Abstract
One approach to circumvent barriers to clinical implementation of pharmacogenetics is to employ pre-prescription genotyping that requires interrogation of multiple pharmacogenetic variants using a high-throughput platform. We compared the performance of the DMET Plus array (1,931 variants in 225 genes) with orthogonal genotyping methods in 220 pediatric patients. A total of 1,692 variants had call rates above 98% and were in Hardy Weinberg equilibrium. Of these, 259 were genotyped by at least one independent method and a total of 19,942 SNP-patient sample pairs were evaluated. The concordance was 99.9% with only 28 genotype discordances observed. For those genes deemed most likely to be clinically relevant (TPMT, CYP2D6, CYP2C19, CYP2C9, VKORC1, DPYD, UGT1A1, and SLCO1B1), a total of 3,799 SNP-patient sample pairs were evaluable and had a concordance of 99.96%. We conclude that the DMET Plus array performs well with primary patient samples when compared to multiple other lower-throughput genotyping methods.
INTRODUCTION
Genetic variants that encode for metabolizing enzymes and drug transporters have been demonstrated to affect interindividual drug responses by altering the absorption, distribution, metabolism or elimination (ADME) of drugs.(1–4) Pharmacogenetic testing has the potential to improve medication use through the pre-emptive detection of clinically significant genetic variants, such as single nucleotide polymorphisms (SNPs), insertions, deletions, and duplications that can drive therapy individualization.(2) One of the barriers to clinical implementation of pharmacogenetics includes a lack of affordable, array-based genotyping technologies that can be performed in a CLIA-certified laboratory. Array-based genotyping would allow for interrogation of a sufficient number of polymorphisms related to the ADME of medications, suggesting that pharmacogenetic testing could become pre-emptive, with test results being available on a per-patient basis to the clinician at the time prescribing is contemplated, rather than waiting for a test result on a drug-by-gene-test basis.
The Affymetrix DMET Plus array is one such technology that types 1,931 variants in 225 ADME genes.(5, 6) The platform contains both rare and common variants, and the array interrogates biallelic and triallelic SNPs, copy number variations, and insertion/deletions.(7, 8) The array uses molecular inversion probes (MIPs) that amplify and hybridize independently of genomic sequences as well as universal primers and tag sequences.(7) The platform uses a single-sample genotype calling method that compares each marker to an expected signal distribution defined by large training sets at Affymetrix.(9)
Few data are available on the performance characteristics of DMET Plus genotyping in patient samples or on its concordance with orthogonal genotyping methods. Herein, we report on the performance characteristics of a high-throughput pharmacogenetics array, assayed in a clinical genetics laboratory, using patient samples, with emphasis on those genes that are most clinically relevant.
RESULTS
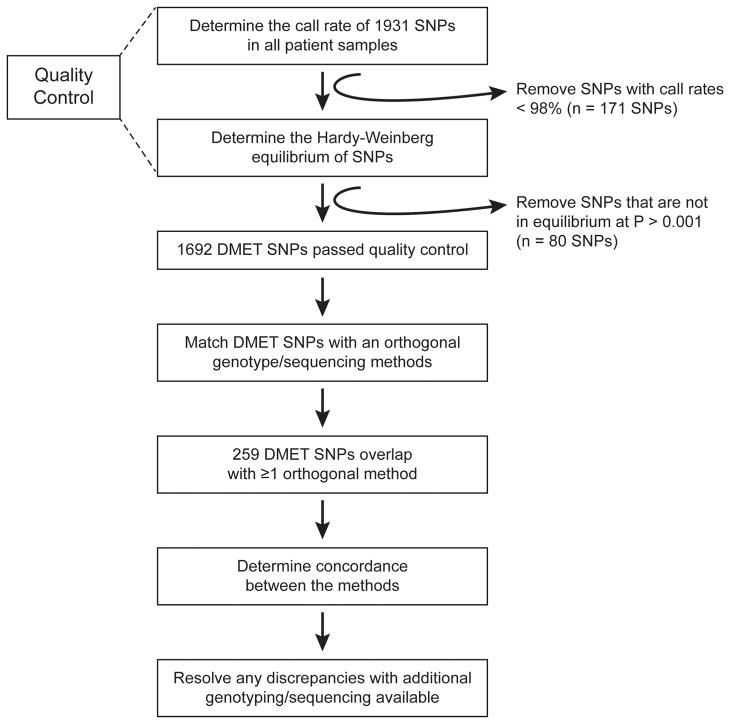
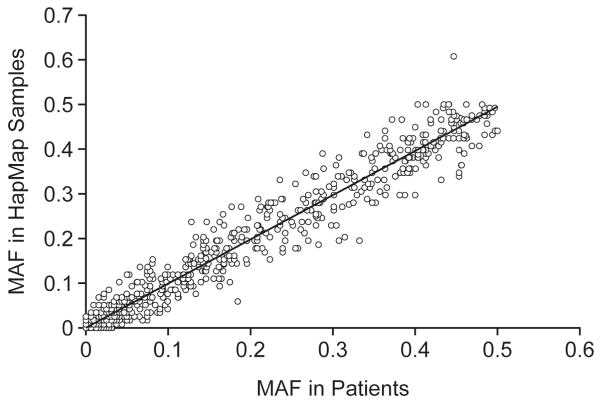
The concordance between the DMET Plus genotypes and all orthogonal genotypes available (Table 1) was determined for all evaluable DMET variants that passed quality control (Figure 1). Quality control filters applied to DMET genotypes showed that of the 1,931 SNPs included in the DMET array, 1,760 SNPs (91.1%) met the call rate criterion and 1,851 met the HWE criterion (Figure 1, Supplemental Table 1). The MAF of SNPs that passed QC in patients of European ancestry was highly correlated to that in HapMap CEU samples (Figure 2, R2 = 0.9711).
Table 1.
Concordance between DMET genotyping and orthogonal methods.
| Genotype Platform | Number of SNPs that overlapped with those typed on DMET Plus | Number of samples typed by both methods | Total Number of SNP-patient samples typed | Number of discordant genotypes | % Concordant |
|---|---|---|---|---|---|
| Beckman Coulter GenomeLab SNPstream, DNA Print Genomics | 7 | 22–34 | 192 | 5 | 97.4% |
| Gene Chip Human Mapping Array (SNP6/500K), Affymetrix | 167 | 88 | 14,637 | 21 | 99.9% |
| Prometheus TPMT Genetics | 3 | 215 | 644 | 2 | 99.7% |
| Sanger Sequencing, Affymetrix | 26 | 67 | 1,736 | 0 | 100% |
| Custom-designed Golden Gate Array, Illumina | 78 | 32 | 2,492 | 0 | 100% |
| iplex Gold MAssARRAY platform, Sequenom | 8 | 31 | 241 | 0 | 100% |
Figure 1. Performance of the DMET Plus array.
The Affymetrix Drug Metabolizing Enzymes and Transporters (DMET) genotyping platform scans 1,931 different variants. The accuracy of the platform was determined for SNPs that passed two quality control criteria. The first quality control filter removed any SNPs with call rates below 98%, and the second filter removed any SNPs that were not in Hardy-Weinberg equilibrium (HWE) at a P-value < 0.001. A total of 1,692 SNPs passed QC and the concordance of genotypes was determined between the DMET array and those available from orthogonal genotyping. A total of 259 SNPs were assessed by DMET and at least one additional genotyping method.
Figure 2. Concordance of DMET MAF with HapMap CEU MAF.
The minor allele frequency (MAF) of all SNPs that passed quality control was determined in patients of European ancestry (x-axis) and compared to the corresponding frequency in HapMap CEU samples (y-axis). Linear regression between the DMET and HapMap allele frequencies suggest that the values between the populations are highly correlated (R2 = 0.9711).
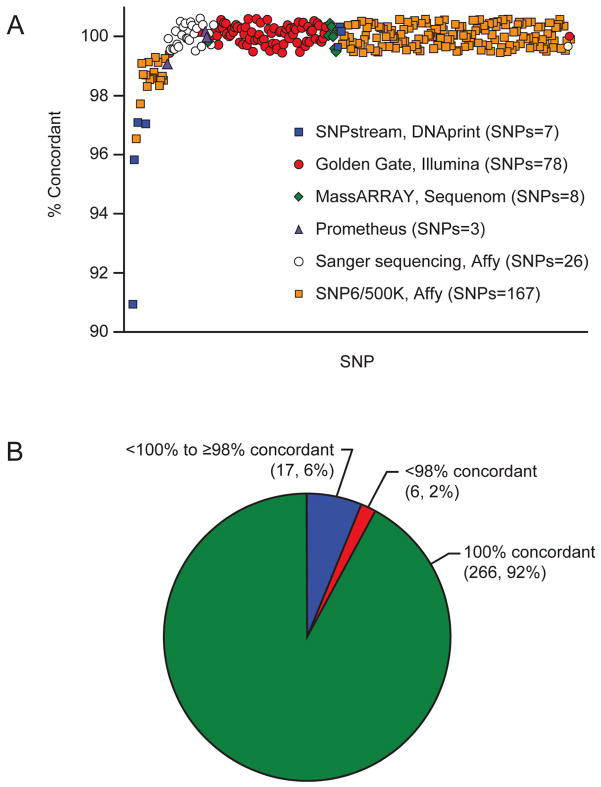
A total of 259 SNPs passing quality control were assessed by the DMET array and at least one orthogonal method. Of the 259 SNPs, 233 SNPS were assessed by one orthogonal method, 22 SNPs were assessed by two orthogonal methods, and 4 SNPs were assessed by three orthogonal methods, for a total of 289 tests of concordance between an orthogonal method and a DMET SNP call. A total of 19,942 SNP-patient pairs were evaluable for concordance with at least one method. Of these, 19,914 SNP calls were concordant (28 discordant) between DMET and the orthogonal methods for an overall concordance of 99.9%.
Genotype concordances were compared with data obtained by several orthogonal methods. Data from the Affymetrix Gene Chip 500K or 6.0 assays for 167 DMET SNPs in 88 patients were compared to results generated by the DMET Plus (Figure 3A, orange squares). The majority of these SNPs had a concordance above 98% (166 SNPs), and only rs2266780 in the FMO3 gene and rs1800440 in the CYP1B1 gene were below the concordance threshold (Supplemental Table 2, 96.5% and 97.7, respectively). Three SNPs in TPMT were genotyped in 215 patients by a reference laboratory (Figure 3A, purple triangles). Genotypes for all three SNPs were highly concordant: both rs1142345 and rs1800462 (contained in the *3A, *3C, *3D and *2 alleles) were 100% concordant and rs1800460 (contained in the *3A,*3B, *3D alleles) had a concordance of 99.1% (Supplemental Table 2). Data obtained from the Illumina Golden Gate assay for 78 SNPs in 32 patients were compared to results from the DMET Plus (Figure 3A, red circles), and there were no SNPs below the concordance threshold of 98%. MassARRAY technology was used to interrogate 8 SNPs in 31 patients (Figure 3A, green diamonds) and each SNP was 100% concordant with the DMET genotyping. Exons of seven genes were sequenced (Sanger) and covered 26 SNPs in 67 patients (Figure 3A, white circles), and these results were 100% concordant with DMET. The SNPs genotyped using the Beckman Coulter Genome Lab SNPstream had the overall lowest concordance with DMET. A total of 7 SNPs were interrogated by SNPstream and DMET (Figure 3A, blue squares), and the number of patients per SNP varied from 22 for rs1800566 to 34 for rs6195. Of these 7 SNPs, only SNPs rs6195, rs10264272, and rs7080681 achieved > 98% concordance. The lowest concordance was for rs1800566 at 91% and the 4 SNPs below the concordance threshold averaged a concordance of 95%. There was a single discordant call responsible for the poor performance of rs2274407, rs1045642, and rs776746, and two discordant calls for rs1800566 (Table 2).
Figure 3. Concordance of the DMET Plus array with other genotyping methods.
The DMET genotyping was compared to all other sequencing and genotyping data available in 220 patients. A total of 259 SNPs were assessed by the DMET array and at least one orthogonal method. Of these, 233 SNPS were assessed by one orthogonal method, 22 SNPs were assessed by two orthogonal methods, and 4 SNPs were assessed by three orthogonal methods, yielding a total of 289 tests of concordance between an orthogonal method and a DMET SNP call. (A) 167 DMET SNPs were in common with the Affymetrix Gene Chip (orange squares), 3 SNPs with Prometheus TPMT genotyping (purple triangles), 78 with the Illumina Golden Gate chip (red circles), 8 with the Sequenom iplex Gold MassARRAY platform (green diamonds), 26 with Sanger sequencing (white circles), and 7 with Beckman Coulter GenomeLab SNPstream (blue squares). (B) A threshold of 98% was used to determine SNPs with poor concordance. SNPs below this threshold (6, 2%) were further investigated, where possible, by comparing the discordant genotypes with any other overlapping genotyping methods.
Table 2.
All DMET discordant genotypes for all SNPs with < 98% concordance between DMET and other orthogonal methods.
| Discordant Patient ID | rsID | Gene | DMET Genotype | SNPstream Genotype | SNP6/500K Genotype | Golden Gate Genotype | Concordant Methods |
|---|---|---|---|---|---|---|---|
| 1 | rs1800566 | NQO1 | C/C | C/T | C/C | C/C | DMET, SNP6/500K, and Golden Gate |
| 2 | rs1800566 | NQO1 | C/T | T/T | C/T | C/T | DMET, SNP6/500K, and Golden Gate |
| 2 | rs1045642 | ABCB1 | C/C | C/T | C/C | None Available | DMET & SNP6/500K |
| 2 | rs776746 | CYP3A5 | A/G | G/G | A/G | A/G | DMET, SNP6/500K, and Golden Gate |
| 3 | rs2274407 | ABCC4 | G/G | T/G | None Available | None Available | None Concordant |
| 1 | rs2266780 | FMO3 | A/A | None Available | G/A | None Available | None Concordant |
| 4 | rs2266780 | FMO3 | A/A | None Available | G/A | None Available | None Concordant |
| 5 | rs2266780 | FMO3 | A/A | None Available | G/A | None Available | None Concordant |
| 6 | rs1800440 | CYP1B1 | A/G | None Available | G/G | None Available | None Concordant |
| 7 | rs1800440 | CYP1B1 | A/A | None Available | G/A | None Available | None Concordant |
Overall, only 6 SNPs in 6 different genes failed to reach the 98% concordance threshold (Figure 3B). A total of 10 genotype calls were responsible for the 6 failed SNPs (Table 2). Additional genotyping by a 2rd and 3th orthogonal method was available for 4 of these samples (Table 2). The two discordant samples for rs1800566 were assessed by 4 genotyping platforms (i.e., DMET, SNP6/500K, Golden Gate, SNPstream), and for both samples, the DMET genotyping was concordant with the SNP6/500K and Golden Gate, but not with SNPstream. Moreover, rs1045642 and rs776746 (also genotyped using SNPstream) were discordant with the additional orthogonal genotyping method (Table 2). Thus, our evidence indicates that of the 10 discordances affecting 6 SNPs, 4 discordances (in 3 SNPs) favor the DMET results being correct, leaving just 3 SNPs unresolved (rs2274407, rs1800440 and rs2266780).
DMET includes 164 SNPs in 8 highly clinically relevant genes (TPMT, CYP2D6, CYP2C19, CYP2C9, VKORC1, DPYD, UGT1A1, and SLCO1B1) that are already subject to testing in some clinical settings. Of these, 148 SNPs (90%) passed the call rate and HWE QC criteria (Table 3), and 46 of those SNPs were genotyped by at least one orthogonal method. A total of 3,799 SNP-patient sample pairs were evaluated within these 8 genes and there was an overall concordance of 99.96% (Supplemental Table 2).
Table 3.
Concordance for the highest priority pharmacogenes.
| Genes | Number of variants on DMET passing QC (call rate + HWE) | Median of call rates for all variants in gene | Methods used in addition to DMET Plus | Number of samples typed by at least 1 orthogonal method | Total Number of SNP-patient samples typed | Number of discordant genotypes | % Concordant |
|---|---|---|---|---|---|---|---|
| TPMT | 8 | 99.9 | Sanger Sequencing, Prometheus, Golden Gate | 67, 215, 32 | 906 | 2 | 99.8% |
| CYP2D6 | 26 | 99.8 | Sanger Sequencing | 67 | 268 | 0 | 100.0% |
| CYP2C19 | 15 | 98.1 | Sanger Sequencing, Golden Gate | 67, 32 | 431 | 0 | 100.0% |
| CYP2C9 | 16 | 98.4 | Sanger Sequencing, Golden Gate | 67,32 | 230 | 0 | 100.0% |
| VKORCI | 21 | 99.2 | SNP6/500K, Sanger Sequencing, Golden Gate | 88, 67, 32 | 608 | 0 | 100.0% |
| DPYD | 15 | 98.5 | Sanger Sequencing, Golden Gate | 67, 32 | 165 | 0 | 100.0% |
| UGT1A1 | 30 | 99.3 | SNP6/500K, Sanger Sequencing, Golden Gate | 88, 67, 32 | 626 | 1 | 99.8% |
| SLCO1B1 | 17 | 99.5 | SNP6/500K, MassARRAY, Golden Gate | 88, 31, 32 | 565 | 0 | 100.00% |
DISCUSSION
In the current study, the performance of the DMET Plus array was assessed in 259 SNPs in 111 different genes using samples from 220 patients and included up to six different orthogonal genotyping methods (Table 1). These genes encoded for 27 P450 enzymes, 44 non-P450 enzymes, and 32 transporters. The concordance in our study varied from 91.0% to 100%, and only 6 variants assessed had a concordance below 98%. All orthogonal methods had a concordance greater than 98%, except for the SNPstream genotyping that had a concordance with DMET of 97.4% (Table 1). The high degree of correlation between the DMET and HapMap CEU allele frequencies in SNPs that passed QC further supports the accuracy of the DMET genotyping (Figure 2). Of the 6 genes carrying SNPs with < 98% concordance between DMET and orthogonal methods, none are currently used routinely for clinical decisions, and three with multiple orthogonal methods favored the DMET genotype being correct.
Of the 225 genes included on the DMET array, 8 genes are high-priority clinical pharmacogenes as identified by the Clinical Pharmacogenetics Implementation Consortium (www.pharmgkb.org).(10) A total of 3,799 SNP-patient sample pairs were assessed within these 8 genes and a concordance of 99.96% was observed, indicating good performance of the array for clinically relevant polymorphisms.
The DMET Plus array has been previously tested for accuracy in two other studies. The larger of the two studies found > 99% concordance for 165 DMET variants across 27 genes in 91 samples (74 Epstein-Barr virus-transformed cell lines and 17 samples from normal donor blood) that had been sequenced at those 165 loci.(11) The second comparison was included in a pharmacogenetics study of docetaxel and thalidomide in patients with prostate cancer using the DMET platform.(12) DMET genotypes were compared in 8 variants across 8 genes in 47 patients using direct sequencing, and the concordance varied from 96 to 100% with an average concordance of 98.6%.(12) Thus, the current study represents the largest number of patients and the broadest spectrum of genotypes for assessing the DMET array against orthogonal genotyping methods. With the development of more comprehensive clinical genetic testing platforms such as we have described herein, and a migration from single gene tests to gene panels or genome sequencing, additional genetic information will be available at the time of testing that may not directly affect the diagnostic question or patient care. Therefore, when deployed in the clinic, these platforms need careful consideration with regard to medical data management, consent for withholding results, and communication to patients and their respective health care providers.
We conclude that the DMET Plus array is a highly accurate method for determining the genotype of a large number of genetic polymorphisms that have potential clinical utility for optimizing drug therapies.
METHODS
Study Population
Genotyping with the DMET Plus array was performed using DNA from 220 patients enrolled on the PGEN5 protocol at St. Jude Children’s Research Hospital. DNA was extracted from blood using the Qiagen Blood and Cell Culture DNA extraction kit. This study was approved by the St. Jude Institutional Review Board, and consent was obtained from the parents of children younger than 18 years of age or from patients older than 18 years of age.
Genotyping and Quality Control Methods
The DMET Plus array uses MIP technology to amplify the sequence-specific targets at each SNP. The PCR products generated with the MIP undergoes enzymatic fragmentation and end labeling followed by hybridization to an array containing allele-specific oligonucleotides used for SNP genotyping. The preparation and processing protocol for patient samples followed the methods described by Burmester et al, (13) and was performed under CLIA-compliant conditions in a clinical genetics laboratory at the Medical College of Wisconsin.
Quality control filters applied to genotypes were as follows. Probe sets with call rates below 98% were removed from the analysis of concordance, as were any SNPs that were not in Hardy-Weinberg equilibrium (HWE) (P-value < 0.001). HWE was determined in patients with European ancestry using an exact test included in the R genetics package, and SNPs on the X chromosome were not evaluated (46 SNPs).(14) No patient samples were removed from the analysis due to poor sample call rates (all samples had calls generated for > 95% of the SNPs). Ancestry was estimated using STRUCTURE as described previously,(15) based on the ancestry-informative SNPs interrogated on the DMET array (Table 4). The minor allele frequency (MAF) for DMET SNPs was calculated in those patients with > 95% European ancestry and compared to available allele frequencies determined in 59 CEU samples from the HapMap extended diversity panel.(6, 16) The correlation between the DMET and HapMap allele frequencies values was determined in SNPs that passed QC using linear regression analysis.
Table 4.
Patient characteristics.
| Total number of patients | 220 |
|---|---|
| Ancestry | |
| European | 159 |
| African | 28 |
| Other | 23 |
| Asian | 10 |
| Gender | |
| Female | 87 |
| Male | 133 |
Several orthogonal methods were used to genotype subsets of the samples. These included the Affymetrix Human Mapping 500K Array Set and the Affymetrix Genome-Wide Human SNP Array 6.0, as we previously described.(17) SNP genotyping was also determined using a custom-designed Illumina Golden Gate assay (San Diego, CA) that was performed at the SNP Center at Johns Hopkins University(18), the iPLEX Gold assay on the MassARRAY platform from Sequenom (San Diego, CA) that was performed at the University of Chicago’s Genetic Services Laboratories (19, 20), and using the Beckman Coulter GenomeLab SNPstream (Fullerton, CA) that was performed by DNAprint.(21) The major non-functional alleles of TPMT (*2, *3A and *3C) were determined using a TaqMan assay by a clinical reference lab (Prometheus Labs, San Diego, CA). Sanger sequencing of exons on 7 genes (TPMT, CYP2D6, CYP2C19, CYP2C9, VKORC1, DPYD, and UGT1A1) was performed by Polymorphic DNA Technologies (Alameda, CA). Concordance between methods was assessed by comparing whether the genotype calls gave identical results for each sample assessed by the methods. A genotype concordance for each variant greater than or equal to 98% between the two methods was considered acceptable performance.
Supplementary Material
STUDY HIGHLIGHTS.
What is the current knowledge on the topic?
Genetic testing for pharmacogenetic genes is currently conducted on a gene by gene basis. With the development of higher throughput platforms, expanding this to a panel of relevant genes becomes an attractive and cost effective approach.
What question this study addressed?
This study evaluates the performance characteristics of a genotyping array for the analysis of a large number of pharmacogenetics genes.
What this study adds to our knowledge?
This study shows for the first time the performance characteristics of a novel genotyping platform. This method is highly accurate and can provide a cost effective approach for comprehensive pharmacogenetic gene analysis.
How this might change clinical pharmacology and therapeutics?
These results should be relevant to clinicians who are interested in pharmacogenetics as well as clinical labs which offer genotyping for genes relevant in pharmacogenetics. Ultimately, the results from this report provide support in favor of a more comprehensive genotyping approach.
Acknowledgments
Supported by NCI grants CA 142665, CA 36401, and CA 21765 and the NIH/NIGMS Pharmacogenomics Research Network (U01 GM92666), and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Authorship Contributions:
Contribution: U.B., M.V.R. and C.A.F. conceived, designed the project, interpreted the data and drafted the manuscript. W.Y. and C.S. performed the statistical analyses; N.K. managed data. All authors contributed to the writing of the manuscript.
Conflict of Interest/Disclosure:
M.V.R and W.E.E: receive a portion of the income St. Jude receives from licensing patent rights related to TPMT and GGH polymorphisms.
Affymetrix paid for Sanger sequencing of 7 genes in a subset of samples. All other authors have no financial disclosures.
References
- 1.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–49. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 2.Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8:283–9. doi: 10.1097/00008571-199808000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Dervieux T, Meshkin B, Neri B. Pharmacogenetic testing: proofs of principle and pharmacoeconomic implications. Mutat Res. 2005;573:180–94. doi: 10.1016/j.mrfmmm.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 4.Sakurai A, Tamura A, Onishi Y, Ishikawa T. Genetic polymorphisms of ATP-binding cassette transporters ABCB1 and ABCG2: therapeutic implications. Expert Opin Pharmacother. 2005;6:2455–73. doi: 10.1517/14656566.6.14.2455. [DOI] [PubMed] [Google Scholar]
- 5.Deeken J. The Affymetrix DMET platform and pharmacogenetics in drug development. Curr Opin Mol Ther. 2009;11:260–8. [PubMed] [Google Scholar]
- 6.Affymetrix. DMET™ Plus Premier Pack allele translation reports. Summary of comprehensive drug disposition genotyping into commonly recognized allele names. 2011 Jun 8; http://media.affymetrix.com/support/technical/whitepapers/dmet_plus_translation.pdf.
- 7.Dumaual C, et al. Comprehensive assessment of metabolic enzyme and transporter genes using the Affymetrix Targeted Genotyping System. Pharmacogenomics. 2007;8:293–305. doi: 10.2217/14622416.8.3.293. [DOI] [PubMed] [Google Scholar]
- 8.Sissung TM, English BC, Venzon D, Figg WD, Deeken JF. Clinical pharmacology and pharmacogenetics in a genomics era: the DMET platform. Pharmacogenomics. 2010;11:89–103. doi: 10.2217/pgs.09.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Affymetrix. DMETTM Plus genotyping and copy number methods. 2011 Jun 8; http://media.affymetrix.com/support/technical/whitepapers/dmet_plus_algorithm_whitepaperv1.pdf.
- 10.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly TM, et al. Multiplex assay for comprehensive genotyping of genes involved in drug metabolism, excretion, and transport. Clin Chem. 2007;53:1222–30. doi: 10.1373/clinchem.2007.086348. [DOI] [PubMed] [Google Scholar]
- 12.Deeken JF, et al. A pharmacogenetic study of docetaxel and thalidomide in patients with castration-resistant prostate cancer using the DMET genotyping platform. Pharmacogenomics J. 2010;10:191–9. doi: 10.1038/tpj.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burmester JK, Sedova M, Shapero MH, Mansfield E. DMET microarray technology for pharmacogenomics-based personalized medicine. Methods Mol Biol. 2010;632:99–124. doi: 10.1007/978-1-60761-663-4_7. [DOI] [PubMed] [Google Scholar]
- 14.Warnes G, Leisch F. Population Genetics. R package. (1.1.0) 2004 [Google Scholar]
- 15.Yang JJ, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43:237–41. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansfield E, et al. Extensive population-based variation in drug metabolism is revealed by analyzing HapMap DNA with the Affymetrix® DMET™ Plus Array (abstract) 2009 http://media.affymetrix.com/community/events/2009-ashg/2009-ashg-pdfs/Variation-in-Drug-Metabolism-Revealed-by-DMET-Plus-Array.pdf.
- 17.Trevino LR, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–5. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen R, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Tantisira KG, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13:1353–9. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 20.Jones TS, et al. CRHR1 polymorphisms predict bone density in survivors of acute lymphoblastic leukemia. J Clin Oncol. 2008;26:3031–7. doi: 10.1200/JCO.2007.14.6399. [DOI] [PubMed] [Google Scholar]
- 21.French D, et al. A PAI-1 (SERPINE1) polymorphism predicts osteonecrosis in children with acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2008;111:4496–9. doi: 10.1182/blood-2007-11-123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.