Abstract
Many cellular responses to stimulation of cell-surface receptors by extracellular signals are transmitted across the plasma membrane by hydrolysis of phosphatidylinositol-4,5-bisphosphate (PIP2), which is cleaved into diacylglycerol and inositol-1,4,5-tris-phosphate by phosphoinositide-specific phospholipase C (PI-PLC). We present structural, biochemical, and RNA expression data for three distinct PI-PLC isoforms, StPLC1, StPLC2, and StPLC3, which were cloned from a guard cell-enriched tissue preparation of potato (Solanum tuberosum) leaves. All three enzymes contain the catalytic X and Y domains, as well as C2-like domains also present in all PI-PLCs. Analysis of the reaction products obtained from PIP2 hydrolysis unequivocally identified these enzymes as genuine PI-PLC isoforms. Recombinant StPLCs showed an optimal PIP2-hydrolyzing activity at 10 μm Ca2+ and were inhibited by Al3+ in equimolar amounts. In contrast to PI-PLC activity in plant plasma membranes, however, recombinant enzymes could not be activated by Mg2+. All three stplc genes are expressed in various tissues of potato, including leaves, flowers, tubers, and roots, and are affected by drought stress in a gene-specific manner.
In animal cells many cellular responses to stimulation of cell-surface receptors by extracellular signals are transmitted across the plasma membrane by hydrolysis of PIP2, which is catalyzed by PI-PLC. This reaction generates the two secondary messengers: IP3, a compound soluble in the cytosol that triggers transient increases of the cytosolic Ca2+ level, and DG, a lipid that stays within the plasma membrane and activates PKC (Berridge, 1993). In plants an enzyme with PKC activity has been partially purified from Brassica campestris (Nanmori et al., 1994), and evidence has been obtained for the participation of PKC in the elicitor-induced defense response in potato (Solanum tuberosum; Subramaniam et al., 1997). DG has been shown to induce both ion pumping in patch-clamped guard cell protoplasts and the opening of intact stomata (Lee and Assmann, 1991). Furthermore, it is now well established that IP3-mediated Ca2+ release occurs in plant cells (Drøbak, 1992, 1993; Coté and Crain, 1993, 1994).
Increasing evidence strongly suggests that IP3-mediated signal transduction is functional in guard cells. The phytohormone ABA, which is involved in multiple stress responses, also triggers stomatal closure and inhibits stomatal opening. Extracellular application of ABA or injection of this phytohormone into guard cells of Commelina communis and Vicia faba results in transient increases in cytosolic Ca2+ (McAinsh et al., 1990; Schroeder and Hagiwara, 1990). In addition, Lee et al. (1996) demonstrated that ABA induces an increase in cytosolic IP3 concentration within seconds after the extracellular application of ABA to V. faba guard cell protoplasts, which is accompanied by a rapid turnover of inositol phospholipids. Photolysis of “caged” IP3 within guard cells of C. communis (Gilroy et al., 1990) and V. faba (Blatt et al., 1992) causes an increase of cytosolic Ca2+ and a subsequent stomata-closing reaction. Finally, increasing concentrations of both Ca2+ (Schroeder and Hagiwara, 1989) and IP3 (Blatt, 1992) reduce inward-rectifying K+ currents in plasma membranes of V. faba guard cells. Inward-rectifying K+ channels are believed to be involved in driving the opening of guard cells.
Current evidence supports a model in which the transduction of the ABA signal in guard cells utilizes changes in cytosolic concentrations of IP3 and Ca2+ and ultimately leads to the inactivation of inward-rectifying K+ channels and stomatal closure. However, in C. communis ABA can induce stomatal closure by a second, Ca2+-independent pathway (Allan et al., 1994). In addition, both cyclic ADP-Rib and IP3 independently are able to trigger Ca2+ release from plant vacuoles (Allen et al., 1995). Therefore, it appears that guard cells operate different, and in part complementary, signal transduction pathways that all lead to stomatal closure.
To investigate the role of the IP3-mediated pathway in guard cells from a new angle we decided to clone cDNAs of PI-PLCs and other enzymes of the phosphoinositide-signaling pathway expressed in epidermal fragments and possibly guard cells of potato leaves. Epidermal fragments were used as the source for the isolation of guard cell mRNA, even though it is possible to isolate mRNA from guard cell protoplasts (Nakajima et al., 1995) or from single plant cells (Karrer et al., 1995). These latter methods, however, exhibit inherent analytical problems, as discussed previously (Kopka et al., 1997b).
Phosphoinositide metabolism in higher plants has recently been approached by molecular cloning of cDNAs encoding multiple PI-PLC isoforms from Arabidopsis thaliana (Hirayama et al., 1995, 1997; Yamamoto et al., 1995) and Glycine max (Shi et al., 1995; GenBank accession nos. U41473, U41474, and U41475). Our laboratory has recently cloned CDS from potato and Arabidopsis (Kopka et al., 1997a). An eye-specific CDS in Drosophila melanogaster has been shown to be essential for IP3-mediated signal transduction during light perception (Wu et al., 1995), where the enzyme is required for the regeneration of PIP2 from DG (one of the reaction products of PI-PLC activity) via PA, and thus participates in PI-PLC-signaling cascades. Another enzyme involved in the resynthesis of PIs, DG kinase, has also been cloned recently from higher plants (A. thaliana; Katagiri et al., 1996).
To obtain a probe for heterologous screening of potato PI-PLC cDNAs we initially cloned a novel PI-PLC homolog from A. thaliana ecotype C24, which was identified by a search of the plant database of expressed sequence tags (J. Kopka and B. Müller-Röber, unpublished data). This clone was successfully used to isolate cDNAs coding for PI-PLCs from Nicotiana rustica (Pical et al., 1997). Here we report the isolation of cDNAs representing three PI-PLC isoforms from epidermal fragments of fully expanded potato leaves. We demonstrate gene expression of the three isoforms in a variety of tissues and differential gene regulation under different stress regimes. Using purified recombinant proteins we identified the reaction products produced by the potato PI-PLCs and characterized the cation requirements of the three isoforms.
MATERIALS AND METHODS
Cloning and Sequencing of cDNAs
A novel PI-PLC homolog was cloned from Arabidopsis thaliana (J. Kopka and B. Müller-Röber, GenBank accession no. X85973). Two HindIII fragments (0.7 and 0.8 kb) containing most of the coding region of the novel cDNA were used for heterologous screening of a λ-ZAP II cDNA library (Stratagene), which was prepared from poly(A+) RNA that was isolated from epidermal fragments of potato (Solanum tuberosum L. cv Désiree) leaves (Müller-Röber et al., 1995; Kopka et al., 1997b). DNA fragments were labeled with [α32P]dCTP using a random-primed DNA-labeling kit (Boehringer Mannheim). Hybridization of plaque lifts overnight at 42°C in PEG buffer (Amasino, 1986) was followed by washes at 45°C in 6× SSC and 0.5% SDS for 15 min and in 5× SSC and 0.5% SDS for 15 min. pBluescript II SK plasmids containing target inserts were obtained from hybridizing phages by in vivo excision according to the manufacturer's (Stratagene) protocol. The plasmids containing the longest inserts were manually sequenced (T7 sequencing kit, Pharmacia). Complete nucleotide sequences of full-length cDNAs representing the genes stplc1, stplc2, and stplc3 were submitted to the EMBL nucleotide sequence database. Standard molecular biology methods were performed as described previously (Sambrook et al., 1989).
Computational Analysis of Predicted Amino Acid Sequences
The computational services and options of the Wisconsin package (version 8.1, Genetics Computer Group, Madison, WI) were used with default parameters. Analysis of amino acid sequence homology was performed with the BLAST program (Altschul et al., 1990). Multisequence alignments were created with the PILEUP option. The percentage of sequence identity and similarity was determined by pairwise alignment using the GAP program. Hydropathy plots were generated according to the algorithm of Kyte and Doolittle (1982). Relative molecular masses and pIs were calculated from the deduced amino acid compositions with the Compute pI/Mw tool available at the ExPASy Molecular Biology Web server (Geneva, Switzerland). The secondary structure of plant PI-PLCs was predicted by submitting a multisequence alignment of all known plant PI-PLCs to the SSPRED Web server at the European Molecular Biology Laboratory (Heidelberg, Germany). Subcellular sorting was predicted at the PSORT Web server for analyzing and predicting protein-sorting signals at the Institute for Molecular and Cellular Biology (Osaka, Japan). Analysis of StPLC primary structures for conserved protein domains was performed with the PROFILESCAN program at the Web server of the ISREC Bioinformatics Group (Lausanne, Switzerland).
Plant Material
Potato plants were obtained through Saatzucht Fritz Lange KG (Bad Schwartau, Germany). Plants were grown in soil in individual 3-L pots in a greenhouse under periods of 16 h of light (with additional illumination giving a total light intensity of approximately 100–200 μmol photons s−1 m−2; 22°C) and 8 h of darkness (15°C).
Material for RNA analysis of steady-state mRNA levels in different tissues (Fig. 3) was harvested from well-watered, flowering potato plants at approximately 3 pm, 8 h after the beginning of the light period. Epidermal fragments for this experiment were prepared from source leaves, i.e. fully expanded, nonsenescent leaves from the fifth node downward (Müller-Röber et al., 1995; Kopka et al., 1997b). Sink leaves were immature leaves (<1 cm in length) harvested from the first visible node of nonflowering shoots.
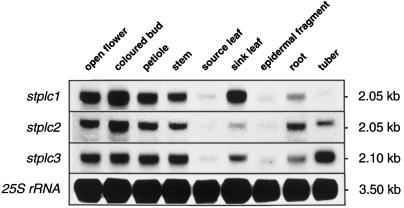
Figure 3.
RNA analysis of stplc steady-state transcript levels in various tissues of potato plants. Epidermal fragments were prepared from source leaves. Total RNA (50 μg per lane) was probed with the complete cDNAs of stplc1, stplc2, and stplc3 and with a cDNA encoding potato 25S rRNA (see Methods). Transcript sizes are indicated.
Leaflets of fully expanded leaves used for analysis of the short-term effect of local wounding on transcript levels (Fig. 4) were crushed once in vertical orientation to the major vein of the leaflet with a 5-cm clamp used to seal dialysis tubes. Total RNA was prepared from these leaves 6 h after application of wound stress. Fast wilting of leaves for RNA analysis of short-term effects on gene expression (Fig. 4) was achieved by air-drying the root system after gentle removal of the pot and adhering soil. Wilted leaves were harvested after 6 h. In this experiment leaf fresh weight was reduced by approximately 20% compared with untreated plants.
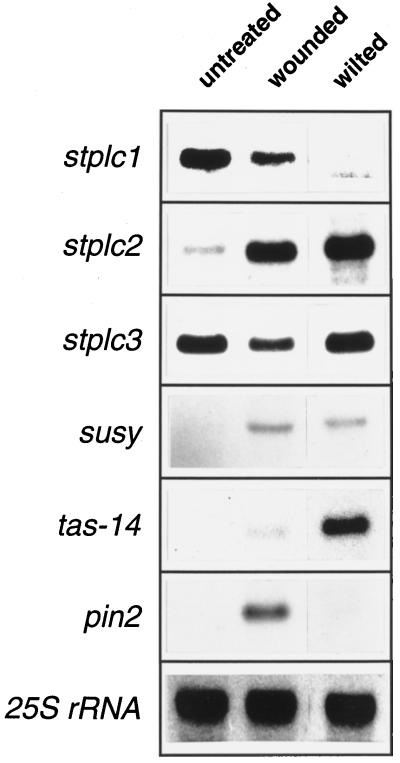
Figure 4.
RNA analysis of stplc steady-state transcript levels in fully expanded leaves of potato plants that were untreated, locally wounded, or wilted by severe short-term drought. Leaves for RNA preparation were detached 6 h after application of stresses at approximately 4:30 pm. RNA preparations (25 μg per lane) were also tested for wound-induced expression of the proteinase inhibitor II gene (pin2), for drought-induced expression of the tas-14 gene, and for induction of Suc synthase (susy) gene expression.
The effect of slowly developing, long-term drought stress was studied on leaves of the upper third to fifth node because most leaves of the lower nodes were subject to fast senescence under the experimental conditions applied. Plants were not watered for 4 d before the leaves were collected, whereas control plants were subjected to a normal watering regime and were kept under otherwise identical growth conditions. Leaves and epidermal fragments were prepared as described by Kopka et al. (1997b).
RNA Analysis
RNA from different tissues of potato plants was prepared according to the method of Logemann et al. (1987), except RNA of epidermal fragments, which was prepared as described by Kopka et al. (1997b). Total RNA was quantitated spectrophotometrically at 260 nm. Total RNA from each sample (25 or 50 μg) was electrophoretically separated in a denaturing 15% (v/v) formaldehyde-1.5% (w/v) agarose gel and blotted onto Hybond-N+ membranes (Amersham). Fixation of RNA to the membrane was achieved by incubation in 50 mm NaOH for 5 min at room temperature and a subsequent wash in 2× SSC (Noonberg et al., 1994). Labeling of cDNA fragments and hybridizations were performed as described for plaque lifts. Washes were performed at 65°C in 3× SSC and 0.5% SDS for 15 min and in 0.2× SSC and 0.5% SDS for 20 min. Blots were exposed to Kodak X-OMAT AR film.
The following DNA probes were used for hybridizations: the SpeI/XhoI fragments containing the complete cDNA inserts of stplc1, stplc2, and stplc3; the 3.5-kb Asp718/BamHI fragment of plasmid efEST G56 coding for 25S rRNA of potato (GenBank accession no. R28706); the 2.7-kb Asp718/BamHI fragment of POTSSYN coding for potato Suc synthase (Salanoubat and Belliard, 1987); the 0.6-kb PstI fragment of PINR5 coding for the wound-inducible potato proteinase inhibitor II (pin2 gene; Peña-Cortés et al., 1996); and the full-length fragment of a tomato cDNA coding for the drought-stress-inducible tas-14 gene (Godoy et al., 1990). Transcript sizes of stplc1, stplc2, and stplc3 were determined in relation to the electrophoretic mobility of rRNA species with a single RNA blot, which was successively probed.
Analysis of Recombinant StPLC Expressed in Escherichia coli
Recombinant StPLC1, StPLC2, and StPLC3 proteins were expressed in E. coli BL21 cells as GST-StPLC fusion proteins using the pGEX-4T-2 vector (Pharmacia), which provides a thrombin cleavage site for removal of the GST part of the fusion proteins. The complete coding regions of stplc1, stplc2, and stplc3 were amplified by PCR using Taq-DNA polymerase. The forward primers were specific for each isoform and introduced BamHI restriction sites, which were used for in-frame cloning of the PCR fragments into the pGEX-4T-2 vector. The reverse primer was the T7 primer, which anneals to the T7 promoter region of pBluescript II SK plasmids. The second restriction site for cloning of PCR fragments was XhoI, which was the cloning site that was used for construction of the epidermal fragment λ-ZAP II cDNA library (Müller-Röber et al., 1995) and therefore immediately follows the poly(A+) tail of the cDNA inserts. The resulting expression vectors were named pGEX-StPLC1, pGEX-StPLC2, and pGEX-StPLC3.
The cloning strategy preserved the native stop codons of StPLCs but introduced an N-terminal Gly-Ser dipeptide to thrombin-cleaved, recombinant StPLCs in place of the native Met residue. E. coli BL21 was transformed with pGEX-StPLC1, pGEX-StPLC2, or pGEX-StPLC3. Five to 10 transformants per isoform, each containing a vector derived from an independent PCR fragment, were screened for protein expression using a small-scale purification protocol. E. coli BL21 transformants (3-mL cultures) were grown at 28°C to A600 = 0.6 and were subsequently induced for 2 h with 2 mm isopropyl-β-d-galactopyranoside. Fusion protein was purified using a glutathione-Sepharose resin according to the manufacturer's (Pharmacia) instructions, and GST- and PIP2-hydrolyzing activities were determined in the affinity-purified fractions. Fusion protein isolated from most transformants had similar, high ratios of PI-PLC activity compared with GST activity; only a few did not exhibit PI-PLC activity, which may have been due to errors induced during DNA amplification or cloning.
A single E. coli BL21 transformant carrying either pGEX-StPLC1, pGEX-StPLC2, or pGEX-StPLC3 and showing high PI-PLC activity compared with GST activity was selected for a large-scale production of recombinant StPLC1, StPLC2, and StPLC3. Thrombin-cleaved StPLCs and uncleaved fusion proteins were purified from 1-L cultures according to the manufacturer's recommendations using PBS buffer (10.1 mm Na2HPO4, 1.8 mm KH2PO4, 0.14 m NaCl, and 2.7 mm KCl, pH 7.3). Thrombin cleavage was performed overnight at room temperature with fusion protein that was bound to a glutathione-Sepharose column.
Purification was monitored by protein determination (Bradford, 1976), by determination of GST activity with 1-chloro-2,4-dinitrobenzene as substrate (Habig et al., 1974), by determination of PI-PLC activity with PIP2 as substrate (see below), and by SDS-PAGE (Laemmli, 1970). Protein purity and molecular masses were estimated by densitometry of Coomassie brilliant blue G-250-stained SDS-PAGE gels. Yield of a representative purification from a 1-L E. coli culture was 0.38 mg of GST-StPLC fusion protein or 0.15 mg of recombinant StPLC. Purity of recombinant StPLCs was greater than 90% and GST activity was not detectable.
Biochemical Analysis of Recombinant StPLCs
The PI-PLC assay was performed according to the method of Melin et al. (1992) and Drøbak et al. (1994). The reaction mixture contained 50 mm Tris/maleate, pH 6.25, 0.2 mm of 3H-head-group-labeled PI or PIP2 at approximately 5000 dpm nmol−1 (Amersham) and varying amounts of Ca2+ in a volume of 50 μL. A 0.6 mm micellar lipid stock solution was prepared by mixing unlabeled (PI, PIP2; Sigma) and 3H-labeled lipid in chloroform:methanol (2:1, v/v), evaporation of solvent under a stream of nitrogen, addition of 166 mm Tris/maleate, pH 6.25, and sonication for 10 min. The reaction was performed at 25°C for 15 min. Substrate consumption did not exceed 15%. The reaction was stopped by the addition of 1 mL of chloroform:methanol (2:1, v/v). Phases were separated by adding 0.25 mL of 1 n HCl, and liquid scintillation counting of water-soluble reaction products was carried out as described previously (Melin et al., 1992).
Cations were added to the reaction mixtures as chloride salts. The standard reaction mixture contained 10 μm Ca2+. Ca2+ concentrations ≤10 μm were buffered with 1 mm EGTA (Owen, 1976). Studies of the effect of Al3+ and Mg2+ on plant PI-PLC activity were performed in the presence of 10 μm Ca2+. Recovery of IP3 from standard reaction mixtures in the presence of Al3+ and Mg2+ was determined with [3H]IP3 (5000 dpm nmol−1; Amersham).
Analysis of Plant PI-PLC Reaction Products
Standard reactions were performed in 50 μL with 0.1 μg μL−1 recombinant StPLC, 10 μm Ca2+, and 0.2 nmol μL−1 phosphatidyl[2-3H]inositol-4,5-bisphosphate (5000 dpm nmol−1). Reactions were stopped by chloroform:methanol (2:1, v/v), and lipophilic and water-soluble reaction products were separated as described above.
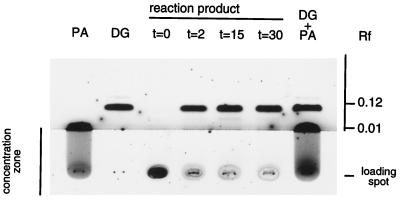
The chloroform phase was concentrated under a stream of nitrogen and applied quantitatively onto silica gel plates with a concentration zone (type Si 250 PA, J.T. Baker, Deventer, The Netherlands). TLC was performed with hexane:diethylether:acetic acid (9:1:0.5, v/v) to analyze neutral lipids or with chloroform:methanol:acetic acid:water (25:15:4:2, v/v) for separation of PA. Lipids were visualized by iodine staining. The standard lipid mixtures contained 10 μg of DG and 10 μg of PA (both from Sigma). PIP2 was immobile in both solvent systems.
The aqueous phase of the chloroform:methanol extraction was neutralized with 2 n NaOH and concentrated by freeze-drying. The reaction products were analyzed on a 25-cm Partisil SAX HPLC column (Whatman, Maidstone, UK). The chromatographic conditions of Brearley and Hanke (1996) were applied. The columns were eluted at 0.5 mL min−1 applying a linear gradient of 0 to 2.5 m NaH2PO4 with a slope of 0.416 m min−1. 3H was monitored with a flow-through counter. Recovery and retention time of IP3 was determined with authentic [3H]IP3 (50,000 dpm per reaction mixture). [3H]Inositol-bisphosphate was prepared by limited alkaline phosphatase treatment of [3H]IP3 (Brearley and Hanke, 1996). Separation of inositol phosphates on the Partisil SAX column was possible but limited by severe peak tailing and did not meet the superior quality of Partisphere SAX columns (Brearley and Hanke, 1996).
RESULTS
Cloning and Sequence Analysis
Using the coding region of an A. thaliana cDNA encoding a novel plant PI-PLC homolog as a molecular probe, we cloned three PI-PLC isoforms from a λ-ZAP II cDNA library, which was prepared from poly(A+) RNA isolated from epidermal fragments of potato leaves (Müller-Röber et al., 1995). The 1981-bp cDNA of stplc1 codes for a 596-amino acid protein; the cDNA of stplc2 has 1940 bp with an open reading frame representing 565 amino acids, and stplc3 encodes a 585-amino acid polypeptide within a cDNA of 2009 bp. All clones were full length, as was shown by determining transcript sizes corresponding to these stplc genes (compare with Fig. 3). The primary structures of StPLC1, StPLC2, and StPLC3 (Fig. 1) were similar to those of currently known plant PI-PLC homologs. Relative molecular weights of the potato proteins were between 64,294 and 67,766 and the pIs ranged from 5.27 to 6.19. The homologies between the coding regions of stplc1, stplc2, and stplc3 were 68 to 81%, and the deduced proteins showed 63 to 77% identity at the amino acid level.
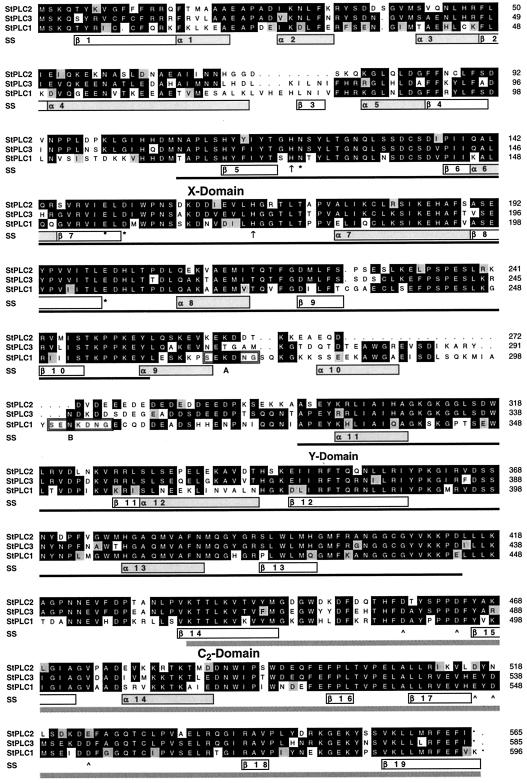
Figure 1.
Multisequence alignment of the amino acid sequences of three PI-PLC isoforms, StPLC1, StPLC2, and StPLC3, as deduced from corresponding cDNA clones isolated from a potato epidermal fragment cDNA library. Amino acid residues conserved in at least two of the three sequences are shaded in black; conservative substitutions are marked in gray. Predicted domains of conserved secondary structure (SS) are indicated by bars (α, α-helix; β, β-sheet). Conserved functional domains are underlined in black (X and Y domain) or gray (C2-domain; PROSITE database at the ExPASy Molecular Biology Web server; see Essen et al., 1996). His residues of the reaction center, which are invariable within the X domains of all known enzymatically active PI-PLCs, are marked by arrows. Ca2+-binding amino acids within the active site are marked by asterisks (*). Putative Ca2+-binding amino acids within the C2-like domain are indicated by arrowheads. Note that the linking region between X and Y domains of StPLC1 contains a short sequence repeat (A and B), which is highlighted by frames.
Analysis of homology between the potato PI-PLC proteins (not shown) indicated that StPLC2 and StPLC3 are more closely related to each other than to any other plant PI-PLC. StPLC1 appeared to be as distantly related to StPLC2 and StPLC3 as to PI-PLCs from other plant species. In general, homology among plant PI-PLCs was higher than 52.0% (identical amino acids). In addition to high amino acid homology, plant PI-PLCs also appeared to share common features on the secondary structure level. Using the SSPRED program for the analysis of conserved structural elements (see Methods), we identified 14 α-helix-forming and 19 β-sheet-forming domains in a multisequence alignment based on all presently known plant PI-PLCs. The positions of these putative structural elements are shown in Figure 1. StPLC isoforms do not contain obvious subcellular sorting signals or transmembrane-spanning domains and therefore these proteins are most likely located in the cytosol and, considering their presumed role, may be associated with the plasma membrane.
Compared with other PI-PLCs, plant PI-PLCs seem to form a distinct subgroup. All PI-PLCs identified so far exhibit high homology within three domains: X and Y, which together constitute the catalytic domain of these enzymes, and a C2-like domain (Fig. 1). The C2-like domain of StPLCs was identified by the C2 domain profile (PS50004) of the PROSITE database. C2 domains are Ca2+-dependent protein-phospholipid interaction domains, which could mediate membrane attachment of the amphipathic plant PI-PLCs. C2 domains were first identified in Ca2+-regulated PKC isoforms (Hug and Sarre, 1993) and are present in a variety of mammalian enzymes involved in transmembrane signaling, such as phospholipase A2 (Clark et al., 1991), PI-PLC (Essen et al., 1996), synaptotagmin (Perin et al., 1991), and rabphilin-3A (Shirataki et al., 1993).
The C2-like domains of StPLCs align to the eight-stranded β-sandwich structure of the C2 domain in rat PLCδ1 (Essen et al., 1996). The predominant predicted secondary structure of StPLCs within this region is β-sheet conformation (Fig. 1, β14–β19). The location of most of these β-sheet domains is in agreement with the two known crystal structures of C2 domains within rat PLCδ1 (Essen et al., 1996) and rat synaptotagmin I (Sutton et al., 1995). Moreover, StPLCs contain the polybasic core region K-(K,R)-T-K typical for C2 domains (Fig. 1, α-14) and five putative Ca2+-binding residues (Fig. 1, arrowheads), which correspond to the Ca2+-interacting amino acids identified in crystallized C2 domains (Sutton et al., 1995; Essen et al., 1996; Shao et al., 1996).
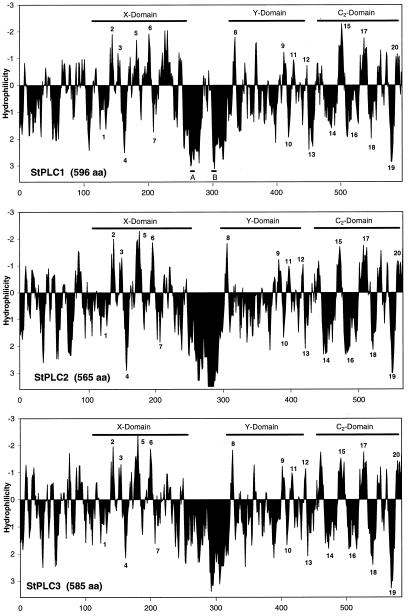
Hydropathy plots of StPLC isoforms illustrate the pattern of sequence conservation and diversity. Within both amphipathic X and Y domains, a large number of conserved hydrophilic and hydrophobic subdomains can be identified in all three StPLC isoforms (Fig. 2). N termini of StPLC isoforms are also amphipathic. In this region, StPLC2 and StPLC3 exhibit almost identical hydropathy patterns, whereas StPLC1 is more variable. The linking region between the X and Y domains is highly hydrophilic and appears to differ between isoforms in length and hydrophilicity. A bipartite hydrophilic linking region, which contains a short sequence repeat that is not present in the single hydrophilic domains of StPLC2 and StPLC3, is characteristic of StPLC1 (Figs. 1 and 2).
Figure 2.
Hydropathy plots of the deduced amino acid sequences of StPLC1, StPLC2, and StPLC3. The total number of amino acids (aa) constituting each isoform is listed. The relative positions of the X and Y domains, the C2 domain, and the sequence repeat in StPLC1 (A and B) are indicated by bars. The conserved hydropathy pattern of StPLC isoforms is marked by 20 individually numbered hydrophilic and hydrophobic domains. Note that significant differences between isoforms are present at the N terminus and within the highly hydrophilic linker region between the X and Y domains.
RNA Expression
Analysis of RNA expression of the three potato PI-PLC isoforms was performed on various tissues from potato plants (Fig. 3). Steady-state mRNA levels of all isoforms were high in flower, petiole, and stem. In contrast, fully expanded source leaves and a preparation of epidermal fragments contained low amounts of stplc transcripts. Epidermal fragment RNA is 90% pure guard cell RNA and may be contaminated only by RNA from trichomes (Kopka et al., 1997b). As judged by phosphorimaging analysis, intensities of hybridization signals observed with RNA isolated from epidermal fragments and source leaves were approximately equal (data not shown). Expression in sink leaves and in tubers differed between isoforms and, in the case of Stplc1, was approximately 10-fold higher than in fully expanded source leaves. Stplc2 and stplc3 transcript levels were also elevated in sink leaves but showed only an approximately 2- to 3-fold difference compared with fully expanded leaves (for definitions of leaf stages, see Methods).
The Stplc1 transcript was hardly detectable in preparations of tuber RNA. In contrast, transcript levels of stplc2 and stplc3 in tubers were equal or slightly higher than in stems. Cross-hybridization between the three stplc probes and their corresponding mRNAs in this and the following experiments can largely be excluded because successive hybridizations of the probes to a single membrane revealed slightly different transcript sizes (compare with Fig. 3) or exhibited differential response of stplc transcript levels to environmental signals (see below; compare with Fig. 4). Furthermore, no cross-hybridization between stplc cDNAs was observed in DNA-blot experiments performed under the same hybridization conditions as used here for the RNA analysis (not shown).
Gene expression of stplc isoforms in source leaves showed different short-term responses to local wound stress and air drying. Whereas stplc1 exhibited strong reduction of its transcript level in response to both stresses, stplc2 mRNA was equally strongly induced by both treatments (Fig. 4). Local wounding induced only a minor, short-term alteration in the expression of stplc3. Analysis of transcript levels in this experiment was performed 6 h after application of wound or drought stress, respectively (see Methods). The effect of the applied stresses on mRNA composition was verified by monitoring wound-induced pin2 gene expression (Peña-Cortés et al., 1996) and the drought-induced increase in tas-14 mRNA levels (Godoy et al., 1990; Harms et al., 1995). In addition, application of both wound and drought stress could also be monitored by an increase in gene expression of Suc synthase (Fig. 4; Kopka et al., 1997b).
We also compared long-term and short-term drought effects (see Methods) on stplc transcript levels in RNA isolated from whole leaves. Under both conditions, stplc1 and stplc2 showed the same behavior as described above, whereas stplc3 transcript levels increased only after long-term drought stress. The same observations were made with RNA extracted from epidermal fragments (not shown). Previously, Hirayama et al. (1997) reported transient induction by drought of the AtPLC1S gene in Arabidopsis rosette plants but constitutive expression of the AtPLC2 gene.
Analysis of Recombinant StPLC Isoenzymes
Recombinant StPLC isoenzymes were expressed in E. coli BL21 cells as GST-StPLC containing a thrombin recognition site between the N-terminal GST-tag and StPLC. Recombinant fusion proteins were affinity purified on glutathione-Sepharose 4B and either eluted with reduced glutathione to yield GST-StPLC fusion proteins or cleaved while bound to the column with thrombin to prepare recombinant StPLC proteins. These preparations contained proteins that showed the expected electrophoretic mobilities for the relative molecular weights predicted from the cDNA sequences. Recombinant StPLCs were at least 90% pure and, as judged by SDS-PAGE and enzyme assays, contained no residual GST (not shown). Preparations of StPLC3 contained approximately equal amounts of two polypeptides, one polypeptide being smaller than expected. Because of differences in protein degradation, the specific enzyme activities of recombinant StPLC isoenzymes could not be compared.
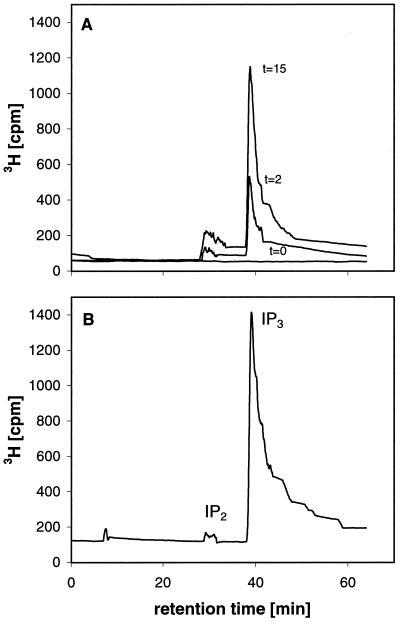
We characterized the nature of the PIP2 hydrolysis products in reaction mixtures incubated under standard conditions with StPLC1, StPLC2, and StPLC3. All StPLC isoenzymes produced the same reaction products from PIP2. As shown for StPLC1, both an unlabeled lipid that co-migrated on TLC plates with authentic DG (Fig. 5) and a 3H-labeled, water-soluble product that co-eluted in HPLC chromatograms with authentic IP3 (Fig. 6) accumulated with increasing reaction time. Under the chosen conditions hydrolysis was complete in 15 min. The amount of product at this time was approximately 10 μg of lipid (Fig. 5), and recovery of 3H into the aqueous phase was approximately 90%. Because of severe peak tailing (Fig. 6), recovery of IP3 from HPLC columns was not determined. Our data indicate complete conversion of the initial amount of educt PIP2 (Mr = approximately 1020 g mol−1) into DG and IP3.
Figure 5.
Analysis of the lipid products of PIP2 hydrolysis catalyzed by recombinant StPLC1. Reactions were performed under standard reaction conditions (see Methods) in 50 μL with 0.1 μg μL−1 StPLC1, 10 μm Ca2+, and 0.2 nmol μL−1 PIP2 (5000 dpm nmol−1). Reactions were started with PIP2, and lipophilic reaction products were extracted with chloroform:methanol (2:1, v/v) at 0, 2, 15, and 30 min. Lipids were concentrated under a stream of nitrogen, applied quantitatively onto TLC plates, separated by hexane:diethylether:acetic acid (9:1:0.5, v/v), and visualized by iodine staining. The standard lipid mixtures contained approximately 10 μg of DG and 10 μg of PA. PIP2 was immobile in the chosen TLC solvent system (autoradiogram not shown).
Figure 6.
Analysis of the water-soluble products of PIP2 hydrolysis, which was catalyzed by recombinant StPLC1. Reactions were performed in 50 μL, as described in Methods. Reactions were started with PIP2 and stopped at 0, 2, and 15 min by extraction with chloroform:methanol (2:1, v/v). Reactions were complete at 15 min (see Fig. 5). The aqueous phase of the chloroform:methanol extractions was neutralized and concentrated by freeze drying. A, A volume representing one-half of the reaction products was applied onto a 25-cm HPLC column. Chromatography was performed as described in Methods. 3H was monitored. B, To demonstrate product recovery, authentic [3H]IP3 (50,000 dpm) was added to a reaction mixture, extracted at 0 min, and analyzed by applying the same conditions. IP2, Inositol bisphosphate.
All reaction products analyzed after 0 to 30 min of incubations contained a minor, nonaccumulating substance that co-migrated with authentic PA when analyzed by TLC with a solvent system for the separation of polar lipids, i.e. chloroform:methanol:acetic acid:water (25:15:4:2, v/v; not shown). The origin and chemical nature of this lipid was not further investigated. The water-soluble reaction product contained a minor but accumulating compound that co-eluted with inositol-1,4,5-bisphosphate in HPLC chromatograms. Given the fact that PA did not accumulate, inositol-1,4,5-bisphosphate can only result from the cleavage of IP3. The origin of the underlying phosphatase activity in our preparations of recombinant StPLC was not investigated.
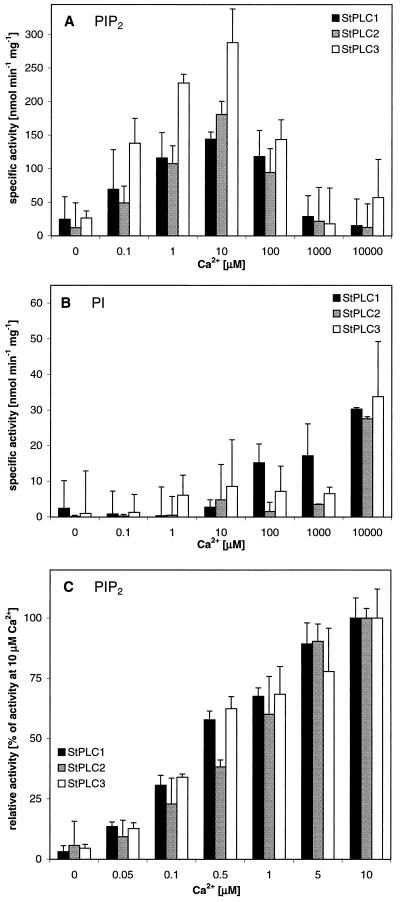
A comparative study of the cation requirements for activation of phosphoinositide hydrolysis by StPLC isoenzymes was performed. All recombinant StPLCs showed a maximum activity of PIP2 hydrolysis at 10 μm Ca2+ and a lower activity at higher Ca2+ concentrations (Fig. 7A). A detailed investigation of the activation of StPLC by low concentrations of Ca2+ normalized relative to 10 μm Ca2+ showed no significant difference in the activation pattern between StPLC isoenzymes. Only at 0.5 μm Ca2+ did StPLC2 show a tendency for lower activity compared with the other isoforms (Fig. 7C).
Figure 7.
PI-PLC activity in preparations of recombinant StPLC1, StPLC2, and StPLC3 in the presence of varying amounts of free Ca2+. In all experiments, 2-3H-labeled, water-soluble product was determined after lipids were removed with chloroform:methanol (2:1, v/v). A, PI-PLC substrate was PIP2 (n = 3). B, PI-PLC substrate was PI (n = 3). C, Activation of PIP2 hydrolysis at low concentrations of Ca2+ (n = 8). Data presented in A and B are specific enzyme activities, whereas data in C are expressed as percentages of maximum activity at 10 μm free Ca2+.
We also investigated the hydrolysis of PI, a metabolic lipid precursor of PIP2, under standard PI-PLC reaction conditions in the presence of recombinant StPLCs (Fig. 7B). All StPLC isoenzymes showed a minor PI-hydrolyzing activity at 10 μm Ca2+, which was substantially increased at 10 mm Ca2+. At this Ca2+ concentration preferential cleavage of PIP2 was lost and the specific activities of all StPLC isoenzymes were approximately the same with PI or PIP2 (Fig. 7, A and B). In contrast to StPLC2 and StPLC3, PI hydrolysis by StPLC1 was already significantly increased at 100 μm Ca2+.
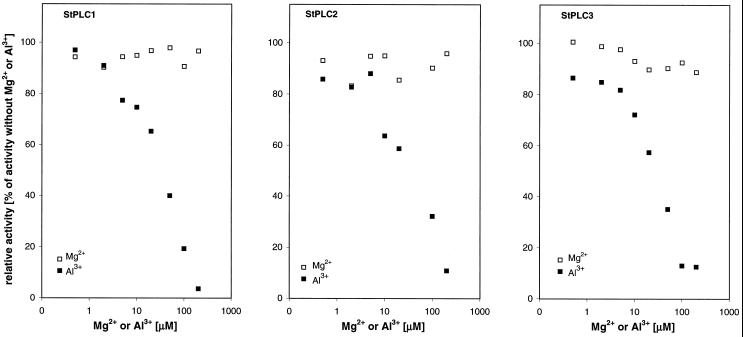
Competition experiments with increasing amounts of Mg2+ and Al3+ were performed in the presence of 10 μm Ca2+. Significant inhibition of PIP2 hydrolysis starting at equimolar concentrations of Al3+ was observed for all StPLC isoenzymes. In contrast, within the range of applied concentrations, Mg2+ did not affect the activity of recombinant StPLC (Fig. 8). The recovery of [3H]IP3 from standard reaction mixtures after extraction with chloroform:methanol (2:1, v/v) was determined in the presence of 0.01 μg μL−1 recombinant StPLC1 and 0.5, 1.0, 5.0, 10, 50, 100, and 200 μm Mg2+ or Al3+. Recovery of [3H]IP3 was approximately 95 ± 2.5% (mean ± sd; n = 3) and, within the range of applied concentrations, was independent of both the concentration and chemical nature of the cation.
Figure 8.
Influence of Mg2+ and Al3+ on PIP2 hydrolysis in preparations of StPLC1, StPLC2, and StPLC3 in the presence of 10 μm free Ca2+ (n = 2). Mg2+ and Al3+ cations were added as chloride salts. Recovery of authentic [3H]IP3 from reaction mixtures after extraction with chloroform:methanol (2:1, v/v) was approximately 95% and independent of the presence of 0.5 to 200 μm Mg2+ or Al3+ (data not shown).
DISCUSSION
The primary structures of all cloned StPLC isoforms show the typical features of PI-PLC enzymes. Based on amino acid sequence homology, plant PI-PLCs, including StPLCs, form a distinct group within the family of eukaryotic PI-PLCs (not shown). This group is closely related to the mammalian PLCδ subfamily (Irvine, 1996). The X domains of StPLCs appear to contain the putative active site, including two conserved His residues. These His residues not only were shown to be located within the active site of crystallized rat PLCδ1 (Essen et al., 1996), but were also essential for enzyme activity, as was demonstrated by site-directed mutagenesis of PLCδ1 (Cheng et al., 1995; Ellis et al., 1995). A similar reaction center with two His residues was also found in the prokaryotic PI-PLC from Bacillus cereus (Heinz et al., 1995).
These observations suggest that the StPLCs encoded by the stplc cDNAs represent enzymatically active PI-PLCs. Previous investigations of cloned plant PI-PLCs demonstrated specificity for phosphoinositides (Hirayama et al., 1995; Shi et al., 1995). However, the fate of the phosphodiester group that links the inositol phosphate head group to the DG part of the glycerolipid was not analyzed; therefore, it could not be ruled out that the cloned enzymes might catalyze a phospholipase D reaction and form PA instead of DG. The latter would be the expected product of an authentic PI-PLC. Partial complementation of a PI-PLC-deficient yeast mutant by a plant PI-PLC cDNA as performed by Shi et al. (1995) is important evidence but could also occur if the PI-PLC activity were only a side reaction catalyzed by the cloned plant enzyme. To confirm the activity of StPLC, we demonstrated that recombinant StPLC almost exclusively forms DG and IP3 from PIP2 (Figs. 5 and 6).
StPLCs, like other plant PI-PLCs (Hirayama et al., 1995; Shi et al., 1995), are activated by Ca2+ concentrations in the nanomolar range (Fig. 7). PI-PLCs from soybean (G. max; Shi et al., 1995) and A. thaliana (Hirayama et al., 1995) have previously been reported to contain EF-hand motifs, i.e. putative Ca2+-binding domains, which, however, are absent from the StPLCs described here. Regulation of plant PI-PLCs by Ca2+, therefore, most likely involves domains other than EF-hands. In accordance with this assumption is the observation that mutating an EF-hand motif in PI-PLC from Dictyostelium discoideum did not affect the Ca2+dependence of this enzyme (Drayer et al., 1995). We suggest that the Ca2+ sensitivity of StPLCs might be brought about by one of two alternative mechanisms. A Ca2+ ion could bind to the charged residues in the reaction center, which are conserved in all PI-PLCs (i.e. N127, E156, D158, and E206 of StPLC1; Fig. 1) and might aid to position the inositol phosphate head group within the active site. The presence of Ca2+ within the active site of rat PLCδ1 and interaction with the conserved charged residues (i.e. N312, E341, D343, and E390 of PLCδ1) was shown by a high-resolution crystal structure of a Ca2+- and IP3-containing complex (Essen et al., 1996). A second possibility is that the Ca2+-regulatory domain in PI-PLC could be the C2-like domain that is present C-terminal to the Y domains of all plant PI-PLCs (Fig. 1). As suggested by Essen et al. (1996), the C2 domain of PI-PLC could regulate enzyme activity by mediating membrane interaction of the mainly amphipathic PI-PLC enzymes (Fig. 2) and thus give access to the phosphoinositide substrate.
Mg2+ ions are known to activate PI-PLC activity in the presence of 10 μm Ca2+ in wheat plasma membranes (Pical et al., 1992; Jones and Kochian, 1995). In contrast, Al3+ is a strong inhibitor of PI-PLC activity in preparations of plasma membranes from wheat roots (Jones and Kochian, 1995). We demonstrated that Al3+ inhibited the activity of all recombinant StPLCs and that Mg2+ was not able to stimulate the recombinant enzyme preparations (Fig. 8). Therefore, we propose that Mg2+ does not directly interact with plant PI-PLC but might stimulate the enzyme via an as-yet-unidentified PI-PLC-binding component that could be present in preparations of plant plasma membranes. In contrast, Al3+ appears to act directly on PIP2 hydrolysis. Two explanations might apply to explain the inhibition of PIP2 hydrolysis: (a) inhibition could be caused by inactivation of the enzyme because of displacement of an essential Ca2+ ion from the putative binding sites at the reaction center or the C2-like domain of the PI-PLC protein (see above), or (b) an Al3+/PIP2 complex could be formed that might not be cleavable by PI-PLC. It has been reported that Al3+ can substitute for Ca2+ in a liposome complex with a different phospholipid, i.e. phosphatidylcholine, due to 560-fold higher affinity of Al3+ for the glycerolipid (Akeson et al., 1989), and it was proposed that Al3+ binds to the phosphodiester group of phosphatidylcholine (Hunter and Etherton, 1989).
We found that, as in other plant species, multiple PI-PLC isoforms exist in potato plants and do not appear to be expressed in a strict tissue-specific manner (Fig. 3). Although stplc cDNA clones were isolated from an epidermal fragment cDNA library, we found high expression for all three genes in several other tissues (cf. Fig. 3), indicating that phosphoinositide metabolism in guard cells most likely does not require cell-specific isoforms of PI-PLC or high expression levels of the corresponding genes. Moreover, expression of three stplc genes in epidermal fragments under both normal and environmental stress conditions strongly suggests that multiple PI-PLC isoforms might be utilized in a single cell type, i.e. guard cells (not shown). The attempt to prove simultaneous expression of multiple PI-PLC isoforms in guard cells by promoter studies is part of ongoing investigations in our group. Whereas we have not yet proven that multiple isoforms are expressed in guard cells, we have identified one PI-PLC isoform in potato (J. Kopka and B. Müller-Röber, unpublished results) and one isoform in N. rustica (Pical et al., 1997), for which no RNA is detectable in epidermal fragments and therefore do not appear to be expressed in guard cells.
We performed a comparative analysis at the levels of protein structure, gene expression, and biochemical properties of the StPLC isoforms isolated from epidermal fragments. StPLC isoforms show high sequence homology and conservation of hydropathy patterns within the X and Y domains (Figs. 1 and 2), which are involved in enzymatic function. In agreement with this observation we were not able to demonstrate significant catalytic differences between the recombinant enzymes at physiological Ca2+ concentrations (≤10 μm; Figs. 7 and 8). However, specificity toward PIP2 started to decline at 100 μm Ca2+ in the case of StPLC1, whereas the other isoforms lost specificity only at higher Ca2+ concentrations (compare A and B in Fig. 7, 100–10,000 μm Ca2+). Whether these differences are of physiological relevance remains an open question.
In contrast to the catalytic domains, StPLC isoforms show rather high sequence diversity in regions that in mammalian PI-PLC subfamilies are known to contain regulatory domains, i.e. the linking region between X and Y domains and the N terminus. We were not able to identify pleckstrin-homology domains or the src-homology domains SH2 and SH3 in plant PI-PLCs. However, the linking regions of StPLCs contain a high number of charged amino acids (Fig. 2). The pattern of hydrophilicity in this region appears to be characteristic for each StPLC isoform. StPLC1, for example, contains a unique bipartite hydrophilic domain, each part of which contains a short sequence repeat (Fig. 2). The structural and biochemical analyses strongly indicate that StPLC2 and StPLC3 belong to one group, and StPLC1 represents a less closely related form of plant PI-PLC isoform. This observation was also substantiated by RNA-expression analysis. Even though there appears to be no distinct tissue specificity in the expression of these genes (Fig. 3), the regulation of transcript levels in response to short-term (Fig. 4) and long-term drought stress (not shown) is markedly different. After long-term stress, stplc2 and stplc3 showed an increase in mRNA levels, but, in contrast, stplc1 mRNA level decreased under identical conditions. Taken together, these results indicate that adaptation to drought might involve changes in the relative levels of PI-PLC isoforms.
In conclusion, in this paper we present the cloning and comparative analysis of three phosphoinositide-specific StPLC isoforms, which are expressed in epidermal fragments and at the mRNA level are regulated in a differential manner. Previously, we reported the isolation of the first cDNA coding for a plant CDS, which among other tissues appears to be also expressed in potato epidermal fragments (Kopka et al., 1997a). The product of the CDS reaction is the initial substrate for the resynthesis of PI from DG produced in plant plasma membranes (Wissing et al., 1992). Thus, we are now able to modulate in planta biosynthesis as well as cleavage of PIP2, which is the lipid precursor of the second messenger IP3. Transgenic approaches should allow us to modulate expression of PI-PLC by attempting guard cell-targeted antisense inhibition studies under the control of cell-specific promoters (Müller-Röber et al., 1994).
ACKNOWLEDGMENTS
We wish to thank Prof. Lothar Willmitzer for his support in recent years. We thank Karsten Harms and Hugo Peña-Cortés for providing cDNA probes for RNA hybridizations.
Abbreviations:
- CDS
CDP-DG synthase
- DG
diacylglycerol
- GST
glutathione S-transferase
- IP3
inositol-1,4,5-trisphosphate
- PA
phosphatidic acid
- PI
phosphatidylinositol
- PIP2
phosphatidylinositol-4,5-bisphosphate
- PI-PLC
phosphoinositide-specific phospholipase C
- PKC
protein kinase C
Footnotes
The European Molecular Biology Organization and the Biotechnology and Biological Science Research Council are acknowledged for their support in providing a short-term fellowship to C.P. and a research grant to J.E.G.
LITERATURE CITED
- Akeson MA, Munns DN, Burau RG. Adsorption of aluminum to phosphatidylcholine vesicles. Biochim Biophys Acta. 1989;986:33–40. doi: 10.1016/0005-2736(89)90269-1. [DOI] [PubMed] [Google Scholar]
- Allan AC, Fricker MD, Ward JL, Beale MH, Trewavas AJ. Two transduction pathways mediate rapid effects of abscisic acid in Commelinaguard cells. Plant Cell. 1994;6:1319–1328. doi: 10.1105/tpc.6.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Muir SR, Sanders D. Release of Ca2+ from individual plant vacuoles by both InsP3and cyclic ADP-ribose. Science. 1995;268:735–737. doi: 10.1126/science.7732384. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amasino RM. Acceleration of nucleic acid hybridization rate by polyethylene glycol. Anal Biochem. 1986;152:304–307. doi: 10.1016/0003-2697(86)90413-6. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- Blatt MR. K+channels of stomatal guard cells. Characteristics of the inward rectifier and its control by pH. J Gen Physiol. 1992;99:615–644. doi: 10.1085/jgp.99.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt MR, Thiel G, Trentham DR. Reversible inactivation of K+ channels of Viciastomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature. 1992;346:766–769. doi: 10.1038/346766a0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Inositol phosphates in the duckweed Spirodela polyrhizaL. Biochem J. 1996;314:215–225. doi: 10.1042/bj3140215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HF, Jiang MJ, Chen CL, Liu SM, Wong LP, Lomasney JW, King K. Cloning and identification of amino acid residues of human phospholipase Cδ1 essential for catalysis. J Biol Chem. 1995;270:5495–5505. doi: 10.1074/jbc.270.10.5495. [DOI] [PubMed] [Google Scholar]
- Clark JD, Lin LL, Kriz RW, Ramesha CS, Sultzman LA, Lin AY, Milona N, Knopf JL. A novel arachidonic acid-selective cytosolic PLA2 contains a Ca2+-dependent translocation domain with homology to PKC and GAP. Cell. 1991;65:1043–1051. doi: 10.1016/0092-8674(91)90556-e. [DOI] [PubMed] [Google Scholar]
- Coté GG, Crain RC. Biochemistry of phosphoinositides. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:333–356. [Google Scholar]
- Coté GG, Crain RC. Why do plants have phosphoinositides? Bioessays. 1994;16:39–46. [Google Scholar]
- Drayer AL, Meima ME, Derks MWM, Tuik R, van Haastert JM. Mutation of an EF-hand Ca2+-binding motif in phospholipase C of Dictyostelium discoideum: inhibition of activity but no effect on Ca2+-dependence. Biochem J. 1995;311:505–510. doi: 10.1042/bj3110505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak BK. The plant phosphoinositide system. Biochem J. 1992;288:697–712. doi: 10.1042/bj2880697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drøbak BK. Phosphoinositides and protein phosphorylation in plant signal transduction. Cell Biol. 1993;4:123–130. doi: 10.1006/scel.1993.1015. [DOI] [PubMed] [Google Scholar]
- Drøbak BK, Watkins PAC, Valenta R, Dove SK, Lloyd CW, Staiger CJ. Inhibition of plant plasma membrane phosphoinositide phospholipase C by the actin-binding protein, profilin. Plant J. 1994;6:389–400. [Google Scholar]
- Ellis MV, Katan SU, Katan M. Mutations within a highly conserved sequence present in the X region of phosphoinositide-specific phospholipase C-δ1. Biochem J. 1995;307:69–75. doi: 10.1042/bj3070069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- Gilroy S, Read ND, Trewavas AJ. Elevation of cytoplasmic calcium by caged calcium or caged inositol trisphosphate initiates stomatal closure. Nature. 1990;346:769–771. doi: 10.1038/346769a0. [DOI] [PubMed] [Google Scholar]
- Godoy JA, Pardo JM, Pintor-Toro JA. A tomato cDNA inducible by salt stress and abscisic acid: nucleotide sequence and expression pattern. Plant Mol Biol. 1990;15:695–705. doi: 10.1007/BF00016120. [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- Harms K, Atzorn R, Brash A, Kühn H, Wasternack C, Willmitzer L, Peña-Cortés H. Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic potato plants but not to a corresponding activation of JA-responding genes. Plant Cell. 1995;7:1646–1654. doi: 10.1105/tpc.7.10.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz DW, Ryan M, Bullock TL, Griffith OH. Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 1995;14:3855–3863. doi: 10.1002/j.1460-2075.1995.tb00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Mitsukawa N, Shibata D, Shinozaki K. AtPLC2, a gene encoding phosphoinositide-specific phospholipase C, is constitutively expressed in vegetative and floral tissues in Arabidopsis thaliana. Plant Mol Biol. 1997;34:175–180. doi: 10.1023/a:1005885230896. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1995;92:3903–3907. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug H, Sarre TF. Protein kinase C isoenzymes: divergence in signal transduction? Biochem J. 1993;291:329–343. doi: 10.1042/bj2910329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D, Etherton B. A model for the binding of aluminum to the phospholipid components of biological membranes (abstract no. 265) Plant Physiol. 1989;89:S-45. [Google Scholar]
- Irvine R. Taking stock of PI-PLC. Nature. 1996;380:581–583. doi: 10.1038/380581a0. [DOI] [PubMed] [Google Scholar]
- Jones DL, Kochian LV. Aluminum inhibition of the inositol 1,4,5-trisphosphate signal transduction pathway in wheat roots: a role in aluminum toxicity? Plant Cell. 1995;7:1913–1922. doi: 10.1105/tpc.7.11.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer EE, Lincoln JE, Hogenhout S, Bennett AB, Bostock RM, Martineau B, Lucas WJ, Gilchrist DG, Alexander D. In situisolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries. Proc Natl Acad Sci USA. 1995;92:3814–3818. doi: 10.1073/pnas.92.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Mizoguchi T, Shinozaki K. Molecular cloning of a cDNA encoding diacylglycerol kinase (DGK) in Arabidopsis thaliana. Plant Mol Biol. 1996;30:647–653. doi: 10.1007/BF00049339. [DOI] [PubMed] [Google Scholar]
- Kopka J, Ludewig M, Müller-Röber B. Complementary cDNAs encoding eukaryotic-type cytidine-5′-diphosphate-diacylglycerol synthases of two plant species. Plant Physiol. 1997a;113:997–1002. doi: 10.1104/pp.113.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Provart NJ, Müller-Röber B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. Plant J. 1997b;11:871–882. doi: 10.1046/j.1365-313x.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee Y, Assmann SM. Diacylglycerols induce both ion pumping in patch-clamped guard-cell protoplasts and opening of intact stomata. Proc Natl Acad Sci USA. 1991;88:2127–2131. doi: 10.1073/pnas.88.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Choi YB, Suh S, Lee J, Assmann SM, Joe CO, Kelleher JF, Crain RC. Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 1996;110:987–996. doi: 10.1104/pp.110.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:21–26. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Brownlee C, Hetherington AM. Abscisic acid-induced elevation of guard cell cytosolic Ca2+precedes stomatal closure. Nature. 1990;343:186–188. [Google Scholar]
- Melin PM, Pical C, Jergil B, Sommarin M. Polyphosphoinositide phospholipase C in wheat root plasma membranes. Partial purification and characterization. Biochim Biophys Acta. 1992;1123:163–169. doi: 10.1016/0005-2760(92)90107-7. [DOI] [PubMed] [Google Scholar]
- Müller-Röber B, Ellenberg J, Provart N, Willmitzer L, Busch H, Becker D, Dietrich P, Hoth S, Hedrich R. Cloning and electrophysiological analysis of KST1, an inward rectifying K+channel expressed in potato guard cells. EMBO J. 1995;14:2409–2416. doi: 10.1002/j.1460-2075.1995.tb07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber B, La Cognata U, Sonnewald U, Willmitzer L. A truncated version of an ADP-glucose pyrophosphorylase promoter from potato specifies guard cell-selective expression in transgenic plants. Plant Cell. 1994;6:601–612. [PMC free article] [PubMed] [Google Scholar]
- Nakajima N, Saji H, Aono M, Kondo N. Isolation of cDNA for a plasma membrane H+-ATPase from guard cells of Vicia fabaL. Plant Cell Physiol. 1995;36:919–924. doi: 10.1093/oxfordjournals.pcp.a078839. [DOI] [PubMed] [Google Scholar]
- Nanmori T, Taguchi W, Kinugasa M, Oji Y, Sahara S, Fukami Y, Kikkawa U. Purification and characterization of protein kinase C from a higher plant, Brassica campestrisL. Biochem Biophys Res Commun. 1994;203:311–318. doi: 10.1006/bbrc.1994.2183. [DOI] [PubMed] [Google Scholar]
- Noonberg SB, Scott GK, Hunt CA, Benz CC. Highly sensitive northern hybridization of rare mRNA using a positively charged nylon membrane. Biotechniques. 1994;16:1075–1078. [PubMed] [Google Scholar]
- Owen JD. The determination of the stability constant for calcium-EGTA. Biochim Biophys Acta. 1976;451:321–325. doi: 10.1016/0304-4165(76)90282-8. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Prat S, Atzorn R, Wasternack C, Willmitzer L. Abscisic acid-deficient plants do not accumulate proteinase inhibitor II following systemin treatment. Planta. 1996;198:447–451. [Google Scholar]
- Perin MS, Johnston PA, Jahn R, Francke U, Südhof TC. Structural and functional conservation of synaptotagmin (p65) in Drosophilaand humans. J Biol Chem. 1991;266:615–622. [PubMed] [Google Scholar]
- Pical C, Kopka J, Müller-Röber B, Hetherington AM, Gray JE. Isolation of 2 cDNA clones for phosphoinositide-specific phospholipase C from epidermal peels (accession no. X95877) and guard cells (accession no. Y11931) of Nicotiana rustica. (PGR 97-086) Plant Physiol. 1997;114:747. [Google Scholar]
- Pical C, Sandelius AS, Melin PM, Sommarin M. Polyphosphoinositide phospholipase C in plasma membranes of wheat (Triticum aestivum L.): orientation of active site and activation by Ca2+ and Mg2+ Plant Physiol. 1992;100:1296–1303. doi: 10.1104/pp.100.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat M, Belliard G. Molecular cloning and sequencing of sucrose synthase cDNA from potato (Solanum tuberosumL.): preliminary characterization of sucrose synthase mRNA distribution. Gene. 1987;60:47–56. doi: 10.1016/0378-1119(87)90212-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schroeder JI, Hagiwara S. Cytosolic calcium regulates ion channels in the plasma membrane of Vicia fabaguard cells. Nature. 1989;338:427–430. [Google Scholar]
- Schroeder JI, Hagiwara S. Repetitive increases in cytosolic Ca2+ of guard cells by ABA activation of nonselective Ca2+permeable channels. Proc Natl Acad Sci USA. 1990;87:9305–9309. doi: 10.1073/pnas.87.23.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Davletov BA, Sutton RB, Südhof TC, Rizo J. Bipartite Ca2+-binding motif in C2domains of synaptotagmin and protein kinase C. Science. 1996;273:248–251. doi: 10.1126/science.273.5272.248. [DOI] [PubMed] [Google Scholar]
- Shi J, Gonzales RA, Bhattacharyya MK. Characterization of a plasma membrane-associated phosphoinositide-specific phospholipase C from soybean. Plant J. 1995;8:381–390. doi: 10.1046/j.1365-313x.1995.08030381.x. [DOI] [PubMed] [Google Scholar]
- Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M, Takai Y. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 related to synaptotagmin. Mol Cell Biol. 1993;13:2061–2068. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam R, Després C, Brisson N. A functional homolog of mammalian protein kinase C participates in the elicitor-induced defense response in potato. Plant Cell. 1997;9:653–664. doi: 10.1105/tpc.9.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Davletov BA, Berghuis AM, Südhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- Wissing JB, Grabowski L, Drewitz E, Hanenberg A, Wylegalla C, Wagner KG. Plasma membrane preparations from suspension cultured plant cells contain the enzymes for the recycling of phosphatidic acid and diacylglycerol. Plant Sci. 1992;87:29–37. [Google Scholar]
- Wu L, Niemeyer B, Colley N, Socolich M, Zuker CS. Regulation of PLC-mediated signalling in vivoby CDP-diacylglycerol synthase. Nature. 1995;373:216–222. doi: 10.1038/373216a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto YT, Conkling MA, Sussex IM, Irish VF. An ArabidopsiscDNA related to animal phosphoinositide-specific phospholipase C genes. Plant Physiol. 1995;107:1029–1030. doi: 10.1104/pp.107.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]