Abstract
The camarodont echinoderms have five distinct mineralized skeletal elements: the embryonic spicules and mature test; spines, lantern stereom and teeth. The embryonic spicules are transient structural elements of the larval skeleton whereas the spines and test plates are permanent structural elements. The teeth are continuously growing structures, matching wear at the incisal adoral end to the rate of new production at the aboral plumula. The mineral in all cases is a high magnesium calcite, but the magnesium content, crystal shape and growth pattern is different in each type of skeletal element. The crystal shape and organization into macro structures depends on the presence of an organic matrix which creates the spaces and controls the environments for crystal initiation and growth. The detailed mechanisms of crystal regulation are not known, but much work has been done on defining the proteins which appear to be involved. Phosphorylated matrix proteins may be of special importance. Biochemical isolation of proteins, construction and analysis of cDNA libraries, and most recently high-throughput proteomic analysis in conjunction with the sequencing of the complete genome have yielded a detailed list of protein components likely to be involved in the mineralization processes. However, the proteome-genome analyses have not yet provided insight into the mechanisms of crystallization, calcite composition, and orientation applicable to all skeletal elements. Although the embryonic pluteus and their spicules are the best studied system, it appears that spicule is not representative of the mature skeletal elements. Now armed with the compositions of most of the proteins involved, the next phase of research will have to focus on the specific localization of the proteins and individual biochemistries of each system with regard to mineral content and placement.
2. INTRODUCTION
Echinoderms have long been a subject of study of special interest to students of genetics and developmental biology and an extensive literature has been developed. In 2006 the Sea Urchin Sequencing Consortium [1] published the complete genome of the Strongylocentrotus purpuratus. While most of the genes were involved with the developmental, metabolic and immunological aspects of functions of the urchins, several hundred genes were putatively ascribed to the processes of skeletal mineralization [2], a unique function in echinoderms as compared to other mineralized invertebrates, but possibly shared with the skeletogenesis in vertebrates. The possible correlation of echinoids with vertebrates stemmed from the realization that the echinoderms were on the same evolutionary branch of deuterostomes as the vertebrates, although one could not fit the echinoderms and vertebrates into the classical morphological scheme of evolution [3]. Adoutte et al. [4] proposed that the evolutionary development was more appropriately mapped by the ribosomal RNA, and on that basis, the echinoderms were more appropriately related to the deuterostome vertebrates than to other protostome invertebrates such as molluscs or brachiopods. Although my laboratory had been focusing on mineralization related proteins of the vertebrate teeth and bone, we had come to the hypothesis that the same kinds of mineral-related proteins might exist in the mineral-related skeletal elements of invertebrates [5] even though these mainly presented carbonate-based mineral. Our first experimental exploration of that hypothesis in 1986 [6], led us to determine that antibodies prepared to and specific to the phosphoproteins of bovine dentin, were cross-reactive with similar acidic proteins from the teeth of Lytechinus variegatusthe common green urchins of the Gulf of Mexico. If not evidence of related evolution, it was at the least evidence that common strategies were developed in the regulation of biomineralization. The work was little noted or appreciated at that time, but as this review will hopefully show, the vertebrate and invertebrate worlds do use many similar mechanisms and protein components to regulate biomineralization processes.
Echinoderms have amazingly complex body plans and a variety of skeletal elements that become mineralized by the deposition of a high magnesium calcite, called magnesian calcite. However, each of the principal skeletal elements may utilize a distinct pathway for regulation of the mineral deposition, and the Mg ion concentration can vary greatly in different skeletal elements of the same animal. In the sea urchin, the important distinct mineralized elements are the transient embryonic spicules of the pluteus, and the more permanent mineral of the spines, dermal skeletal plates, stereom and teeth of the adult animal. The presence of the stereom skeleton, appearing in the Early Cambrian about 520 Ma, is the distinctive major echinoderm synapomorphy [7]. This review is focused on the role and nature of the organic matrix components within or surrounding these skeletal elements. However, to understand the varied kinds of mineralized structures and the particulars of their formation, we need to briefly examine the body plan of the echinoderm and the morphologies of the skeletal elements. Since more is known about the genome and proteome of the camarodont sea urchins, their body plan and structure will be the focus of this discussion.
3. BODY PLAN AND SKELETAL ELEMENTS OF THE MATURE ECHINOID, LYTECHINUS VARIEGATUS (Lv)
3.1 Teeth and Masticatory Structure
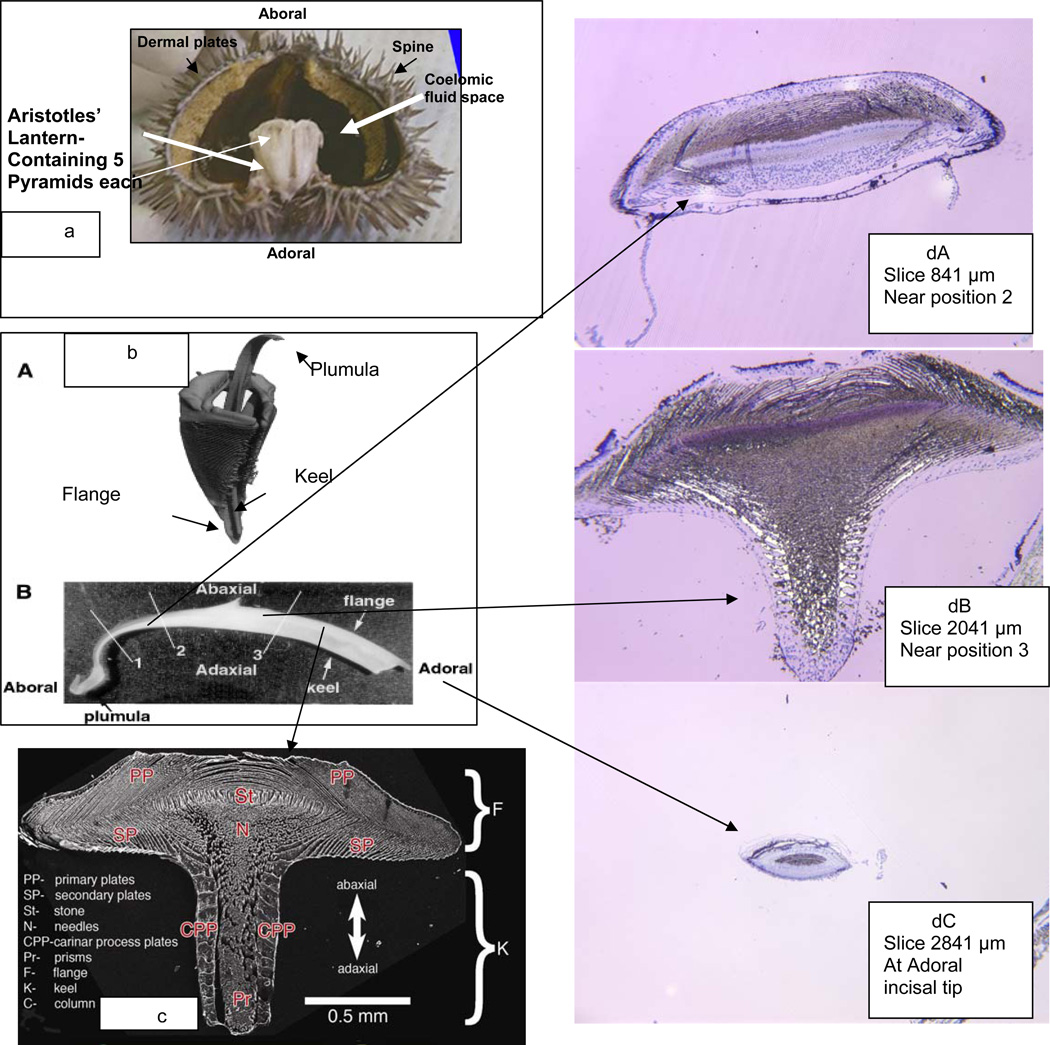
Figure 1a shows a hemisection of a mature L.v. illustrating the placement of the main mature calcified elements: spines, dermal plates and the masticatory structure consisting of five pyramids constructed from stereom, with each pyramid housing a tooth. The incisal edges of these teeth are at the adoral position. The teeth move within their pyramids in concert with each other extending a few millimeters below the oral cavity and taking in food by an abaxial-adaxial scraping action. The pyramid stereom structures are bathed in the coelomic fluid which contains the primary mesenchymal cells (PMC) that assemble and give rise to the stereom and the tooth structures. A more detailed view of a single pyramid and tooth is shown in Figure 1b [8, 9]. In Figure 1bA the most aboral, lightly mineralized portion of the plumula has fallen away and only the heavily mineralized tooth and pyramid are visualized in the microCT scan. About 2/3 of the tooth is within the pyramid. The pyramid stereom is highly fenestrated and porous. The tooth plumula is entirely visible in Figure 1bB, and as will be shown in more detail shortly, is highly cellular and has a lower density than the remainder of the tooth as imaged. In an adult L.v. the tooth may be 25 - 30 mm in length. As a camarodont, the L.v. tooth is flanged in a T-shape. A cross-section view of a tooth such as that shown in Figure 1bB, at the position marked 3, would have the appearance shown in the SEM of a cut and polished section in Figure1c.[10] This view provides only the mineral components and one can see the complex number of structures of different thickness and spatial arrangement. The primary plates at the top of the flange form first and they grow independently from lateral edges of the flange. As illustrated in Figure 1dA, a 1 µm histological section stained with toluidine blue, the primary plates growing from one side do not fuse with the plates advancing to the mid point (the umbo region) from the other side, rather upon close inspection of the syncytia under the umbo they appear to interdigitate. In between the mineralizing plates are the sheets of the multinuclear cellular syncytia. These primary plates form first and the mineral is deposited as guided by the cellular layers. The secondary plates on the underside of the keel are essentially continuous with the primary plates but are less tightly stacked. The carinar process plates are thickened calcite structures which interlock to provide high strength to the keel. Detailed descriptions of the tooth organization and its setting within the pyramid stereom are given by Stock et al. [8, 9]. An important point made evident in Figure 1dB is that a living cell syncytium pervades every part of the tooth, even at the most densely mineralized parts, and the syncytial cytosol and sparse cell nuclei persist all the way to the adoral incisal tip, Figure 1dC. The nuclei stained deep blue with toluidine blue within the multinucleated syncytial layers do not appear to divide after they leave the plumula region [11] but they are metabolically active and can produce proteins at all stages.
Fig. 1.
Lytechinus Variegatus Tooth Apparatus. a. Hemi section with intact Aristotles’ Lantern, containing 5 pyramids. b. A. MicroCT scan of a single tooth and pyramid. b.B. An intact tooth, showing the complete mature aboral to adoral length, ~28 mm. c. SEM of a polished cross-section of the tooth at position 3 (b.B.). d. Cross sections of a tooth at various positions from the aboral end, surface surface etched to expose organic contents, stained with toluidine blue. d.A. Cross section at the beginning of keel formation, just aboral to position 2 in 1b.B. d.B. Cross section at position aboral to position 3 in 1b.B.. The white unstained regions show heavy concentrations of mineral not removed by surface etching. d.C. Cross section through the adoral tip. The central gray region is the end on view of the stone part. It is surprising to find that the tip is still coated with a few syncytial cellular layers Reproduced, with permission, from a,b [8,9], c [56].
One of the most interesting aspects of the tooth structure is the development of strengthening interplate columns, or pillars called pillar bridges [12]. We prefer not to use the pillar bridge nomenclature, rather we designate the initial flange primary and secondary plates as first stage mineral and the later forming reinforcing mineral as second stage mineral [13] and emphasize that while both first and second stage mineral are calcite, they are sharply different in Mg content with the first stage mineral, often referred to as high Mg calcite, containing 4 to 10 % Mg and the second stage mineral, called very high Mg calcite, has 30 to 40% Mg. These structural elements clearly have different biomechanical properties. Uniquely, each plate, needle or prism behaves as a single crystal or a polycrystalline array with all crystals having the same crystal c-axis orientation [12, 14, 15], the mechanism by which this is attained is an argument still unresolved
3.2. Spines and Body Wall
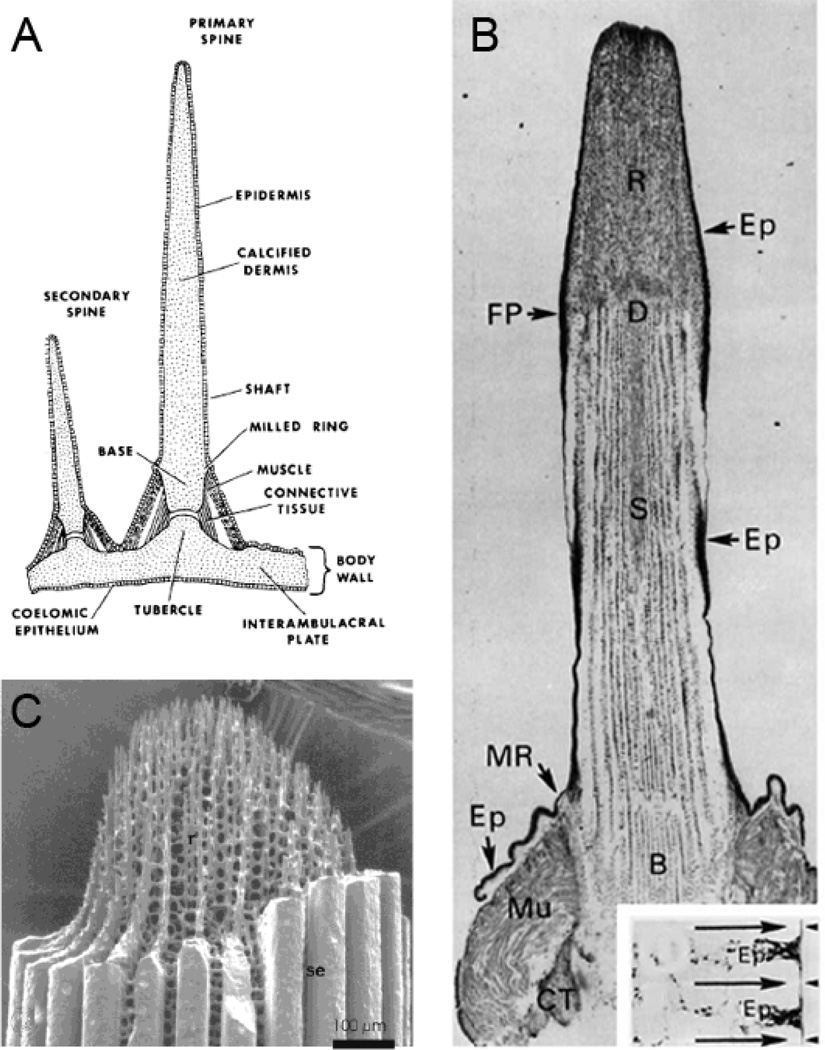
Each plate of the body wall has, as shown in schematic Figure 2A, a primary and secondary spine. The external spines are also heavily mineralized and have much of the character of fenestrated stereom of the pyramids, but motile cells pervade the spine stereom and endow the spines with the ability to regenerate if the growing tip is damaged [15,17]. As pointed out by Heatfield and Travis [16,18], and Kniprath [19] the stereom, teeth and coelomic fluid are populated by cells of a wide variety, distinguished by large numbers of vacuoles and inclusion bodies, and fat droplets and penetrated by mineral spicules which may or may not be bounded by intracellular membranes. As indicated in Figure 2A,B the spines are joined to the body wall by muscle (myocytes) and collagenous connective tissue. In the body wall and the spine fenestrated stereom, Figure 2B,C, the coelomocytes exhibit various functions and activities, including matrix biosynthesis, and biomineralization, as well as phagocytic remodeling and tissue repair, Figure 2C. As in the urchin teeth, mineral spicules appear to take form inside membrane bounded regions, but it is not clear whether the membrane bounded spicules are intracellular or extracellular. A deep consideration of this is not really within the bounds of this review, but is a very important question in considering the role of the matrix proteins in the mechanism of mineral formation [17, 18, 19]. The problem will be discussed better in the next section related to embryonic spicule formation where more studies have been carried out, and have somewhat clarified the problem. While different types of sclerocytes probably follow specific pathways in forming their specific tissues, it is likely that similar basic cellular processes are involved. The sclerocytes in the pyramid stereom, spines and test plates appear to migrate and carry out mineralization independently, they are not compact or of defined shape, but exhibit many fillopodia and lamellae with complex internal membranes. In the tooth the lamellae form multinucleated syncytia [18].
Figure 2.
A. Schematic of the attachment of primary and secondary spines to a test plate [15]. B. A longitudinal section of a decalcified primary spine, revealing the organic matrix of the dermis D, within the base B, shaft S, and regenerate (repaired fracture, at fracture plane, FP) tissue, R. At the base, are the structures called the milled ring, MR, and the Muscle attachment, Mu. The clear spaces within the shaft and regenerate mark the locations of the mineral phase calcite. Ep denotes the epidermis. As shown in the transverse section in the inset, the Ep invaginates between calcite septa at the shaft surface [15]. C. A spine regenerate tip clearly showing the fenestrated calcite, as well as the heavy calcite septa (Se) surrounding the shaft, also shown in the FP cross section in the inset in 2b [16]. Reproduced with permission.
3.3. The Pluteus and Embryonic Spicule Formation
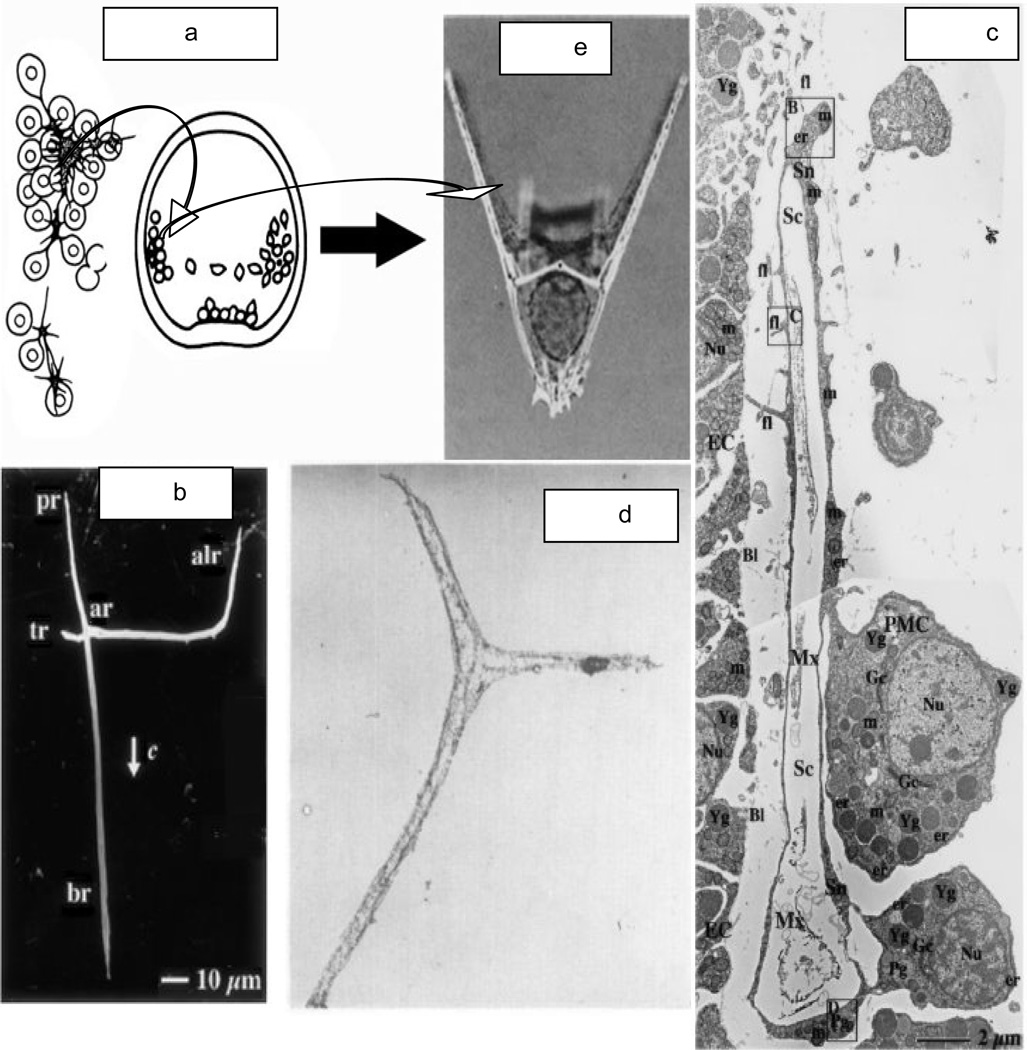
The first mineral deposition within the urchin embryo is detected at the 32–64 cell level of the blastula where one can see the primary mesenchyme cells (PMC) enter the blastocel to cluster near the vegetal pole. The PMC then migrate bilaterally forming two annular clusters, mesenchymal rings, in which the PMC fuse their processes to form independent cell syncytia, Figure 3a, as depicted by Okazaki [20], each of which forms a spicule, Figure 3e, demonstrating the pluteus bilateral symmetry.
Fig. 3.
a. [20] The PMC from the vegetal plate aggregate and migrate along the blastocel wall. Spicule formation begins in the center of the syncytium. b. [22] The triradiate spicule grows along the wall. c. [23] The spicule grows within the syncytial envelope with the connected PMC. d. [23] Upon demineralization the intraspicular organic matrix is revealed. e. [21] The 2 spicules define the structure of the bilateral pluteus. ar-post-oral rod; br-body rod; alr-anteriolateral rod. c-c-axis of entire triradiate spicule. With permissions.
The first mineral spicule appears in the central space created by the annular syncytium, and the very initial mineral deposit is a single crystal of calcite [21]. This crystal grows into the triradiate spicule, Figure 3b, in which the longest structure is the body rod (br) which grows in the direction of the c-axis of the calcite [22]. The cells surrounding the spicule and attached to each other by the spiculogenic syncytial envelope, Figure3c, contain inclusions called polymembranous granules Pg (which contain amorphous CaCO3 that is probably the source of the ions required for spicule growth [22]. How the calcium carbonate is released from the granules and transported into the spiculogenic envelope and added to the calcite of the growing spicule is not understood. Beniash et al. [22] found that the growing spicule was a mixture of calcite and amorphous calcium carbonate (ACC) during growth, but transformed in the solid state to calcite during maturation. The presence of ACC in the spicule is considered to be a factor in the enhanced solubility of the spicules with respect to the much lower solubility and higher stability of more pure calcite. The highly pitted surface of the spicule after exposure to water is in contrast to the smoothness of the calcite of the mature urchin tooth. However, in the in vivo situation, the spicules are covered by the membranes and syncytial matrix of the spiculogenic envelope and the question is how these membranes and macromolecular components of the matrix might affect the stability of the spicules and other skeletal elements, and how the syncytial clustered assembly of PMC at the anonymous rod induces direct calcite formation. Clearly, the spicule is not pure calcite, but contains an embedded proteinaceous matrix, Figure 3d, that is revealed upon demineralization in the presence of a cross-linking agent [23], otherwise the components are solubilized during demineralization. It is interesting in Figure 3d, e] to see that each cross-linked spicule matrix retains the triradiate shape. Perhaps the protein matrix has a role in guiding the shape of the mineral of the spicule, or at least influencing the rates of growth in the axial directions.
Since the spicules are relatively simple in structure and easy to isolate they have been extensively studied in terms of matrix protein content and localization. Collagen had been suggested to be a component of the spicule matrix [24] but a detailed study of the composition of matrix obtained from spicules cleaned from attached cells showed no detectable hydroxyproline (hence no collagen) content [25]. However, properly crosslinked fibrillar collagen within the early embryo is necessary for the proper migration of the PMC and subsequent formation of the PMC syncytia which creates the space and environment for triradiate spicule formation [26]. The soluble spicule matrix proteins had an overall amino acid composition now recognized as typical of invertebrate matrix proteins in general, with a high content of glycine, serine and potential acidic residues and much lower content of basic residues [25]. The question was, and still is"What roles do these spicule proteins play in the mineralization process?”
4. PROTEIN COMPONENTS OF THE SEA URCHIN SKELETAL ELEMENTS AND BIOMINERALIZATION
Biomineralization, the capacity of living systems to sequester mineral deposits, is a widespread phenomenon, and carbonate, silicates and phosphates comprise the principle skeletal elements. The chemistries of the different minerals are clearly different, but from the biological perspective it is possible to ask if the factors regulating the biological controls over mineral deposition are similar or have common mechanisms, even in very diverse life forms. Lowenstam [27, 28] organized thinking on this point by introducing the terms “biologically induced” and “organic matrix-mediated” to express different ways by which the organism in question might control mineralization, but it has become more and more evident that in every case examined, there is a more basic perspective. That is, whether the mineral is an internal or external deposit, the organism must synthesize or capture and deliver a macromolecular component, or components, that control nucleation in localized compartments or spaces in controlled environments. In that sense all biomineralization is matrix-controlled. This applies to vertebrates [5] as well as invertebrates, bacteria or plants. Direct studies of matrix macromolecules influencing mineralization began in vertebrate bone [29] and dentin [30, 31] in the late 1950’s and early 60’s. By the early 1980’s it began to be recognized [5, 27] that the macromolecules involved directly in initiating and organizing biominerals were broadly similar in character with regard to composition, in spite of the evolutionary position of the organism. As noted above, this realization brought my laboratory to the study of sea urchin mineralization proteins [6], but obviously we were preceded by a number of studies on sea urchin mineralization [23, 25, 26, 32–42] emanating from laboratories, all much more expert in urchin developmental biology, especially from the Wilt laboratory. They selected sea urchin spicules from several different species for study because the spicules, as shown in Figures 3b and 3d, could be isolated cleanly from other components of the pluteus. Some problems arose because different laboratories used different protocols for cleaning, collecting and then demineralizing the spicules to obtain the occluded intraspicule proteins and yielded somewhat different distributions of the proteins, a problem common to the isolation of matrix proteins from other mineralized tissues [43]. Further, the mineral of the spicules are of the high magnesium type, about 5% Mg, whereas the non-embryonic magnesian carbonate may include very high, 30 to 40% Mg magnesian carbonate, such as that found in the urchin tooth. It is reasonable to ask if there are other matrix proteins, and perhaps other macromolecular components, within mature urchin mineral in addition to those found with the embryonic spicule mineral.
Since the early work there have been two approaches. The first was to isolate the proteins directly, and then explore their distribution in tissues. This was a problem of protein isolation and purification, followed by preparation of antibodies and immunohistochemistry to determine tissue localization [44], as well as to relate the sequences and compositions of the proteins to their roles in the mineral phase structure and physical properties. The alternate pathway was to examine the genome of the urchin and use proteomics to sort out the overall compositions of the proteins and relate that to potential function. With the complete sequencing of the Strongylocentrotus purpuratus (S.p..) genome this has become the base for relating components with function. Most of the proteins detected have metabolic and immunologic functions. Here we shall concentrate on those proteins which clearly can be designated as mineral-related. However, as we shall also see, there is still the necessity for the direct protein isolation and analysis approach.
5. BIOCHEMICAL AND IMMUNOLOGICAL IDENTIFICATION OF MINERALIZATION RELATED PROTEINS
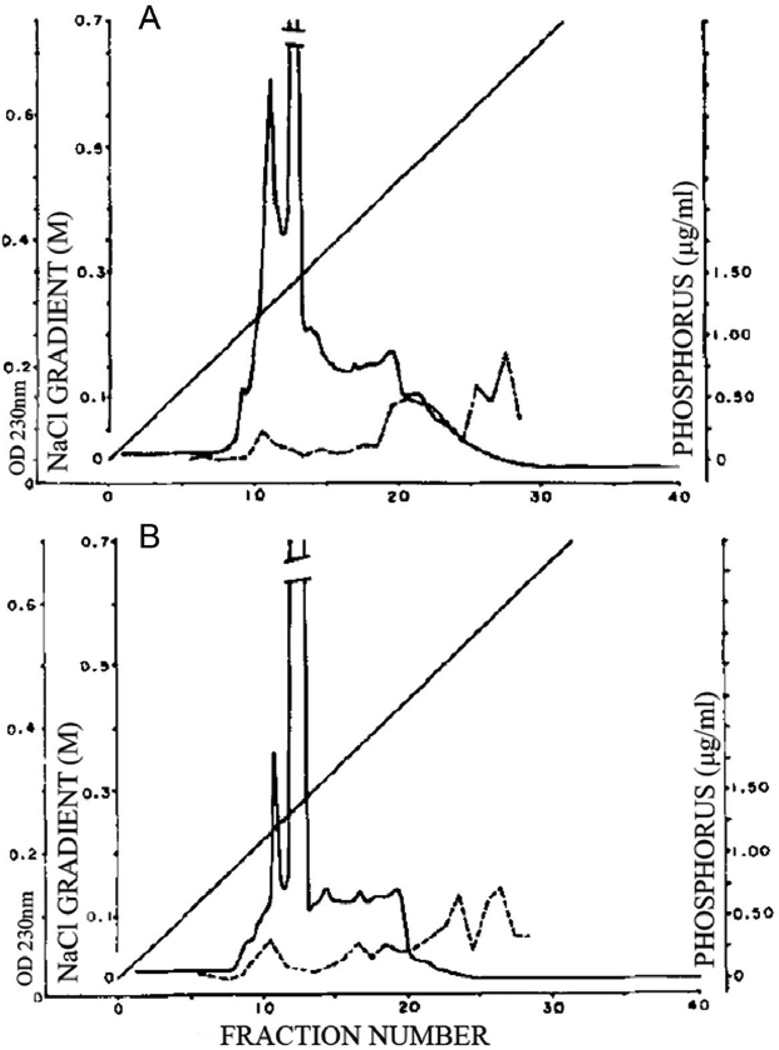
Three very nearly simultaneous papers (1985–86) on the organic components of sea urchin mineralized tissues produced somewhat differing results, but each study used different approaches for protein extraction from different tissues: test plates and teeth [45]; teeth and lantern stereom [6]; and embryonic spicules [25]. In each case the tissue was cleaned, potential enzyme degradation was inhibited, then the tissue was demineralized and separated into soluble components in the demineralized extracts and the residual insoluble material, which was then solubilized by organic solvent or sodium dodecyl sulfate detergent. The resulting proteins were then fractionated on gel filtration columns, reverse phase HPLC, ion exchange chromatography or left unfractionated. The various proteins were subject to amino acid analysis. Benson et al. [25] examined detergent-washed, hypochlorite cleaned embryonic spicules that were demineralized by either EDTA or dilute acid. The amino acid composition of the entire soluble protein obtained was analyzed. Weiner [45] had similarly extracted both skeletal plates and teeth obtained from mature urchins and also analyzed the amino acid composition of the soluble proteins. In addition, the proteins were passed over Sep-Pak C18 reverse phase HPLC columns which in effect passed the most hydrophilic proteins and retained the more hydrophobic. The HPLC profiles clearly showed the teeth to have a much higher content of early eluting hydrophilic components than the skeletal plates. Further, and most strikingly, there was a clear distinction between the compositions of the test plates and the teeth fractions. Veis et al.[6] collected both the mature teeth and Aristotle’s lantern stereom from the same tooth-lantern complexes, extensively cleaned and washed with protease inhibitors prior to EDTA extraction and demineralization. The soluble extracts, after extensive dialysis and solvent changes, were examined by gel electrophoresis, and then fractionated by HPLC DEAE ion exchange chromatography. The amino acid contents of the soluble and insoluble tooth (T-I, T-II) and lantern (L-I, L-II) proteins were determined. The amino acid composition data, summarized in Table 1, are especially interesting in showing that the mineral in the spicule, skeletal plates, and lantern stereom are distinctly different from those of the teeth in having proteins with the ratios of Asp/Glu > 1 in the teeth, but ≤1 in the stereom, plates and spicules, indicating that there may be different proteins involved with tissue specific minerals, just as bone and dentin in the vertebrates have different non-collagenous protein distributions. Another important finding revealed upon direct analysis of the fractionated proteins, was the presence of phosphorylated components [6], surprising in that it had not been expected that the calcite mineral might retain phosphorylated proteins. The phosphorylated proteins were not present in great amount, but were highly charged and tightly bound to the DEAE column matrix, Figure 4.
Table 1.
Selected Amino acid Contents of Urchin Mineralized Tissues. Mole %
| Specimen examined | Asx | Glx | Gly | Ser |
|---|---|---|---|---|
| Benson et al. [24] | ||||
| Larval spicule | 11.0 | 12.1 | 21.3 | 12.5 |
| Weiner [44] | ||||
| Tooth, Total Soluble | 17.1 | 9.0 | 25.3 | 11.6 |
| Tooth , Soluble Sep-Pak A2 | 20.6 | 9.1 | 29.0 | 16.8 |
| Plates ,Total soluble | 13.0 | 11.4 | 17.2 | 11.6 |
| Plates, Soluble Sep-Pak A2 | 18.0 | 17.0 | 23.8 | 13.6 |
| Veis et al. [6] | ||||
| Teeth, Total Sol, T-I | 17.3 | 7.5 | 20.4 | 8.9 |
| Lantern, Total Sol., L-I | 10.4 | 14.2 | 12.6 | 7.3 |
Fig. 4.
DEAE-chromatography of the soluble protein from the mineralized portion of a L. variegatus tooth. Solid line, optical density at 230nm. Dashed line, the phosphate content, directly determined from each fraction [6]. A. Proteins extracted from the most adoral, highly mineralized half of the tooth. B. Proteins extracted from the lightly mineralized aboral half. From [6], with permission.
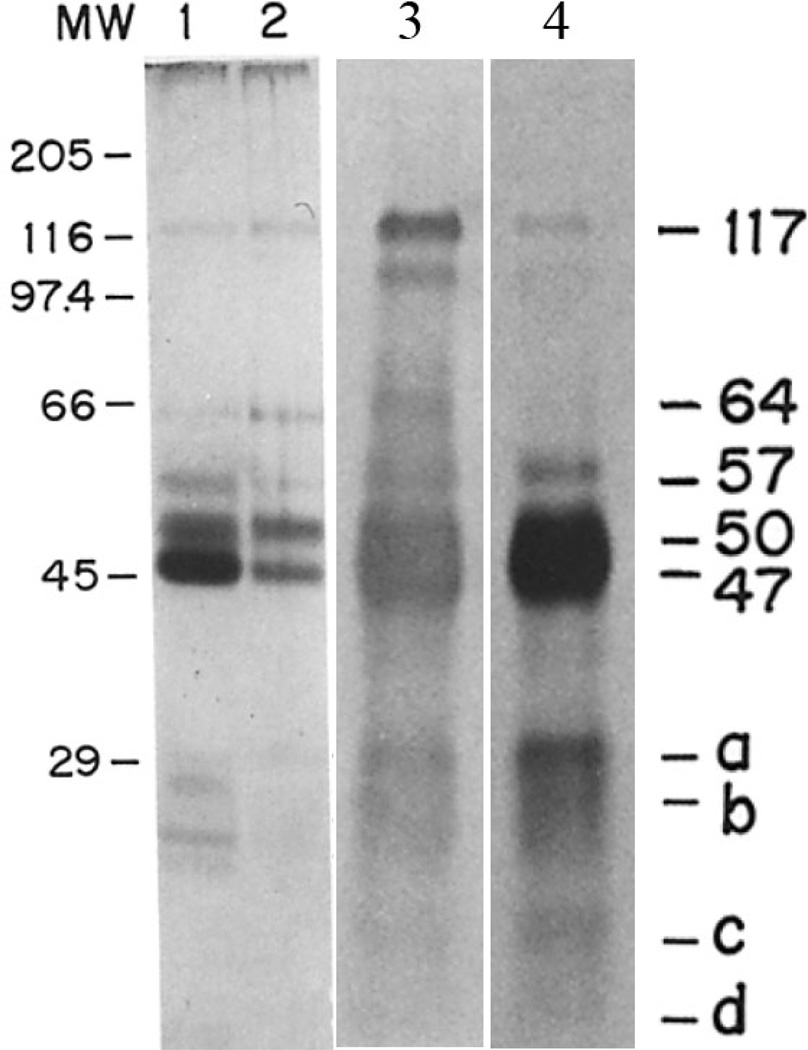
In addition to determining the amino acid content of the mixture of spicule matrix proteins, Benson, et al.[25] isolated the spicule protein components on SDS-gels and determined that a range of differentially stained proteins were present in the isolated, cleaned and hypochlorite washed spicules. These proteins were not stained well with the commonly used Coomassie Blue but were visualized by a modified silver stain procedure [46]. As shown in Figure 5, bands were detected all the way from <10 kDa to 117 kDa with prominent silver stained bands at 47, 50, 57 and 64 kDa. There were variations in band intensities from one preparation to the next, but it was shown that all bands were sensitive to pronase digestion. Similarly, treatment with endoglycosidases reduced the apparent masses of the protein bands indicating that some were acidic glycoproteins. An important step was the preparation of a polyclonal polyspecific-spicule matrix (SM) antibody. It is crucial to emphasize that antigen used to create this antibody was the mixture of all extracted mineral-related spicule proteins as defined above. The relative specificities and intensities of the constituent protein-antibodies pairs were not determined. This general SM antibody enabled much of the subsequent work on tissue localization of these particular antigens. However, all of the proteins detected with the polyspecific SM antibody [25] were clearly distinct from each other. Some were detected by the silver stain, while others were not silver stained. More importantly, the availability of this SM antibody allowed the screening of an expression vector cDNA library from S.purpuratus embryos [32], and the identification of a principal component, the 50 kDa protein named SM50. The structure of the gene and the sequence and structure of the protein were determined [47]. The first report contained a single base frameshifting error in the cDNA and consequently an erroneous protein sequence, but this was corrected shortly thereafter by Katoh- Fukui et al. [36]. Originally, SM50 was erroneously thought to be a typical extracellular matrix acidic protein containing a potential N-glycosylation site. Neither of these was correct. SM50 has a basic pI (pH ~ 12.9), and no predicted glycosylation site was detected. The NCBI nucleotide and protein reference sequences of the S.purpuratus SM50 can be found at NM_214610.2 and NP_999775.1, respectively, in PubMed, and Figure 6. SM50 is a single copy gene.
Fig. 5.
SDS- gels, in12.5 % acrylamide, of the soluble spicule matrix protein. Lanes 1 and 2 are identical silver stained gels, but represent independent extracts showing the variability of tissue extraction and washing procedures. Lane 3 is another comparable extract as in lane 2, but from spicules labeled in vivo with [35S] Met, but stained with the SM-antibody. Lane 4 is the gel autoradiogram from the same extract. From [25] with permission.
Fig. 6. Sequences of selected Sp SM and Cell surface msp proteins.
The amino acid sequences of spicule matrix proteins Sp SM50, Sp SM30 and Sp msp130. The various sequence domains are color coded as noted for each protein. The PubMed sequence reference numbers are given.
George et al.[38] recognized that there were spicule matrix proteins in addition to the SM50 and proceeded to clone them by similar methods, including use of the total anti-SM antibody. A 30.6 kDa protein named SM30 was determined to be a product of the embryonic PMCs. SM30 was distinctly different in structure and sequence from SM50, with a confirmed acidic isoelectric point (NM_214601.1; NP_999766.1). The gene structure was definitively determined [39] and compared with that of SM50. When the complete upstream regulatory sequences of the genomic clone were determined it was evident that there was a family of tandemly arranged SM30 genes, now designated SM 30-alpha and SM30-beta, and the possibility was raised that there might be even further family members. Different upstream regulatory controls were thought to account for the differences in SM30s in various tissues. A more specific SM30 antibody showed the SM30 protein band as a doublet on Western blots. SM30 mRNA was found to be approximately 3 times more abundant than the SM50 mRNA. The relationship between SM30 and SM 50 sequences will be taken up later.
cDNA libraries were prepared from the PMCs of spicules and examined in detail [48,] and led to the identification of an entirely different gene and protein exclusive to sea urchin PMCs [49], designated as msp130 (mesenchyme specific protein 130). The msp130 is a lipid anchored, sulfated cell surface glycoprotein [50]. The protein sequence does not include any membrane spanning sequence, nor any relationship to SM30 or SM50, but, as shown in Figure 6, it has a number of unique features. In the amino terminal region there is a Gly-rich domain spanning Sp residues 39–70, the central domain has a sequence from residue 261 to residue 350, rich in Gly- and Gln including several 13-residue repeats of QPG[F/M/W]G[N/G]QPG[V/M]GG[RQ] while the C-terminal domain has several N-glycosylation sites. Overall, there are 19 QPG triplets. The direct functions of msp130 remain to be determined.
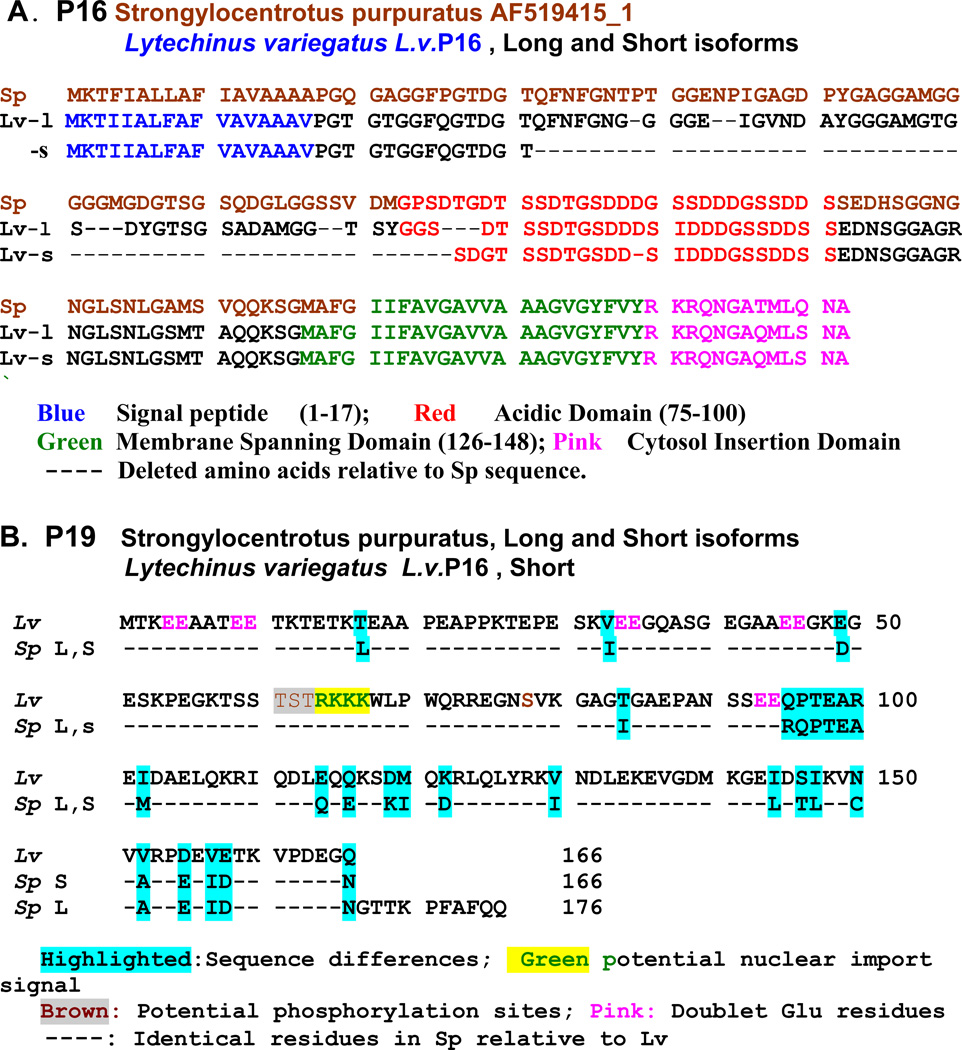
In a large scale analysis of the mRNAs expressed by PMCs of Strongylocentrotus purpuratus gastrula stage embryos, Illies et al.[42] found additional mineralization-related proteins not related to the SM50, SM30, or msp130 protein families. Most notable were the proteins designated as SpP19 and SpP16. At the gastrula stage, SpP19 mRNA was among the most highly represented messages, with only cytochrome c oxidase subunit I and msp130 messages being more abundant. The SpP19 mRNA differed from the other putative mineralization-related SM proteins in that it encoded neither a signal peptide domain, nor a transmembrane domain, but did encode a prominent –RKKK- potential nuclear localization sequence. There were at least two potential alternatively spliced isoforms differing in C-terminal sequence. In S.p. the putative long form contained 176 amino acids, with a mass of 19.5 kDa while the short form was comprised from 166 amino acids with a mass of 18.4 kDa, Figure 7B. In situ hybridization studies showed intense P19 staining of PMCs at all stages of embryogenesis. More recently, our laboratory cloned and sequenced the equivalent P19 from the mature teeth of L. variegatus (L.v.) [51]. In L.v.P19 only one mRNA species was detected, 166 amino acid residues in length identical to the short form of the S.p.P19. There are 26 sequence amino acid substitutions in comparing S.p. and L.v. short forms, most of which are quite conservative, but only 3 are within the N-terminal half of the molecule. There are 5 –EE- sequences in the amino-terminal portion but none in the C-terminal sequence. The nuclear import signal -RKKK-, at residues 64–67, along with the lack of a signal peptide, suggest that P19 is not secreted from the cell syncytia but may have a regulatory role within the cell nuclei. This is consistent with the prediction that the P19 may have one or more threonine or serine phosphorylation sites, most prominently at the 61TST63 site adjacent to the nuclear localization signal. Thus, the P19 may be one of those proteins not directly involved in the extracellular mineralization process, but is occluded within the syncytia, inaccessible to extraction prior to demineralization. The sequence comparison of S.p.P19 and L.v.P19 is shown in Figure 7B. The S.p. P16 mRNA was less abundant than P19 mRNA, but encoded a protein with 172 amino acids [42]. The L.v. P16 was also isolated and sequenced [51]. The Sp and Lv sequences are highly homologous, Figure 7A. The protein was predicted to have a signal sequence, an N-terminal region rich in G residues and other hydrophyllic and uncharged residues likely to create a relatively unstructured region, now frequently described as an intrinsically disordered protein (IDP) [52–54], followed by a distinctly acidic domain rich in Ser and Asp residues, also likely to be an IDP sequence. A predicted transmembrane domain sequence, followed by a concluding C-terminal sequence predicted to be a cytosolic domain [51], Figure 7A, led to the suggestion that P16 was a membrane anchored protein with a short cytosolic component and a longer relatively unstructured extracellular surface which included a potential mineral-binding acidic sequence that might locate extracellular calcite deposition along the syncytial membrane surface, or within other folded internal membrane bound compartments [19, 20]. Following this thought, an antibody was prepared to recombinant rLvP16 and used to locate P16 within the mineralizing primary and secondary plates of the growing urchin tooth. Figure 8 shows that indeed, the anti-rLvP16 outlines the surfaces of both primary and secondary calcite plates in the urchin tooth.
Fig. 7.
The amino acid sequences of the P16 and P19 proteins from Strongylocentrotus purpuratus and Lytechinus variegatus. A. P16, Reference sequence, Strongylocentrotus purpuratus P16 from embryonic pluteus compared with Lytechinus variegatusL.v.P16 long (L) and short (S) isoforms from mature teeth. The putative domains are color coded. Dashes represent absent amino acid residuesThe L.v. Acidic Domain alignment of the first 4 residues of the Short isoform is arbitrary, somewhat different from the initial alignment presented in [51]. The signal peptides and immediate N-terminal residues are highly conserved, and the C-terminal domain, including the acidic domain, membrane spanning domain and cytosol insertion domains are virtually identical, or have only very conservative substitutions. The major sequence differences are in the N-terminal half of the P16 molecules. B. P19, Reference sequence is from Lytechinus variegatus long isoform. Dashed lines represent sequence identities in all 3 sequences. The spliced isoforms in this case appear only at the C terminus. P19 does not have a signal sequence, but does contain a central nuclear import sequence.
Fig. 8.
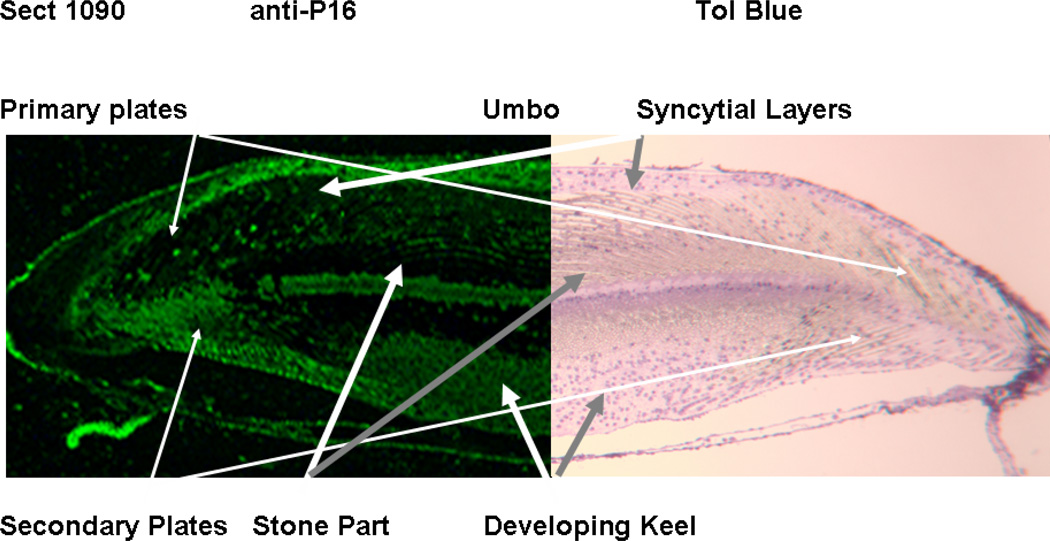
Immunocytochemical localization of P16 within a growing not completely mineralized portion of a Lytechinus variegatus tooth. The sections were cut from an undemineralized tooth, glutaraldehyde treated, then embedded in Epon, The section was etched with ethylene oxide prior to staining with rabbit anti-rLvP16 ( Left half). The corresponding control section (Right half) was stained with secondary anti-rabbit antibody only and counter stained with toluidine blue. The fluorescence outlines the cell syncytial membrane surrounding the calcite mineral. The staining is heaviest within the secondary plates where active primary high Mg mineral deposition is taking place at this stage. The primary plates in this area had mineralized earlier and had begun to form secondary stage very high Mg mineral bridges between plates. Adapted from [52
Unlike the P19, the S.p. P16 distribution appeared to be closely linked to the developmental stage of the embryo and its mineralization. Illes et. al. [42] had reported that P16 were most prominent in the PMC syncytia at the earliest pluteus stage before ingression, but expression was subsequently down regulated and not found in mature urchin tissues. Cheers and Ettensohn [55] microinjected morpholino antisense oligonucleotides (MOs) for P16 into fertilized S.p. and L.v. eggs, blocking the synthesis of the targeted mRNAs during subsequent development. They showed that P16 was not required for the formation of skeletal rudiments, PMC specification, ingression, migration and fusion. But at a late stage of skeletogenesis P16 was essential for skeletal enlargement and growth.
The morpholinos used did not completely inhibit skeletogenesis, but it was argued that the morpholinos used were not 100 percent efficient in the knockdown of P16 synthesis. Since we had isolated P16 from mineralizing teeth, well beyond the embryonic stages, it was not entirely correct to restrict P16 to function only within the embryonic development period. However, in our first studies of the distribution of P16 within the teeth [10, 51, 56], still in progress, we find that the distribution of the P16 is not uniform and is linked to the amount of viable syncytial content where protein synthesis might still be a factor. This is an area of active investigation and requires further detailed examination.
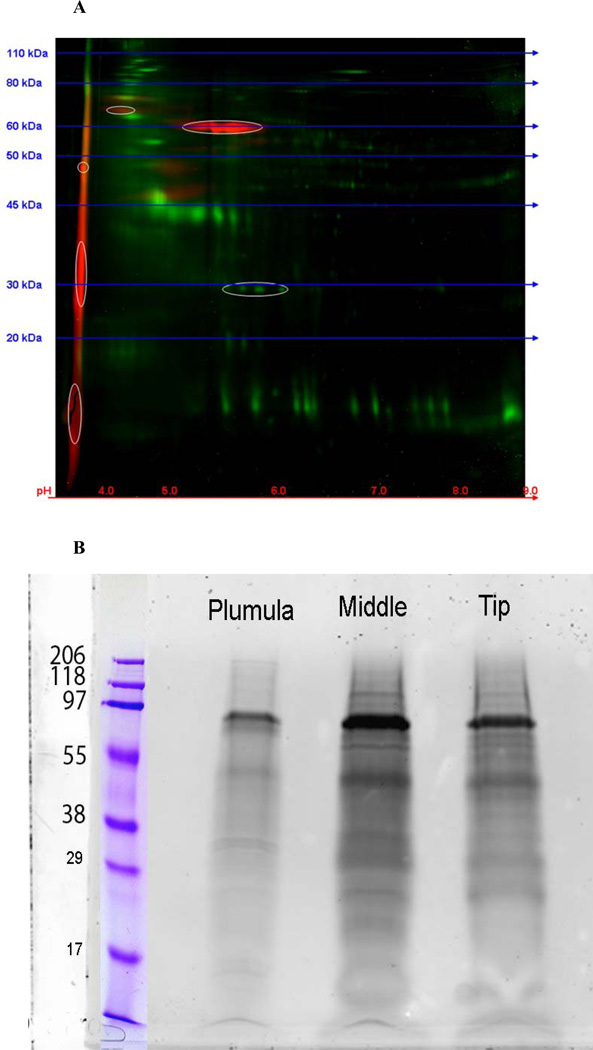
Because of our earlier finding [6], shown in the chromatograms in Figure 4, that some anionic components were phosphorylated, we explicitly used fractionation schemes and protocols designed to sensitively determine the presence of phosphoproteins [10, 51, 56] among the proteins extracted during demineralization. Figure 9A shows a 2-D gel electrophoresis of an HCL extract of the mineralized portion of L.v. teeth, stained for both protein (green) and phosphate (red). P16 and P19 are evident, with isoelectric points pI < pH 4, and there are several others in the same pI range, apparently of higher mass and more heavily phosphorylated. There are faintly stained bands at the upper left region of the gel hinting that some other high mass phosphorylated proteins are present. The total HCL extract was run on a regular 1-D SDS- gel to separate the components by mass and the phosphoproteins were visualized. Figure 9B shows that additional higher mass phosphoproteins are present. Their compositions are the subjects of further study. Importantly, it is evident that there are many phosphorylated proteins in the urchin tooth, and their distribution is not uniform along the length of the continuously growing tooth. The mineralization-related proteins are present at different levels and mixtures at different stages in tooth growth.
Fig. 9.
Gel electrophoresis of the total HCL extract of Gdn.HCl washed L. v. teeth The red stains are for bound phosphates, green for other proteins. A. A 2-D gel of the HCl extract of the total mineralized portion of an urchin tooth. Red- phospho-stain, Greentotal protein. The phosphoproteins are mainly very acidic, with pI <4.0. Development of the 2-D gel was stopped before the most acidic components could run off the left edge of the gel. B. Standard 1-D SDS-gels. The urchin tooth was cut into sections comprising the plumula, the highly mineralized midsection, and the incisal tip and the HCl extracts were prepared separately. After routine SDS-gel electrophoresis, they were stained with a phospho-specific dye. Protein loading was equivalent in the Plumula, Middle and Tip labeled lanes. These data show the non-uniform distribution of mineral-related phosphoprotein components in the different parts of the tooth. The molecular size markers were different in A and B and serve only as guides to the relative values.
The pI of each of the sequenced proteins described in this section have been determined, and the molecular charge at pH 7 computed, Table 2. It is easy to see that there are distinct classes of proteins, but it will be more instructive to consider this from the perspective developed by proteomic analysis coupled with the complete mapping of the sea urchin genome.
TABLE 2.
Isoelectric points and Charge of Urchin Mineralized Tissues
| Protein | Isoelectric Pt. | Net Charge | Accession No. |
|---|---|---|---|
| Sp16 | 3.54 | −13.91 | NM_214646 |
| Lv P16L | 3.62 | −11.08 | DQ0584210 |
| LvP16S | 3.72 | −7.09 | NA |
| Phosphodontin | 3.77 | −109.6 | Glean3_18919 |
| SpP19 | 4.72 | −11.03 | GQ340974 |
| Msp 130 | 4.93 | −15.28 | SUSMSP130B |
| Sp C-type lectin | 5.04 | −7.26 | AF519418 |
| SM 30A | 6.49 | −1.38 | NM_214601 |
| PM27 | 7.89 | +2.35 | U18132 |
| SM 32 | 8.09 | +2.11 | NM_214638 |
| SM 29 | 9.06 | +7.57 | AF519417 |
| SM 37 | 10.35 | +16.18 | AF068737 |
| SM 50 | 10.84 | +15.78 | NM_214610 |
6. CATEGORIZING THE MINERALIZATION - RELATED GENOMIC SEQUENCES
The complete genome of Strongylocentrotus purpuratus was published in 2006. In the introductory paper describing the sequencing effort and some of the resulting insights [1] a number of cogent findings were summarized. The number of genes was amazingly large, much greater than the number of genes in the more complex vertebrates: 23,300 were present and encompassed almost all the known vertebrate gene families. Most important for our present discussion were the finding that some genes thought to be restricted to vertebrates were present in the sea urchin (deuterostome specific), while other genes were present in the urchin, but not in the chordate lineage. Thus, the transition from the basal node of deuterostomes to chordates may have apparently involved the loss or silencing of these genes as vertebrates developed. The classical morphological evolutionary concept of gain of new functional genes as vertebrates developed bones and teeth, has to be considered in the light of loss of many genes. In fact it was pointed out that “distinct genes for biomineralization exist in sea urchins and vertebrates” [2]. The most important point with regard to biomineralization was the selection and categorization of those genes relating to the mineralized tissues. The Livingston et al. paper [2] presented a superbly concise but complete description of the basis of the methods chosen to categorize the genes. At the risk of plagiarizing that description in part, and without copying the “miniprint” list in detail, five points were used to analyze the whole S. purpuratus genome: (1) The shotgun assembly (WGS) Blast [57] search procedures were used to identify closely related proteins. (2) Since sea urchin and vertebrate (the Sibling) biomineralization genes were physically clustered on their DNA scaffold, all urchin DNA scaffolds known to contain one of the known biomineralization genes were examined to identify any closely linked genes that might be new members of the class. Candidate genes with some predicted sequence similarity were identified. (3) New spicule matrix proteins were identified by searching for all predicted proteins that contained a signal sequence and at least one C-type lectin (CTL) carbohydrate binding domain. (4) A cDNA library was generated from gastrula stage S.p. PMCs and a collection of 51,000 expressed sequence tags (ESTs) was produced. The ESTs were mapped to a list of genes expressed by PMCs when mineralization was taking place. Several other sets of ESTs were assembled from other developmental stages. “A substantial enrichment the number of ESTs matching a particular gene model was taken as a reliable estimate of the abundance of the transcript and a predictor of PMCrestricted expression. Finally, (5) a comparison was made between these data and that collected on a comprehensive list of biomineralization proteins from vertebrates. The details of the predicted urchin genomic sequences have been collected in the Strongylocentrotus purpuratus annotated database (Glean3) ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Spurpuratus/fasta/Annotation [1] and http://goblet.molgen.mpg.de/cgi-bin/seaurchin-genombase.cgi. The use of these databases is explained in Poustka et al. [58].
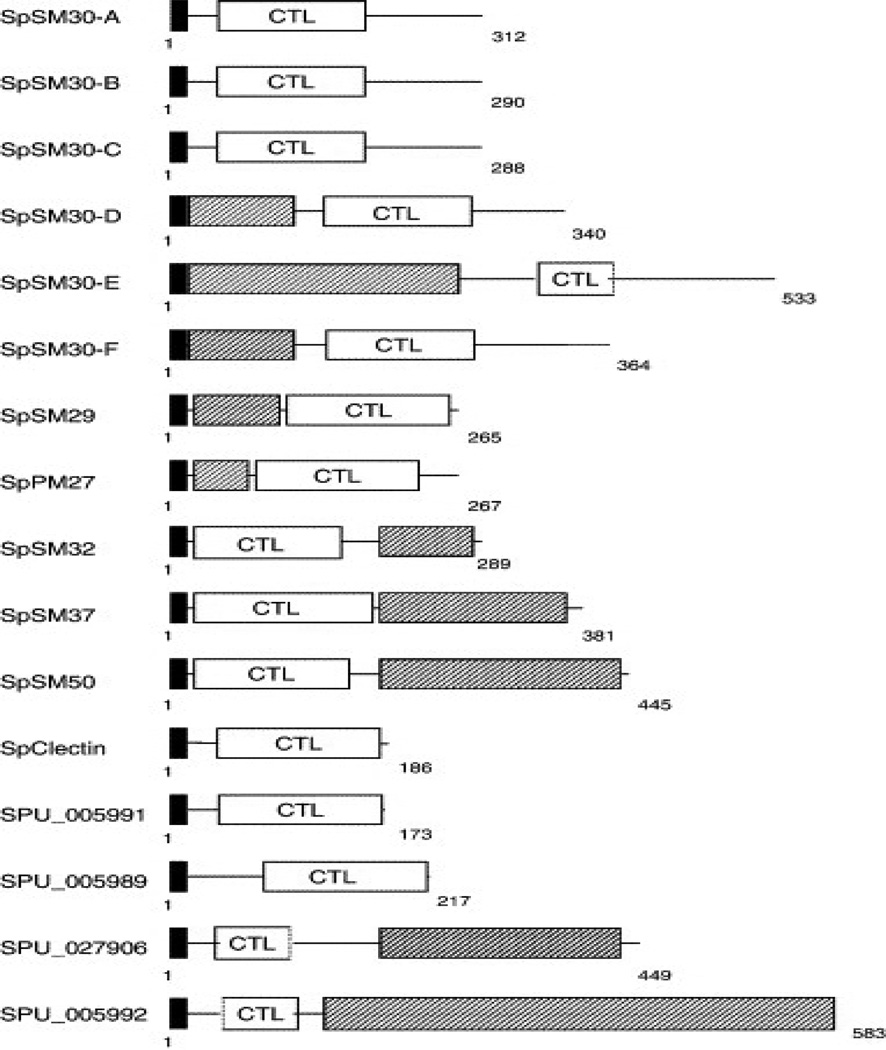
By the time of publication of the Strongylocentrotus purpuratus genome sequence seven proteins had been described as spicule matrix proteins, restricted to their gene expression in Primary Mesenchyme cells: PM27, SM29, SM30alpha, SM32, SM37, SM50 and SpC-lectin (PM refers to Primary Mesenchyme, SM to spicule matrix). Although the initially defined SM 50 and SM30 spicule proteins appeared to have distinct distributions and probably different functions [44], all of these proteins have subsequently been predicted to have multi-domain structures with some related domain region sequences. The genomic analyses also predicted that there were families of SM30-like and SM50-like proteins. Each of the proteins had signal sequences, and hence all were to be considered as secreted extracellular matrix proteins. Each protein had a C-type lectin-like domain (CTLLD), but further analysis of the sequences showed that there were at least two families of CTLLD, those related to SM50 and those related to SM30, distinguished by the observation that the SM50 proteins had a basic isoelectric point (pI), whereas the SM30 proteins had acidic pI’s. Table 2, Figure 10. Whereas these studies all required extensive biochemical and immunological procedures for isolation and preparation of antibodies, the proteomic approach took an entirely different path.
Mann et al. [59–62] carried out an in depth, high accuracy analysis of the S.p..proteome of the spicules, test, spines and teeth. For total sea urchin tooth proteome analysis [60] the urchins were frozen, and the internal skeletal elements, presumably the intact lantern-tooth complexes as shown in Figures 1A & 1B, were collected and immediately sonicated in 6–14% Na hypochlorite to digest all exposed organic matter. After thorough washing in water and air drying, 300 teeth were collected, and powdered in a mortar. The powdered teeth were digested again with hypochlorite. The washed tooth powder was suspended in 5% acetic acid, and the turbid suspension was dialyzed with a Mr 1000 cut-off membrane, the total turbid retentate was lyophilized. The matrix proteins were carbamidomethylated, then cleaved with lysyl endopeptidase followed by trypsin, or with lysyl endopeptidase followed by endoprotease Asp-N and trypsin. In the later work [61–62] phosphopeptides in these mixtures were enriched by binding to TiO2 beads. The bound peptides were eluted with NH4OH, and then dissolved in trifluoroacetic acid for C18 reverse phase chromatography and mass spectrometry.
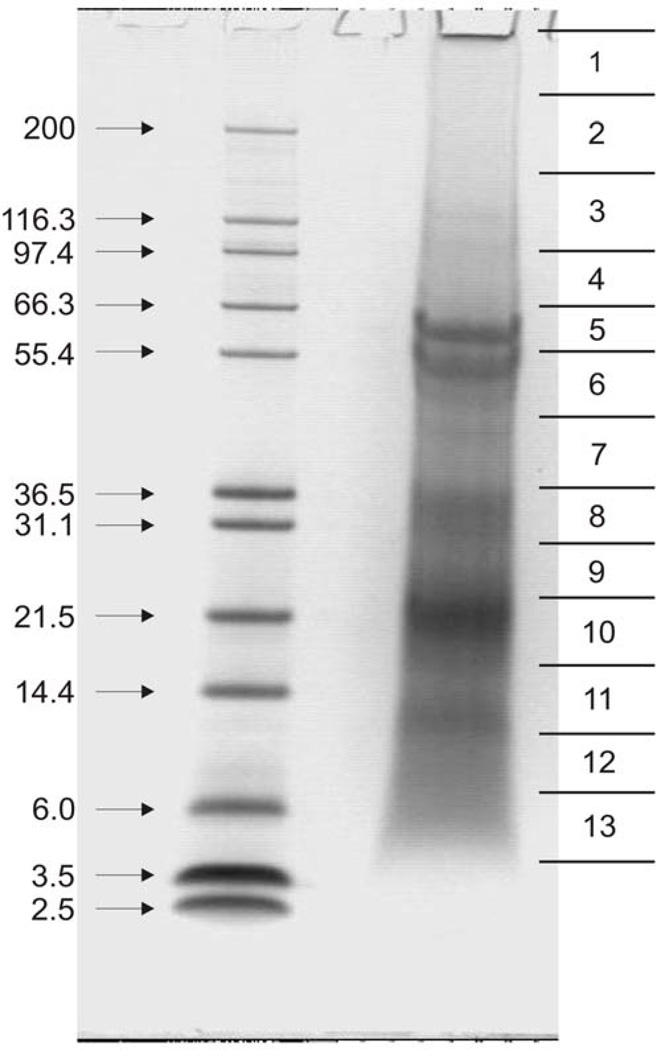
Spicule matrix proteins were obtained from cleaned, hypochlorite treated spicules demineralized with 50% acetic acid, then the suspension was dialyzed into 5% acetic acid, and the mixture was lyophilized. The protein was then dissolved in SDS sample buffer in preparation for gel electrophoresis. Centrifugation of the sample removed an insoluble precipitate. The soluble supernate was then electrophoresed. The recovered protein yield was ~ 0.007 mg/mg of spicule. Following electrophoresis, the gels were stained with colloidal Coomassie and several identical gel lanes were cut into 10 to 16 slices whether the slice was stained or not (Figure 11) [62] in any given slice. The slices from 3 identical lanes were combined and used for in-gel digestion. Thus, every part of each gel lane was subject to trypsin digestion, irrespective of stain intensity or the presence of any visible staining. The preparative details have been described to emphasize the point that either by combining the solubilized components and insoluble residues remaining after the initial hypochlorite treatment, or by examining the contents of every part of the SDS-gels independently of their component staining properties, one obtains a more complete picture without bias. This was a primary problem in the biochemical approach wherein the more acidic protein components stained poorly with the usual protein stains and were more easily lost during fractionation. The same was true of the chromatographic separation schemes, again since the acidic components, and those rich in Gly, Ala and Pro, had no strongly uv absorbent bands in the usual protein spectral ranges.
In the first of the Mann group proteome studies, the organic matrix of hypochlorite treated S.p. spines and test matrix [59] was examined. No attempt was made to include phosphorylation or any other post translational protein modifications. A total of 110 proteins were shown to be present, of these, 40 were present in both test and spines, while the others were restricted to one tissue or the other. The same proteins were also among those present in both tissues at the highest relative protein concentrations. Remarkably, according to the emPAI (exponentially modified protein abundance index score [63], a measure of relative quantitation of the proteins in a mixture), the spicule matrix protein SM50 (Glean 3:18811) was far and away present at the highest concentration in the test as compared to the spines, and in comparison with all other protein components detected. The complexity of the SM 50 family was illustrated in that SM37 (Glean 3:18813), SM32 (Glean 3:18810), and SM29 (Glean 3:05990) were also found in lesser amounts in both test and spines. SM29-like-1 (Glean 3:05991) and SM29-like-2 (Glean 3:05992) were differently distributed: SM29-like-1 was found only in the spines whereas SM29-like-2 was found only in the test. PM 27, also a member of the SM50 family, determined to be present by immunological studies, was not detected in the MS/MS study. It was suggested that the PM27, might have been on the surfaces of the mineral elements, and was degraded during the hypochlorite washes. C-type lectin domains were present in all the SM50 family proteins, but several C-type lectin proteins were found without any relation to SM-50 protein sequences.
The distribution of members of the SM30 family was likewise complex. As illustrated in Figure 10, SpSM30- A-F had been described in spicule studies. In the test and spines SM30-A,B,C and D were not present. SM30-E was abundant in both test and spine, while SM30-F was of low abundance in the spine matrix, in spite of being highly expressed at the transcript level by RT-PCR in adult spines [64]. The nomenclature for the 6 members of the SM30 family is confused by the initial findings of SM30-related genes designated as SM30-alpha and SM30-beta [2]. Now, with more data, SM30- alpha is the same as SM30-B, and SM30- beta is SM30-C [61].
As indicated in Figure 10 all of the SM50 and SM30 related proteins have a C-type lectin domain. All of the SM50 members have an additional domain of a proline-rich repeat nature, but each family member has a unique Pro-rich domain with a different repeat. SpSM50 has two repeat motifs, PG(M/V/F) G(GR/N)Q or PNN; SpSM37 has repeats G(A/G)G(A/G)GGAGAGAGGRWNPQ; while SpSM32 has PNQ, SpSM29 has PN(N/Q), and SpPM27 has PGMG repeats. In contrast SpSM30-A, SM30-B and SM30-C do not have any P-rich domains, but SM30-D, -E and –F do [2, 44]. The C-type lectins of these proteins are all different, but there are two additional proteins with a predicted C-lectin domain: Glean3:13825 and Glean3:11163 with no similarity to the SM50 or 30 families [59].
The membrane-associated glycoprotein msp130 also proved to be a member of a family of proteins, msp130 (Glean3:13821/02088) and msp130-related-1 (Glean3:13822), -2(Glean3:16506/21385),-3 (Glean3:13823),-4 (Glean3:114496),- 5 and -6. Spicule components msp130-related 5 and 6 were not found in test or spines. Figure 12 compares the sequences of spicule msp130 (Glean3:06387) and the predominant test msp130-related-3 protein (Glean3:13823). The main differences were in the amino-terminal portion of both molecules whereas in the carboxy terminal portion of the molecule there was near identity in a PN-rich region of 475 residues, although there were several GE rich sequences in this domain as well.
The majority of the proteins certified as present could be matched to typical vertebrate cellular proteins, enzymes, regulatory proteins and immunoglobulins. There were urchin specific modifications, but it was clear that the basic processes of cell function and metabolism were not unique to the echinoderms, but rather followed the same mechanisms. Carbonic anhydrases, which catalyze the reversible hydration of CO2 to carbonate, needed for calcite formation, were encoded in 19 S.p. genes. The carbonic anhydrase (Glean3:012518) was the only carbonic anhydrase detected in both spine and test. Only a small amount of collagen was detected in the urchin spines and teeth, much less than expected compared to the total amount of collagen determined directly. It was assumed that the collagen, and many other proteins were in the spaces or matrix surrounding the calcified mineral elements, and not occluded within the calcite. Thus they were presumed to be destroyed and removed by the hypochlorite.
It was with this problem in mind that the tooth extraction protocols described earlier were compared [60]. Teeth were washed in hypochlorite, dried, ground to a fine powder, and then extracted again with hypochlorite. The residual calcitic powder was presumed to only contain organic components occluded in the mineral phase. In the second preparation intact teeth were simply washed in hypochlorite. Both preparations were subsequently completely demineralized in acetic acid, and the resulting solubilized organic matter designated here as “tooth powder matrix, PTM” or “matrix of intact teeth, ITM”. The PTM yielded approximately 138 identified proteins, and an additional 52 were identified tentatively from unique peptides that were sequenced more than once from good quality spectra but could not be confirmed by MS3 [MS3: Fragmentation of tryptic peptides by tandem MS/MS, further fragmented by ion trap instruments, and analyzed again by MS (MS/MS/MS). The C-terminal ions of the peptides provide low background MS3spectra [65] even at low subfemtomole levels. Fifty six of the confirmed PTM proteins were also found in the test and spine. Similar analyses of the ITM yielded 144 positively identified and 37 tentatively identified proteins. Of the 144 identified proteins 110 were present in the PTM. Members of the SM50 family: SM50, 37, 32, 29 were present and among the most abundant proteins, whereas SM30-E was the only SM30 family protein in that group. PM 27 and SM30-F were not identified. Table 3 shows the relative distribution of SM50 and SM30 family members in the mineralized structural elements. MSP130, MSP130-related -1, -2 and -3 were identified in the PTM, but MSP130-related-4 was absent.
TABLE 3.
Relative presence of SM50, SM30 and Sp-Clectin family members in skeletal elements.
| Protein | Glean3 No | Tooth | Spicule | Spine | Test |
|---|---|---|---|---|---|
| SM 30 FAM | |||||
| SM30-A | 00825 | − | −, (++) | − | − |
| SM30-B/C | 00826 | − | + | −,(++) | − |
| SM30-D | 00828 | ++ | − | ++ | + |
| SM30-E | 04867 | ++ | ++ | ++ | ++ |
| SM30-F | 04869 | − | − | + | − |
| Sp-Clect | 00164 | + | ++ | (+) | − |
| SM50 FAM | |||||
| SM50 | 18811 | ++ | ++ | ++ | ++ |
| PM27 | 30147 | ++ | ++ | ++ | ++ |
| SM29 | 05990 | ++ | ++ | ++ | ++ |
| SM32 | 18810 | ++ | ++ | ++ | ++ |
| SM37 | 18813 | ++ | ++ | ++ | ++ |
| Sp-Clect-13 | 05989 | ++ | ++ | ++ | ++ |
| Sp-Clect-25 | 11163 | (+) | ++ | ++ | ++ |
| Sp-Clect-76 | 13825 | ++ | ++ | + | ++ |
An important observation was the presence of P16, previously identified as a PMC specific protein in embryonic spicules [2, 44, 47], but isolated as abundant and wide spread in mature teeth [51] was identified in two forms in PTM (Glean3: 18406 and 18407). The form Glean3:18406 did not show any signal sequence whereas 3:18407 did. These proteins had emPAI scores placing them as comparable in abundance to SM37 and MSP130. They were recovered in Gel slices 7–11, in the 14<Mr <40 range, Figure 11. The 2-D gel shown in Figure 9A confirms this low molecular weight range, and points out that the P16 is likely to be phosphorylated and its concentration highest in the fully developed and mineralized central portion of the tooth, Figure 9B. P19, which had been isolated from mature teeth [51] and sequenced along with P16, was not detected in the PTM but was found [60] in the ITM proteome, confirming that P19 is probably an intracellular protein not directly involved with mineral deposition. As suggested from its sequence and content of a nuclear localization sequence, it is most probably localized in the nucleus. The fact that the P19 sequences were found in the very high molecular mass portion of a gel (comparable to bands 1–3, Figure 11) indicates that ~ 19 kDa P19 protein is complexed into stable aggregates with other nucleoproteins.
In our first attempts at characterizing the digests of proteins isolated from the L.v. teeth [10], we had obtained partial sequences of peptides with Ala- and Pro-rich PA, PAA, and PQ repeats, or acidic peptides with ESSG, GSSSSEGSSD and SDSTSD repeats, but at that time no data bases were available to help sequence completion. It was gratifying to see that the proteome of the intact tooth matrix did contain some similar Ala- and Pro-rich sequences, as well as acidic Gly-rich motifs [60] one of which appears to correspond to the sequences of the protein unique protein presented as Glean3:17589, Figure 13. The protein shown was the most abundant of a group of proteins of similar domain structure. The blue-highlighted sequence was a Kazal-type serine protease inhibitor sequence. In fact, several related proteins were described with similar compositions. This is a set of proteins that should be examined closely. The presence of domains with a high tyrosine content provides an interesting way to get into a study of their conformation, properties and potential cross-linking reactions via glycation.
Another component of interest was mortalin, a member of the HSP70 family that we had found primarily in the plumula region of the tooth, at the point of differentiation and formation of the syncytial layers in the newly forming plumula [66]. Mann et al. have reported [60] that they could not find the mortalin, but the plumula is the most exposed and least mineralized part of the tooth, thus particularly susceptible to oxidation by the hypochlorite digestion. This again shows how the preparation methods used must be considered in determining the presence of any particular component in a complex, manycompartment tissue. In this case, however, it is not likely that mortalin has any role in mineral formation.
7. THE POTENTIAL ROLE OF PHOSPHOPEPTIDES IN CALCITE MINERALIZATION PROCESSES
It has been a little strange to propose that phosphate moieties on the matrix proteins might be important in regulating the in vivo formation of some calcites, but our initial findings of phosphoproteins in calcite mineralizing systems, by the same methods that had been applied in dentin, emboldened my laboratory to pursue this point even in the face of what one might modestly say was disregard. Recently Mann et al. [61, 62] repeated their study of test, tooth and spicule matrix by virtually the same methodology for protein isolation as they had used before [59,60], with the exception that specific attention was devoted to the question of phosphorylation. This involved concentrating the tryptic or other phosphopeptides by adsorption to TiO2 then using a neutral loss activation method followed by quadrupole ion trap MS [67] analysis of the phosphopeptides. The improved sensitivity of the ion trap procedures brought the total list of spicule matrix proteins to 231, of which 218 proteins were identified with high confidence [62]. The majority of the proteins had been previously identified, but most of these related to cellular enzyme activities, to resident proteins of the endoplasmic reticulum or Golgi, or transmembrane constituents, but were present in greater abundance than previously seen. These data suggested that the spicule matrix contained many occluded proteins of cellular origin that had been trapped within the mineral during the rapid and dynamic mineral development, but not as direct participants in the mineralization process. There were no surprising phosphoproteins detected within the spicule matrix. However, when the same protocols were applied to the mature test and tooth matrix, 34 additional phosphoproteins were detected. While many of these phosphoproteins were orthologs of mammalian kinases and signal transduction proteins, with conserved phosphorylation sites, there were some sea urchin specific phosphoproteins. Most notably and not previously described was an acidic and highly phosphorylated protein that was specific to the tooth proteome and absent in the test, hence termed phosphodontin. A second, unique non-acidic but phosphorylated protein was Glean3: 20139, a threonine and proline-rich protein. However, it was present in the test and not in the tooth. It was also determined that SM30-E was phosphorylated at several sites that were predicted to be kinase substrates for phosphorylation, but phosphorylation of those SM30-E sites was only partial.
According to Mann et al. [61, 62] phosphodontin is the principal phosphorylated protein of the mature tooth matrix. This highly phosphorylated protein was missed in previous tooth proteome analysis because the specific search for phosphopeptides had not been included, although a non-phosphorylated peptide, Glean3:18919, had been reported. In the new study in which a specific search for the phosphopeptides was carried out, it was proposed that phosphodontin might be the second most abundant protein of the tooth matrix, second only to the SM50 protein. The composition of phosphodontin, with a calculated mass of 49,717.67 Da and pI of 3.77 in the non-phosphorylated form is presented in Table 4. Because of the overwhelming content of Asp and Glu residues the net charge at pH 7 was calculated to be -109. Figure 13 shows the unique nature of phosphodontin in several respects. First, all of the S residues, except for 6S and 30S in the most amino terminal sequence, are in SS pairs, many of these embedded in a sequence EXSSGEG where X=I, M or V. This is a classic casein kinase 2 substrate for at least one of the S resides. However, all three possibilities have been detected (pSS, SpS, and pSpS). Thus, there is an obvious phosphorylation micro-heterogeneity based on both phosphodontin sequence and presence of kinase activities. This requires much more study. If only one S in each SS pair is phosphorylated, and with the pKa of the phosphate monoesters at ~ 7, the added charge per phosphate would be 1.5, so the net charge per molecule at pH 7 would increase to at least -150. If both members of an SS pair were phosphorylated, as frequently appears to be the case, the net molecular charge would be increased further. Phosphodontin has not been isolated as yet for detailed study as protein, but as the gels shown in Figure 9 demonstrate, there are phosphoproteins in the phosphodontin molecular weight range in the mineralized tooth matrix. Importantly, our preliminary data show that the phosphoproteins are not distributed evenly in amount throughout an urchin tooth.
TABLE 4.
The amino acid composition of phosphodontin, residues/molecule
| AA | # | % | ||
|---|---|---|---|---|
| A | Ala | 9 | 1.29 | 1.94 |
| D | Asp | 29 | 6.71 | 6.26 |
| N | Asn | 8 | 1.84 | 1.73 |
| G | Gly | 58 | 6.66 | 12.53 |
| E | Glu | 116 | 30.13 | 25.05 |
| Q | Gln | 32 | 8.25 | 6.91 |
| P | Pro | 35 | 6.84 | 7.56 |
| K | Lys | 32 | 8.25 | 6.91 |
| R | Arg | 3 | 0.94 | 0.65 |
| S | Ser | 72 | 12.61 | 15.55 |
| I | Ile | 21 | 4.78 | 4.54 |
| L | Leu | 5 | 1.14 | 1.08 |
| F | Phe | 2 | 0.59 | 0.43 |
| H | His | 1 | 0.28 | 0.22 |
| V | Val | 12 | 2.39 | 2.59 |
| M | Met | 18 | 4.75 | 3.89 |
| T | Thr | 6 | 1.22 | 1.30 |
| W | Trp | 0 | 0.00 | 0.00 |
| Y | Tyr | 4 | 1.31 | 0.86 |
| C | Cys | 0 | 0.00 | 0.00 |
| 373 | ||||
The distribution of the glutamic acid residues is also of special interest. Within the central region of phosphodontin, from 34E to 423E, there are 16 EE pairs or longer stretches of (En, n≥3) sequence. There does not appear to be any regular structure in this very hydrophilic region. Thus the phosphodontin may be best modeled as a random chain molecule or as an intrinsically unstructured protein, dependent on interaction with other molecules or ions for specific ordered structure formation in vivo. The insertion of a Pro residue every 10 to 13 residues also mitigates against any long range order. Phosphodontin is clearly a protein comparable to phosphophoryn in vertebrate teeth [5, 30, 31] and mollusk shell acidic proteins [43] and merits detailed study. The absence of phosphodontin in the urchin spicule matrix is especially interesting. Perhaps the mineral stabilizing effect of proteins such as phosphodontin and phosphophoryn, and similar vertebrate “Sibling” proteins is important for long lived tissues, but not essential to the transient life of the embryonic urchin spicule.
The lightly phosphorylated test matrix threonine-proline rich protein, Glean3:20139, Figure 14 [61], presents a very different profile. The protein has an unmodified mass of 57,462 D and a pI = 10.53. It is a very hydrophilic protein with a positive charge of 10.5 at pH 7. It has 4 confirmed phosphorylated groups, three within acidic domains 295FVQDD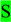 E
E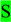 NEADED308,339NVAEAAGL
NEADED308,339NVAEAAGL NEVTQVK355, and one within the Q-rich domain, 370QQQPLPF
NEVTQVK355, and one within the Q-rich domain, 370QQQPLPF EQQQEY383. There is no specific data on the localization or function of this test matrix specific protein.
EQQQEY383. There is no specific data on the localization or function of this test matrix specific protein.
P19, identified in L.v. teeth [42] as a mineralization-related protein, and subsequently shown to be a phosphorylated protein [51], was predicted to have a nuclear import sequence. The P19 did not have a signal sequence, but was confirmed [61] to be phosphorylated on threonine rather than serine. The P19 function is not known, but as a nuclear protein we predict it to have an indirect and probably regulatory role in the mineralization process. P16 [42, 51, 55] is an enigmatic protein, clearly phosphorylated and abundant as a transmembrane protein [51] in L.v. with phosphorylation in an acidic sequence external to the syncytial membrane, at the mineral face, has not been seen in the proteome analyses. Mann et al.[62] have suggested that this may be because of the lack of Lys in the sequence, thus not providing small peptides. Although only 16 kDa in size, P16 is still too large for conventional MS/MS sequencing.
8. PROSPECTS FOR THE FUTURE
The combination of biochemical isolation of the protein components of the various skeletal elements, the isolation of mRNA and creation of cDNA libraries, and the availability of high through-put sequencing from mass spectrometry coupled with the knowledge of the complete genome of the model Strongylocentrotus purpuratus has provided a catalog of the majority of proteins of the sea urchin, applicable to other members of the echinoderm family. It is encouraging that many of the proteins detected were orthologs of most of the cellular proteins of other systems, including the vertebrates so that the basic echinoid cellular biochemistry must not be particularly unique.
Nevertheless there are several distinct echinoid proteins that may be linked somehow to the production of the urchin mineral. These were identified early on as members of the large SM50 and SM30 spicule matrix protein families. These proteins are also present in mature skeletal tissues, very widespread in distribution. They are characterized as mineralization-related proteins because they cannot be extracted and collected in most cases without first demineralizing the skeletal element. Yet there is no specific mineralization function related to the presence of these proteins that can be suggested from any data presently available. For example SM30-A is present exclusively in the spicule, while SM30-D is absent in the spicule, but present and abundant in the tooth, test and spine. On the other hand SM30–F is present only in the spine. SM30-E is present in all: spicule, tooth test and spine. Similarly, as shown in Table III, all members of the SM50 family are present in each tissue. In unpublished immunolocalization experiments currently in progress (Alvares and Veis), we find that within the L.v. tooth plumula the SM30 and SM50 proteins are distinctly different in distribution and appear to be most abundant in the matrix not associated with the mineral. In the same experiments and as seen earlier [51] earlier, anti-P16 is concentrated at the syncytial membranes and is prominent at the point of initial mineral deposition. Thus, Cheers and Ettensohn [55] were not only correct in pointing out that P16 was essential for spicule formation in the embryo, but P16 is likely to be directly involved in the nucleation of mineral at a syncytial membrane surface in the mature tooth syncytium.
As discussed above, Mann et al, [61] argued that they could not find P16 within their proteomic studies. The clear evidence from the cDNA-based cloning of the tooth mRNA, and isolation of the P16 from mature tooth tissue, and the observation that P16 is phosphorylated shows that one must be very cautious in interpreting the proteomic data. Finding the protein present is positive evidence of its presence, but lack of MS/MS derived evidence does not mean that a particular protein may not be present. Peptides may be missed based on peptide size, absence of cleavage sites or abundance. Similarly, proteins may be missed because of post translational modifications, as in the case of phosphorylation. Clearly, phosphorylation is an important attribute of some urchin mineral-related proteins. Sulfation and glycosylation also need detailed study. Further, protein abundance may not be a reliable indication of the importance of a particular protein involvement.
As defined above, the terms mineral-related and mineral-occluded protein need to be clarified. The mineral occluded proteins are those actually trapped within the calcite crystals as defects, probably a result of rapid calcification within a confined space simply enclosing the organic components resident within the space as described by Robach et al. [68], while the mineral –related proteins are those possibly at the surface or held between densely packed crystals, and thus inaccessible to solvents without dissolution of the crystals. In operational terms since both types of proteins are obtained only after mineral dissolution both are present in the various extracts and no function is ascribed to those proteins. A clear conceptual distinction between these mineral-related or mineral-occluded proteins and their role, if any, and that of the specific involvement of the mineral-interactive proteins in the nucleation of the crystallization process, the stabilization of the crystals, or the limitations on growth along particular crystal faces needs to be considered and examined. We hypothesize that it is these the mineral-interactive proteins that are important in regulating the processes of calcite formation. Thus future studies must deal with the specific roles of individual component mechanisms in crystal initiation and calcite growth. For those interested in using these data for planning biomimetic reconstruction of calcitic tissues it will also be of importance to investigate the role of the matrix proteins in controlling the composition of the high magnesium calcites and their effect on mineral solubility and other properties. A great deal of detailed study lies ahead.
Fig. 10.
Domain structures of spicule matrix family proteins. Black box = signal sequence, hatched box = proline-rich repeat domain, CTL = C-type lectin domain. Livingston et al. With permission [2].
Fig. 11.
SDS Page of hypochlorite washed PTM acetic acid extract. Marks along the right side denote how the equivalent unstained gels were cut. The protein identified within bands 7–11 contained Sp P16. Compare with the phospho-stained gels of Figure 9. From Mann et al [62], with permission..
Fig. 12. Alignment of  [Glean3:13823] and
[Glean3:13823] and  [Glean3:06387] sequences.
[Glean3:06387] sequences.
The sequence alignment comparison of the test Sp MSP130-3 (yellow highlight and spicule MSP130 (green highlight). There are clearly some major domains in each molecule not duplicated in the other, but both have PN-rich (red font) and PQ-rich domains, which are most prominent from test residue 292 to 396. The positions of sequence identity are indicated by an underline of the spicule sequences. The N-terminal region, from test residue 11 to 123 is identical to spicule residues 2 to 115. A much longer region of identity is in the C-terminal sequence domains where test residue 292 to 777 is virtually identical to spicule msp130 resdues 471 to 957. The choice of using the test sequence as the reference sequence is arbitrary.
Fig. 13.
The Amino Acid Sequence of Phosphodontin. Glean3:18919. The color coding point out the repetitive placement of SS and EE or longer sequences.
Fig. 14.
The Amino Acid Sequence of Test Matrix Protein. Glean3:20139 Threonine and Proline rich protein. The color coding marks the phosphorylated S residues  , an extended Q-rich domain
, an extended Q-rich domain  , and separate T,P-rich domain
, and separate T,P-rich domain  . The functions of these domains and the protein are yet to be determined.
. The functions of these domains and the protein are yet to be determined.
9. ACKNOWLEDGEMENTS
I am pleased to acknowledge that all of the work cited from my laboratory has been supported by Grant DE 001374 from the National Institute of Dental and Craniofacial Research, NIH.
I am also pleased to acknowledge the support and participation of Dr. Keith Alvares, Dr. Stuart Stock and Ms. Elizabeth Lux in much of the work presented, and especially to Keith Alvares for critically reading and helping to revise this manuscript.
Footnotes
This is an, un-copyedited, author manuscript that has been accepted for publication in the Frontiers in Bioscience". Cite this article as appearing in the Journal of Frontiers in Bioscience. Full citation can be found by searching the Frontiers in Bioscience (http://bioscience.org/search/authors/htm/search.htm) following publication and at PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed) following indexing. This article may not be duplicated or reproduced, other than for personal use or within the rule of "Fair Use of Copyrighted Materials" (section 107, Title 17, U.S. Code) without permission of the copyright holder, the Frontiers in Bioscience. From the time of acceptance following peer review, the full final copy edited article of this manuscript will be made available at http://www.bioscience.org/. The Frontiers in Bioscience disclaims any responsibility or liability for errors or omissions in this version of the un-copyedited manuscript or in any version derived from it by the National Institutes of Health or other parties.
REFERENCES
- 1.Sea Urchin Genome Sequencing Consortium. The Genome of the Sea Urchin Strongylocentrotus purpuratus . Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livingston BT, Killian CE, Wilt F, Cameron A, Landrum MJ, Ermolaeva O, Sapojnikov V, Maglott DR, Buchanan AM, Ettensohn CA. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus . Dev Biol. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 3.Donoghue PCJ, Sansom IJ. Origin and early evolution of vertebrate skeletonization. Microsc Res Tech. 2002;59:352–372. doi: 10.1002/jemt.10217. [DOI] [PubMed] [Google Scholar]
- 4.Adoutte A, Balavoine G, Lartillot O, Prud’homme B, de Rosa R. The new animal phylogeny: Reliability and implications. PNAS. 2000;97:4453–4456. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veis A, Sabsay B. Bone and Tooth Formation. Insights into Mineralization Strategies. In: Westbroek P, deJong EW, Ed , editors. Biomineralization and Biological Metal Accumulation. Dordrecht, Holland: D. Reidel Pub. Co.; 1983. [Google Scholar]
- 6.Veis DJ, Albinger TM, Clohisy J, Rahima M, Sabsay B, Veis A. Matrix Proteins of the Teeth of the Sea Urchin Lytechinus variegatus. J Exptl Zool. 1986;240:35–46. doi: 10.1002/jez.1402400106. [DOI] [PubMed] [Google Scholar]
- 7.Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314:956–959. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 8.Stock SR, Barss J, Dahl T, Veis A, Almer JD. X-ray absorption microtomography (microCT) and small beam diffraction mapping of sea urchin teeth. J Struct Biol. 2002;139:1–12. doi: 10.1016/s1047-8477(02)00500-2. [DOI] [PubMed] [Google Scholar]
- 9.Stock SR, Nagaraja S, Barss J, Dahl T, Veis A. X-ray MicroCT study of the pyramids of the sea urchin Lytechinus variegatus . J Struct Biol. 2003;141:9–21. doi: 10.1016/s1047-8477(02)00554-3. [DOI] [PubMed] [Google Scholar]
- 10.Veis A, Dixit SN, Barss J, Robach J, Alvares K, Malone J, Lux E, Stock S. Mineral-Matrix Relationships in the Developing Tooth of the Sea Urchin, Lytechinus variegatus . In: Arias JL, Fernández MS, editors. Biomineralization: From Paleontology to Materials Science. Santiago, Chile: Editorial Universitaria; 2007. [Google Scholar]
- 11.Holland ND. An autoradiographic investigation of tooth renewal in the purple sea urchin (Strongylocentrotus purpuratus) J Exptl Zool. 1965;158:275–282. doi: 10.1002/jez.1401580304. [DOI] [PubMed] [Google Scholar]
- 12.Killian CE, Metzler RA, Gong YUT, Olson I, Aizenberg J, Politi Y, Wilt FH, Scholl A, Young A, Doran A, Kunz M, Tamura N, Coppersmith SN, Gilbert PUPA. Mechanism of calcite co-orientation in the sea urchin tooth. J Am Chem Soc. 2009;131:18404–18409. doi: 10.1021/ja907063z. [DOI] [PubMed] [Google Scholar]
- 13.Robach JS, Stock SR, Veis A. Structure of first- and second-stage mineralized elements in teeth of the sea urchin Lytechinus variegatus . J Struct Biol. 2009;168:452–466. doi: 10.1016/j.jsb.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towe KM. Echinoderm calcite: Single crystal or polycrystalline aggregate. Science. 1967;157:1048–1050. doi: 10.1126/science.157.3792.1048. [DOI] [PubMed] [Google Scholar]
- 15.Ma Y, Aichmayer B, Paris O, Fratzl P, Meibom A, Metzler RA, Politi Y, Addadi L, Gilbert PUPA, Weiner S. The grinding tip of the sea urchin tooth exhibits exquisite control over calcite orientation and Mg distribution. Proc Natl Acad Sci USA. 2009;106:6048–6053. doi: 10.1073/pnas.0810300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heatfield BM, Travis DF. Ultrastructural studies of regenerating spines of the sea urchin Strongylocentrotus purpuratus I. Cell types without spherules. J Morph. 1975;145:13–50. doi: 10.1002/jmor.1051450103. [DOI] [PubMed] [Google Scholar]
- 17.Dubois P, Ameye L. Regeneration of spines and pedicellariae in echinoderms: A review. Microsc Res Tech. 2001;55:427–437. doi: 10.1002/jemt.1188. [DOI] [PubMed] [Google Scholar]
- 18.Heatfield BM, Travis DF. Ultrastructural studies of regenerating spines of the sea urchin Strongylocentrotus purpuratus II. Cell types with spherules. J Morph. 1975;145:51–72. doi: 10.1002/jmor.1051450103. [DOI] [PubMed] [Google Scholar]
- 19.Kniprath E. Ultrastructure and growth of the sea urchin tooth. Calc Tiss Res. 1974;14:211–228. doi: 10.1007/BF02060296. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki K. Spicule formation by isolated micromeres of the sea urchin embryo. Amer Zool. 1975;15:567–581. [Google Scholar]
- 21.Okazaki K, Inoue S. Crystal property of the larval sea urchin spicule. Dev Growth Differ. 1976;18:413–434. doi: 10.1111/j.1440-169X.1976.00413.x. [DOI] [PubMed] [Google Scholar]
- 22.Beniash E, Addadi L, Weiner S. Cellular control over spicule formation in sea urchin embryos: A structural approach. J Struct Biol. 1999;125:50–62. doi: 10.1006/jsbi.1998.4081. [DOI] [PubMed] [Google Scholar]
- 23.Benson SC, Jones EME, Crise-Benson N, Wilt F. Morphology of the organic matrix of the spicule of the ea urchin larva. Exp Cell Res. 1983;148:249–253. doi: 10.1016/0014-4827(83)90204-5. [DOI] [PubMed] [Google Scholar]
- 24.Pucci-Minafra I, Casano C, La Rosa C. Collagen synthesis and spiculae formation in sea urchin embryos. Cell Differ. 1972;1:157–165. doi: 10.1016/0045-6039(72)90025-5. [DOI] [PubMed] [Google Scholar]
- 25.Benson SC, Benson NC, Wilt F. The organic matrix of the skeletal spicule of sea urchin embryos. J Cell Biol. 1986;102:1817–1886. doi: 10.1083/jcb.102.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler E, Hardin J, Benson S. The role of lysyl oxidase and collagen crosslinking during sea urchin development. Exp Cell Res. 1987;173:174–182. doi: 10.1016/0014-4827(87)90343-0. [DOI] [PubMed] [Google Scholar]
- 27.Lowenstam HA. Minerals formed by organisms. Science. 1981;211:1126–1131. doi: 10.1126/science.7008198. [DOI] [PubMed] [Google Scholar]
- 28.Lowenstam HA, Weiner S. Mineralization by organisms and the evolution of biomineralization. In: Westbroek P, deJong EW, editors. Biomineralization and Biological Metal Accumulation. Dordrecht, Holland: D. Reidel Pub Co; 1983. [Google Scholar]
- 29.Glimcher MJ, Hodge AJ, Schmitt FO. Macromolecular aggregation states in relation to mineralization: the collagen hydroxyapatite system as studied in vitro. Proc Natl Acad Sci USA. 1957;43:860–867. doi: 10.1073/pnas.43.10.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veis A, Schlueter RJ. The macromolecular organization of dentin matrix collagen. I. Characterization of dentin collagen. Biochemistry. 1964;3:1650–1656. doi: 10.1021/bi00899a009. [DOI] [PubMed] [Google Scholar]
- 31.Veis A, Perry A. The phosphoprotein of the dentin matrix. Biochemistry. 1967;6:2409–2416. doi: 10.1021/bi00860a017. [DOI] [PubMed] [Google Scholar]
- 32.Benson S, Sucov H, Stephens L, Davidson E, Wilt FH. A lineage-specific gene encoding a major matrix protein of the sea urchin spicule. I. Authentication of the cloned gene and its developmental expression. Dev Biol. 1987;120:499–506. doi: 10.1016/0012-1606(87)90253-3. [DOI] [PubMed] [Google Scholar]
- 33.Venkatesan M, Simpson RT. Isolation and characterization of spicule proteins from Strongylocentrotus purpuratus . Exp Cell Res. 1986;166:259–264. doi: 10.1016/0014-4827(86)90526-4. [DOI] [PubMed] [Google Scholar]
- 34.Katijma T, Tomita M, Killian CE, Akasaka K, Wilt FH. Expression of spicule matrix protein gene SM30 in embryonic and adult mineralized tissues of sea urchin Hemicentrotus pulchrimus. Dev Growth Differ. 1996;38:687–695. doi: 10.1046/j.1440-169x.1996.t01-5-00012.x. [DOI] [PubMed] [Google Scholar]
- 35.Killian CE, Wilt FH. Characterization of the proteins comprising the integral matrix of Strongylocentrotus purpuratus embryonic spicules. J Biol Chem. 1996;271:9150–9159. doi: 10.1074/jbc.271.15.9150. [DOI] [PubMed] [Google Scholar]
- 36.Katoh-Fukui Y, Noce T, Ueda T, Fujiwara Y, Hashimoto N, Higashinakagawa T, Killian C, Livingston B, Wilt F, Benson S, Sucov H, Davidson E. The corrected structure of the SM50 spicule matrix protein of Strongylocentrotus purpuratus . Dev Biol. 1991;145:201–202. doi: 10.1016/0012-1606(91)90226-s. [DOI] [PubMed] [Google Scholar]
- 37.Killian CE, Wilt FH. The accumulation and translation of a spicule matrix protein mRNA during sea urchin embryo development. Dev Biol. 1989;133:148–156. doi: 10.1016/0012-1606(89)90306-0. [DOI] [PubMed] [Google Scholar]
- 38.George NC, Killian CE, Wilt FH. Characterization and expression of a gene encoding a 30.6 kDa Strongylocentrotus purpuratus spicule matrix protein. Dev Biol. 1991;147:334–342. doi: 10.1016/0012-1606(91)90291-a. [DOI] [PubMed] [Google Scholar]
- 39.Akasaka K, Frudakis TN, Killian CE, George NC, Yamasu K, Khaner O, Wilt FH. Genomic organization of a gene encoding the spicule matrix protein SM30 in the sea urchin Strongylocentrotus purpuratus. J Biol Chem. 1994;269:20592–20598. [PubMed] [Google Scholar]
- 40.Harkey MA, Klueg K, Sheppard P, Raff RA. Structure, expression, and extracellular targeting of PM27, a skeletal protein associated specifically with growth of the sea urchin larval spicule. Dev Biol. 1995;168:549–566. doi: 10.1006/dbio.1995.1101. [DOI] [PubMed] [Google Scholar]
- 41.Lee YH, Britten RJ, Davidson EH. SM37, a skeletogenic gene of the sea urchin embryo linked to the SM50 gene. Dev Growth Differ. 1999;41:303–312. doi: 10.1046/j.1440-169x.1999.413429.x. [DOI] [PubMed] [Google Scholar]
- 42.Illies MR, Peeler MT, Dechtiaruk AM, Ettensohn CA. Identification and developmental expression of new biomineralization proteins in the sea urchin Strongylocentrotus purpuratus. Dev Genes Evol. 2002;212:419–431. doi: 10.1007/s00427-002-0261-0. [DOI] [PubMed] [Google Scholar]
- 43.Gotliv B-A, Addadi L, Weiner S. Mollusk shell acidic proteins: in search of individual functions. Chembiochem. 2003;4:522–529. doi: 10.1002/cbic.200200548. [DOI] [PubMed] [Google Scholar]
- 44.Ameye L, Hermann R, Killian C, Wilt F, Dubois P. Ultracellular localization of proteins involved in sea urchin biomineralization. J Histochem Cytochem. 1999;47:1189–1200. doi: 10.1177/002215549904700911. [DOI] [PubMed] [Google Scholar]
- 45.Weiner S. Organic matrixlike macromolecules associated with the mineral phase of sea urchin skeletal plates and teeth. J Exp Zool. 1985;234:7–15. doi: 10.1002/jez.1402340103. [DOI] [PubMed] [Google Scholar]
- 46.Morrisy J. Silver stain for proteins in in polyacrylamide sels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 47.Sucov HM, Benson S, Robinson JJ, Britten RJ, Wilt F, Davidson EH. A lineage-specific gene encoding a major matrix protein of the sea urchin embryo spicule*1: II. Structure of the gene and derived sequence of the protein. Dev Biol. 1987;120:507–519. doi: 10.1016/0012-1606(87)90254-5. [DOI] [PubMed] [Google Scholar]
- 48.Harkey MA. Aspects of gene expression in the sea urchin micromere-primary mesenchyme cell line. In: Sawyer RH, Showman RM, editors. The Cellular and Molecular Biology of Invertebrate Development. University of South Carolina Press: The Belle W. Baruch Library in Marine Science # 15; 1985. [Google Scholar]
- 49.Anstrom JA, Chin JE, Leaf DS, Parks AL, Raff RA. Localization and expression of msp130, a primary mesenchyme lineage-specific cell surface protein of the sea urchin embryo. Development. 1987;101:255–265. doi: 10.1242/dev.101.2.255. [DOI] [PubMed] [Google Scholar]
- 50.Parr BA, Parks AL, Raff RA. Promoter structure and protein sequence of msp130, a lipid-anchored sea urchin glycoprotein. J Biol Chem. 1990;265:1408–1413. [PubMed] [Google Scholar]
- 51.Alvares K, Dixit SN, Lux E, Veis A. Echinoderm phosphorylated matrix proteins UTMP16 and UTMP19 have different functions in sea urchin tooth mineralization. J Biol Chem. 2009;284:26149–26160. doi: 10.1074/jbc.M109.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 53.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci U S A. 2010;107:14609–1461414. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheers MS, Ettensohn CA. P16 is an essential regulator of skeletogenesis in the sea urchin embryo. Dev Biol. 2005;283:384–396. doi: 10.1016/j.ydbio.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 56.Veis A, Barss J, Dahl T, Rahima M, Stock S. Mineral related proteins of the sea urchin teeth. Lytechinus variegates . Microsc Res Tech. 2002;59:342–351. doi: 10.1002/jemt.10216. [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poustka AJ, Kühn A, Groth D, Weise V, Yaguchi S, Burke RD, Herwig R, Lerach H, Panopoulou G. A global view of sea urchin expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 2007;8:1–18. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mann K, Poustka AJ, Mann M. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Proteome Sci. 2008;6:22–32. doi: 10.1186/1477-5956-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann K, Poustka AJ, Mann M. In depth, high accuracy proteomics of sea urchin tooth organic matrix. Proteome Sci. 2008;6:33–44. doi: 10.1186/1477-5956-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mann K, Poustka AJ, Mann M. Phosphoproteomes of Strongylocentrotus purpuratus shell and tooth matrix: identification of a major acidic sea urchin tooth phosphoprotein, phosphodontin. Proteome Sci. 2010;8:6–19. doi: 10.1186/1477-5956-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mann K, Wilt FH, Poustka AJ. Proteomic analysis of sea urchin (Strongylocentrotus purpuratus) spicule matrix. Proteome Sci. 2010;8:33–45. doi: 10.1186/1477-5956-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Killian C, Croker L, Wilt FH. SpSM30 gene family expression patterns in embryonic and adult biomineralized tissues of the sea urchin, Strongylocentrotus purpuratus. Gene Expression Patterns. 2010;10:135–139. doi: 10.1016/j.gep.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci USA. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alvares K, Dixit SN, Barss J, Lux E, Veis A. The proteome of the developing tooth of the sea urchin, Lytechinus variegatus: Mortalin is a constituent of the developing cell syncytium. J Exp Zool (Mol Dev Evol) 2007;308B:357–370. doi: 10.1002/jez.b.21159. [DOI] [PubMed] [Google Scholar]
- 67.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 68.Robach JS, Stock SR, Veis A. Transmission electron microscopy characterization of macromolecular domain cavities and microstructure single-crystal calcite tooth plates of the sea urchin Lytechinus variegatus . J Struct Biol. 2005;151:18–29. doi: 10.1016/j.jsb.2005.04.001. [DOI] [PubMed] [Google Scholar]