasome degraded the substrates faster, although the 26S reaction did go to completion (inset, Fig. 2B). Apparently, the shorter α-syn, when blocked with GFP, was less accessible to the longer channel of the 26S proteasome. In this regard, the 26S and 20S proteasomes exhibited similar activity toward unprotected α-syn (Fig. 1A) and circular α-syn whose termini were blocked by a peptide bond. For all substrates, degradation was effectively inhibited by MG132 (Fig. 2C), a potent proteasome inhibitor. Finally, the proteasome degraded the substrates blocked at either terminus at equivalent rates (Fig. 2C), indicating that the reported preference for COOH- to NH2-terminal degradation (4) is not a general feature of all substrates.
Fig. 2.
Proteasomal degradation of unilaterally protected p21 and α-syn. (A and B) 100 nM GFP-p21 and GFP–α-syn fusion proteins were incubated with 20 nM 20S or 26S proteasome in buffer A (GFP-p21) or buffer B (GFP–α-syn). Reactions were monitored by immunoblotting (inset) and fluorescence monitoring as in Fig. 1. A precursor-product relationship was demonstrated during the time course of degradation with the use of 20S proteasome (right half of each panel). (C) 100 nM substrates with GFP either NH2-terminal to [as in (A) and (B)] or COOH-terminal to the p21 or α-syn moieties were incubated with 10 nM 20S or 26S proteasome as in (A), and the degradation time course was assessed by immunoblotting. Substrate remaining in the 20S proteasome reactions in the absence (solid lines) or presence (dashed lines) of MG132 was quantitated by densitometry and normalized to the 0 time point.
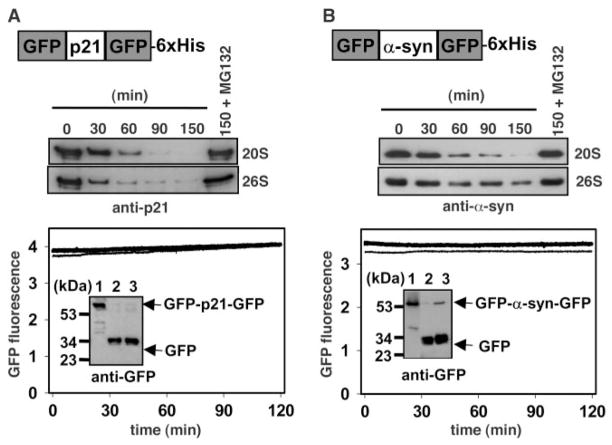
To assess the ability of the proteasome to cleave these substrates at internal sites, we protected both termini of p21 and α-syn by sandwiching them between nondegradable GFP domains (Fig. 3). As assessed by immunoblotting, GFP-p21-GFP and GFP–α-syn–GFP proteins were degraded by the 20S and 26S proteasomes, and degradation was inhibited by MG132. The rate of degradation of bilaterally protected substrates was slower than the degradation of p21 alone, α-syn alone (Fig. 1A), and substrates protected at a single terminus (Fig. 2C), indicating that the endoproteolytic cleavage was less efficient than degradation from the termini. Again, neither GFP-blocking domain was degraded to any measurable extent in the sandwich proteins, as shown by the stable fluorescence (Fig. 3). Thus, free termini are not required for either the 26S or 20S proteasome to efficiently degrade either p21 or α-syn, suggesting that degradation can be initiated at an internal endoproteolytic site.
Fig. 3.
Proteasomal degradation of bilaterally protected p21 and α-syn. Substrates with two GFP domains flanking either an internal p21 (A) or α-syn (B) domain were analyzed as in Fig. 2. For the degradation time course, 100 nM substrate was incubated with 10 nM 20S or 26S proteasome. For fluorescence monitoring, the amount of proteasome was increased to 20 nM.
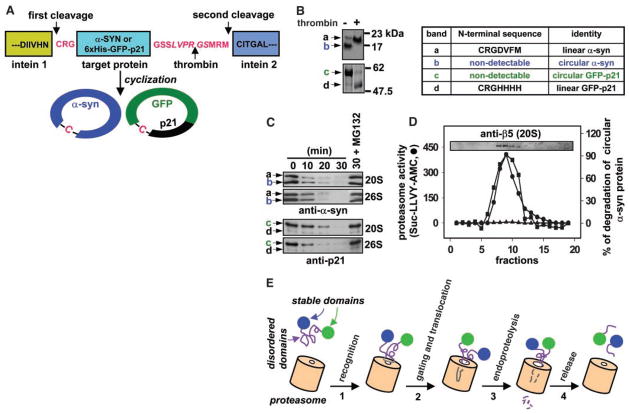
To formally test this possibility, we produced circular α-syn and GFP-p21 proteins that lacked NH2- and COOH-termini with the use of a protein splicing technique (26, 27) (Fig. 4A). Purified α-syn formed two products after the cyclization process (Fig. 4B). The predominant form, band b, was converted to band a after digestion with thrombin, indicating that band b was the circular α-syn. Furthermore, band b produced no signal from NH2-terminal amino acid sequence analysis, consistent with it being the circular variant, whereas the NH2-terminal sequence Cys-Arg-Gly-Asp-Val-Phe-Met for band a indicated that it is the linear α-syn (Fig. 4B). Similarly, band c was circular GFP-p21 and band d was linear GFP-p21 (Fig. 4B). The circular protein composed 80 to 95% of each preparation as determined by Coomassie-stained SDS–polyacrylamide gel electrophoresis (SDS-PAGE). Like linear α-syn, the cyclized α-syn protein showed a random coil conformation (28). Circular α-syn protein migrated faster than linear α-syn on SDS-PAGE, as typically observed (27). In contrast, the linear GFP-p21 protein migrated faster than the circular variant, as previously observed for variants of the maltose-binding protein (27).
Fig. 4.
Proteasomal degradation of circular p21 and α-syn. (A) Circular α-syn and p21 substrates were synthesized and purified with the use of the IMPACT-TWIN system (New England Biolabs, Beverly, MA). Cys-Arg-Gly (CRG) and Gly-Ser-Ser-Leu-Val-Pro-Arg-Gly-Ser-Met-Arg-Met sequences were introduced at the NH2- and COOH-termini, respectively, of both proteins to promote cyclization and encode a thrombin site. In the circular schematic, -C-indicates the cysteine residue of CRG, which becomes the NH2-terminus after intein 1 processing, before cyclization. (B) α-syn (top) and GFP-p21 (bottom) cyclization reaction products were incubated without (−) or with (+) thrombin and analyzed by Coomassie-stained SDS-PAGE. Excised bands from the (−)-thrombin lanes were also subjected to NH2-terminal amino acid sequencing as indicated in the table. (C) 500 nM α-syn or 120 nM GFP-p21 cyclization reaction products (linear plus circular) were incubated with 10 nM 20S or 26S proteasome at 37°C in buffer A (for p21) or buffer B (for α-syn), and the degradation time course was followed by immunoblotting. The α-syn antibody preferentially recognizes the linear protein in the preparation (compare to Fig. 4B). (D) Proteasomes were subjected to 10 to 40% glycerol gradient sedimentation. Proteasome-containing fractions were identified by immunoblotting with antisera to the 20S β5 subunit (inset). Proteasomal peptidase activity (left axis) was assayed with the LLVY substrate as in Fig. 1. Proteasomal circular–α-syn endoproteolysis (right axis) was assayed with (▲) or without (■) MG132 as in (C). Immunoblots were quantitated by densitometry. (E) A model for 20S proteasome action. The 20S proteasome recognizes the disordered domain of a substrate (1). Binding gates the channel, and a substrate loop translocates into the 20S catalytic chamber (2). Degradation initiates by endoproteolysis at an internal site followed by processive degradation of the unstable domain (3). Cleavage ceases, and the substrate is released when a stable domain that cannot enter the annulus is encountered (4).
Purified 26S and 20S proteasomes effectively degraded both circular substrates and were inhibited by MG132 (Fig. 4C), again leaving the GFP moiety of the p21-GFP substrate as in Figs. 2 and 3 (29). This endoproteolysis activity cosedimented with the intact proteasome complex on glycerol gradients (Fig. 4D), excluding the possibility that contaminating proteases or fragments of the proteasome were responsible.
These results call into question several fundamental aspects of currently favored models of proteasome function and regulation. First, they demonstrate that the proteasome can catalyze endoproteolytic cleavage of polypeptide bonds (Fig. 4E). These data differ appreciably from models in which proteasome-mediated degradation is restricted to processive hydrolysis of a polypeptide chain from a free NH2- or COOH-terminus (4, 5) and instead support a model in which a disordered polypeptide chain loop can enter the axial channel to permit initial endoproteolytic cleavage. In this regard, crystal structures of the active proteasome with open access gates (24, 30) could easily accommodate a β-hairpin structure. Studies with some disulfide-trapped substrates (31) but not others (32) suggest that there is space for up to three extended chains within the chamber.
Second, little difference in the rates of endoproteolytic cleavage of these disordered substrates was detected between latent 20S and active 26S proteasomes (Figs. 1 to 4) in which the status of the gate that controls entry to the central axial channel of the proteasome is closed and open respectively (Fig. 1C) (24, 30). Physiological regulators of the proteasome, such as PA700 (19S cap) and PA28, increase proteasome activity in part by opening this gate, thereby increasing access of substrates to the proteasome’s catalytic centers (24, 25). The ability of closed, latent 20S proteasome to catalyze cleavage of these natively disordered, physiological substrates suggests they possess certain features that also promote “gating” of the proteasome (Fig. 4E), features that folded proteins lack. This mechanism suggests a potential role for the free 20S proteasome found in the absence of bound regulatory proteins in many cells (33). It is possible that these inherent signals could target substrates directly for 20S proteasomal degradation without the need for polyubiquitin modification.
Acknowledgments
We thank S. J. Elledge (Baylor College of Medicine) for his generous gift of p21cDNA; R. Nussbaum (National Human Genome Research Institute) for α-syn cDNA; C. Wigley, R. Stidham, and S. Muallem for critical suggestions; and members of our laboratories for helpful comments. This work was supported by grants from the Welch Foundation (P.J.T.), Muscular Dystrophy Association (G.N.D.), and NIH [grants DK46818 (G.N.D.) and DK49835 (P.J.T.)].
Footnotes
References and Notes
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Unno M, et al. Structure. 2002;10:609. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 3.Lowe J, et al. Science. 1995;268:533. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 4.Navon A, Goldberg AL. Mol Cell. 2001;8:1339. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 5.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 6.Lin L, DeMartino GN, Greene WC. Cell. 1998;92:819. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 7.Hoppe T, et al. Cell. 2000;102:577. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 8.Rape M, et al. Cell. 2001;107:667. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 9.Nasmyth K, Peters JM, Uhlmann F. Science. 2000;288:1379. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 10.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr Biochemistry. 1996;35:13709. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 11.Sheaff RJ, et al. Mol Cell. 2000;5:403. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 12.Tofaris GK, Layfield R, Spillantini MG. FEBS Lett. 2001;509:22. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- 13.McGuire MJ, McCullough ML, Croall DE, DeMartino GN. Biochim Biophys Acta. 1989;995:181. doi: 10.1016/0167-4838(89)90078-2. [DOI] [PubMed] [Google Scholar]
- 14.Ma CP, Vu JH, Proske RJ, Slaughter CA, DeMartino GN. J Biol Chem. 1994;269:3539. [PubMed] [Google Scholar]
- 15.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. Nature. 2002;416:763. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 16.Devereaux Q, Ustrell V, Pickart C, Reichsteiner M. J Biol Chem. 1994;269:7059. [PubMed] [Google Scholar]
- 17.Strickland E, Hakala K, Thomas PJ, DeMartino GN. J Biol Chem. 2000;275:5565. doi: 10.1074/jbc.275.8.5565. [DOI] [PubMed] [Google Scholar]
- 18.Braun BC, et al. Nature Cell Biol. 1999;1:221. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 19.Yao T, Cohen RE. Nature. 2002;419:403. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 20.Verma R, et al. Science. 2002;298:611. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 21.Liu CW, et al. J Biol Chem. 2002;277:26815. doi: 10.1074/jbc.M201782200. [DOI] [PubMed] [Google Scholar]
- 22.Materials and Methods are available as supporting material on Science Online.
- 23.Ward WW, Bokman SH. Biochemistry. 1982;21:4535. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 24.Kohler A, et al. Mol Cell. 2001;7:1143. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 25.Whitby FG, et al. Nature. 2000;408:115. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 26.Perler FB. Cell. 1998;92:1. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 27.Evans TC, Jr, Benner J, Xu MQ. J Biol Chem. 1999;274:18359. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 28.The far-ultraviolet circular dichroism spectrum of the mixed population of circular and linear α-syns exhibits a predominant negative absorption at circa 198 nm, characterisitic of a random coil conformation.
- 29.GFP fluorescence was stable during the degradation of the linear and circular GFP-p21 substrates, as in Figs. 1 to 3.
- 30.Groll M, et al. Nature Struct Biol. 2000;7:1062. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 31.Lee C, Prakash S, Matouschek A. J Biol Chem. 2002;277:34760. doi: 10.1074/jbc.M204750200. [DOI] [PubMed] [Google Scholar]
- 32.Wenzel T, Baumeister W. Nature Struct Biol. 1995;2:199. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 33.DeMartino GN, Slaughter CA. J Biol Chem. 1999;274:22123. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]