Abstract
Here we describe the genome of Mesotoga prima MesG1.Ag4.2, the first genome of a mesophilic Thermotogales bacterium. Mesotoga prima was isolated from a polychlorinated biphenyl (PCB)-dechlorinating enrichment culture from Baltimore Harbor sediments. Its 2.97 Mb genome is considerably larger than any previously sequenced Thermotogales genomes, which range between 1.86 and 2.30 Mb. This larger size is due to both higher numbers of protein-coding genes and larger intergenic regions. In particular, the M. prima genome contains more genes for proteins involved in regulatory functions, for instance those involved in regulation of transcription. Together with its closest relative, Kosmotoga olearia, it also encodes different types of proteins involved in environmental and cell–cell interactions as compared with other Thermotogales bacteria. Amino acid composition analysis of M. prima proteins implies that this lineage has inhabited low-temperature environments for a long time. A large fraction of the M. prima genome has been acquired by lateral gene transfer (LGT): a DarkHorse analysis suggests that 766 (32%) of predicted protein-coding genes have been involved in LGT after Mesotoga diverged from the other Thermotogales lineages. A notable example of a lineage-specific LGT event is a reductive dehalogenase gene—a key enzyme in dehalorespiration, indicating M. prima may have a more active role in PCB dechlorination than was previously assumed.
Keywords: lateral gene transfer, thermotogales, mesophilic, temperature adaptation
Introduction
The bacterial order Thermotogales was once thought to exclusively comprise thermophiles and hyperthermophiles. However, we have shown, using PCR amplification and metagenomic methods, that some Thermotogales lineages can be detected in—and likely thrive in—mesothermic environments (Nesbø et al. 2006, 2010). Here, we report the genome sequence of the type strain from the Thermotogales lineage most commonly detected in mesothermic environments—Mesotoga prima strain MesG1.Ag.4.2 (DSM 24739, ATCC BAA-2239) (Nesbø et al. 2012). The strain was isolated from a mesothermic anaerobic 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial enrichment culture inoculated with anaerobic sediments taken from the Northwest Branch of Baltimore Harbor, Baltimore, Maryland (Holoman et al. 1998). Characterization of this isolate showed that it has an optimal growth temperature of 37°C with a temperature range of 20–50°C (Nesbø et al. 2012). A closely related isolate from a mixture of marine sediments and sludge collected from a dump and a wastewater treatment plant in Tunisia also showed growth at a moderate temperature (Ben Hania et al. 2011), confirming the presence of this lineage in anaerobic mesophilic environments.
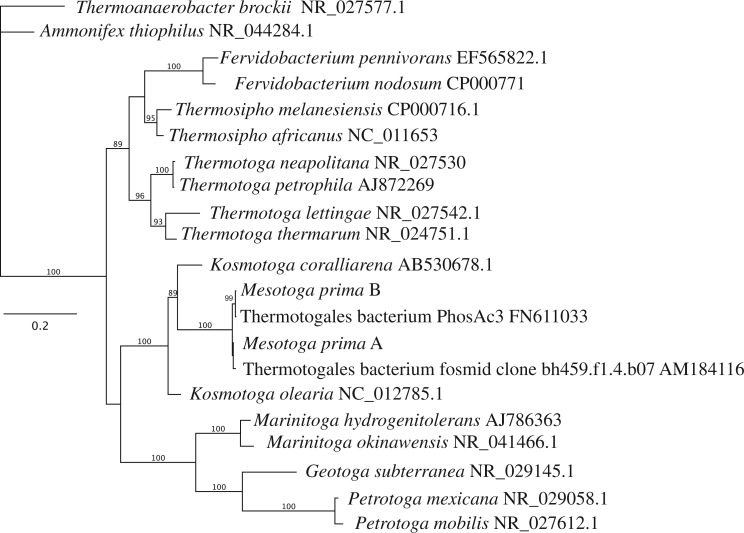
Earlier analyses of GC content of Thermotogales’ 16S rRNA genes and of amino acid composition of protein-coding genes suggested a thermophilic nature of the Thermotogales’ last common ancestor (Zhaxybayeva et al. 2009). This implies there may have been a secondary adaptation of the Mesotoga lineage to a mesophilic environment. According to the 16S rRNA phylogeny, the closest isolated relative of M. prima with a genome sequence available is Kosmotoga olearia (fig. 1), a thermophilic bacterium with optimal growth at 65°C, but with an extraordinarily wide temperature range: it can grow well at temperatures as low as 20°C (Dipippo et al. 2009). Given the mesophilic lifestyle of the Mesotoga lineage, we anticipate that the M. prima genome will provide insights into the evolutionary mechanisms of adaptation to moderate temperatures.
Fig. 1.—
Position of M. prima within the Thermotogales. The maximum likelihood tree of 16S rRNA sequences was reconstructed using the PhyML program (Guindon and Gascuel 2003), under GTR + G + I model with 100 bootstrap replicates. Only bootstrap support values above 70% are shown. The Mesotoga prima genome contains two 16S rRNA genes, labeled A and B. Thermoanaerobacter brockii and Ammonifex thiophilus were used as an outgroup. Sequences were aligned by the NAST aligner at GreenGenes (Desantis et al. 2006). GenBank accession numbers are shown for each sequence.
The Genome of M. prima Is at Least 0.7 Mb Larger Than Any Other Thermotogales Genome
The genome of M. prima MesG1.Ag.4.2 consists of a 2,974,229 bp circular chromosome with a GC content of 45.5%, and of a 1,724 bp plasmid. The chromosome is predicted to contain 2,736 genes, 2,660 of which are protein-coding. Eighty-six genes (3.14%) are predicted to be pseudogenes.
The size of the M. prima genome is considerably larger than those of other sequenced Thermotogales, which range from 1.86 Mb for Thermotoga maritima MSB8 to 2.30 Mb for K. olearia. Although the M. prima genome encodes more proteins, it is also less gene-dense than other Thermotogales: only 84.6% of its DNA is predicted to be located within protein-coding regions, in contrast to 88–97% in other Thermotogales genomes.
The M. prima chromosome contains a large, almost perfect, duplication of ∼80 kb (positioned from approximately nucleotide 1,761,470 to 1,928,740, and spanning ORFs Theba1633–Theba1781; Supplementary fig. S1). The region is flanked by group II introns containing reverse transcriptase domains. Eight additional copies of this intron are scattered across the region, which also contains three ORFs annotated as transposases. The presence of these 13 mobile genetic elements suggests this may be a dynamic region of the genome.
The plasmid is predicted to code for two hypothetical ORFs, the largest of which exhibits significant similarity to plasmid replication initiation factors (gene family Pfam02486). The plasmid shows characteristics of unidirectional replication, and GC, RY, and MK skews were linear (Grigoriev 1998; Saillard et al. 2008), suggesting that the plasmid replicates via rolling circle replication, similar to the only other described Thermotogales plasmid (Harriott et al. 1994).
The M. prima genome contains CRISPR-Cas adaptive immunity systems, which are similar to other Thermotogales (Makarova et al. 2011). Two Type I-B, Type III-B, and Type III (MTH326 variant) CRISPR-Cas systems are encoded in two loci (Supplementary table S1). The presence of seven CRISPR repeat loci suggests both systems are functional.
Mesotoga prima-Specific Genes and Expanded Gene Families Reveal a Larger Repertoire of Proteins with Regulatory Functions
Of 575 genes (21.6%) in M. prima with no detectable homologs in other Thermotogales genomes, the majority (383, or 67%) are annotated as hypothetical or uncharacterized proteins. Only 195 of the 575 genes could be assigned to COG categories, with 93 of those being the poorly characterized protein families (categories S and R). Among the Mesotoga-specific genes with a predicted function are a family of 6 dipeptidases, and 11 genes with “GCN5-related N-acetyltransferase (GNAT)” domains. GNAT-type enzymes are particularly numerous in the genome, with a total of 23 M. prima ORFs containing this domain. These enzymes are involved in a wide range of functions: some catalyze Nε- or Nα-acetylation of proteins controlling the activity of proteins, whereas others inactivate antibiotics through aminoglycoside N-acetylation (Hu et al. 2010).
Among all M. prima protein-coding genes, 1,092 could be grouped into 312 clusters of paralogous or xenologous genes, corresponding to a gene content redundancy of 40%. As observed in other Thermotogales genomes (Nelson et al. 1999; Nesbø et al. 2009; Zhaxybayeva et al. 2009), the largest families encode ABC transporters, GGDEF and HD domain containing proteins, as well as mobile genetic elements: for example, there are 19 copies of two different group II intron-carrying reverse transcriptases, 10 copies of an IS1634 type insertion sequence and 10 copies of an IS256 type insertion sequence. Notably, M. prima has more transcription regulators (89) than other Thermotogales genomes currently available at the Integrated Microbial Genomes (IMG) portal: T. lettingae has 68, whereas the remaining 14 Thermotogales have between 33 and 54 regulators each. This increase in transcription regulators is consistent with a study by Konstantinidis and Tiedje (2004), which reports a positive correlation between the number of transcriptional regulators and genome size.
A Large Proportion of Genes in M. prima Has Been Involved in Lateral Gene Transfer
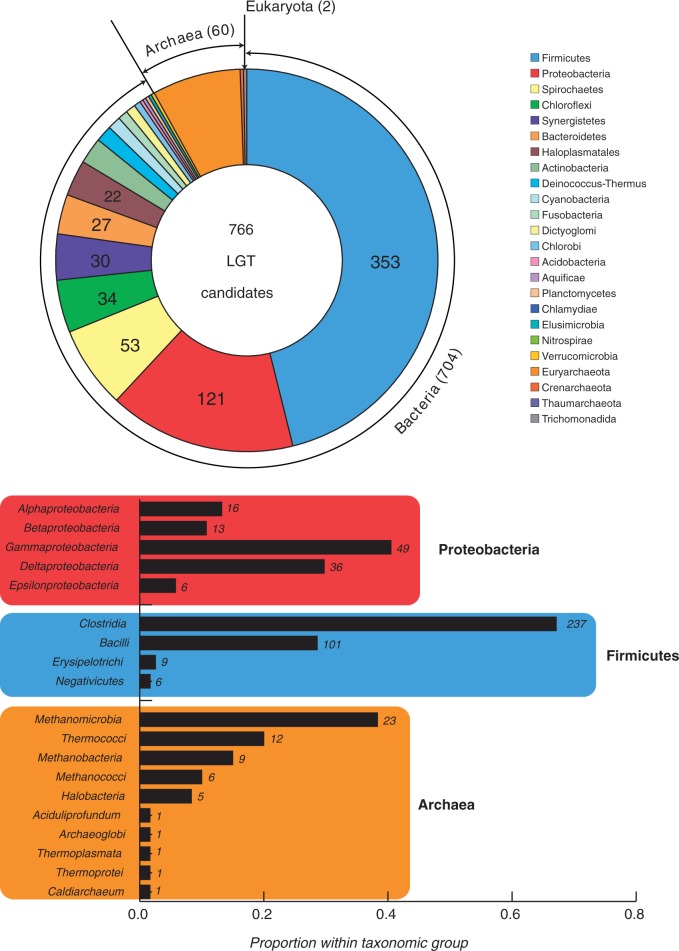
The DarkHorse program (Podell and Gaasterland 2007) predicts that 766 of the M. prima protein-coding genes (32% of genes with significant matches in the nr database) are likely of “foreign” origin (fig. 2). The genes with homologs in other Thermotogales genomes (523 of 766) have most likely been acquired after the M. prima lineage diverged. This subset could harbor genes that have replaced “native” Thermotogales genes and might be important in M. prima’s adaptation to a mesophilic lifestyle.
Fig. 2.—
Taxonomic distribution of putatively laterally transferred genes. The candidate genes were identified using the DarkHorse program (see Methods for details). NCBI Taxonomy database was used to assign organismal classification. The upper panel shows taxonomic distribution of 766 putatively transferred genes at the phylum level, whereas the lower panel details taxonomic distribution within the three most represented taxonomic groups.
Among the 766 putatively transferred genes, 353 are predicted to have been exchanged with Firmicutes—particularly with representatives of the class Clostridia (237 genes)—as has been observed for other Thermotogales genomes (Nelson et al. 1999; Nesbø et al. 2009; Zhaxybayeva et al. 2009). For thermophilic Thermotogales, a large proportion (34–40%) of their Clostridia matches were to thermophilic Thermoanaerobacterales (Zhaxybayeva et al. 2009). In contrast, only 45 of 237 M. prima genes exchanged with Clostridia (19%) belong to Thermoanaerobacterales, and the majority of the remaining 192 Clostridia matches were to mesophiles, many of which are found in the same type of environment as M. prima. Sixty genes (7.7% of putatively transferred genes) in the M. prima genome are predicted to have been exchanged with Archaea. Notably, 38 of these belong to methanogens, the most common archaeal representatives observed in mesothermic environments and cultures where Thermotogales 16S rRNA sequences have been detected (Nesbø et al. 2010). These observations support the genomic-based inferences that prokaryotes tend to exchange genes preferentially with organisms sharing their environmental niche (Zhaxybayeva et al. 2009; Smillie et al. 2011). A notable example of such a transfer is a catalase-encoding gene (Theba_0075) most closely matching a gene in Desulfitobacterium dichloroeliminans, a dehalo-respiring member of the Clostridia. There are no homologs of this gene in other Thermotogales, and catalases have been shown previously to be transferred frequently between microorganisms (Klotz 2003). Catalase is a heme-containing protein (Klotz 2003), and a cluster of transferred genes upstream of the M. prima’s catalase gene (Theba_0063–Theba_0068) could be involved in heme biosynthesis (Supplementary table S2).
Functional Comparison to Other Thermotogales
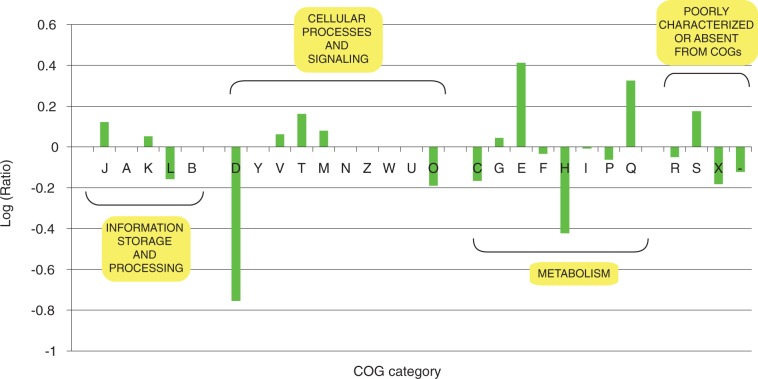
Classification of the putatively transferred genes into functional categories (fig. 3 and Supplementary table S3) indicates that the M. prima genome has, in comparison to that of K. olearia, exchanged a larger number of genes involved in signal transduction mechanisms (COG category T), secondary metabolite biosynthesis (COG category Q), and amino acid transport and metabolism (COG category E) (P < 0.01 in a χ2 two-sample test). Proteins from categories T and Q are involved in cell–cell interactions in the environment (Straight and Kolter 2009), and acquisition of such genes is likely to be advantageous when adapting to a new type of environment. Many of the COG category E genes are annotated as peptidases and proteases. Several of these proteins (e.g., a family of dipeptidases, COG4690) have predicted signal cleavage regions at their N-terminal end, suggesting that some might be extracellular and could potentially participate in interactions with other microorganisms in biofilms.
Fig. 3.—
Comparison of the distributions of transferred genes across functional categories in M. prima and K. olearia genomes. The categories on the X axis are defined according to the COG database (see Methods) and grouped into four super-categories. For each functional category, a value above the Y axis shows the overrepresentation of transferred genes in M. prima in comparison to those in K. olearia genome, whereas the value below Y axis shows the overrepresentation of transferred genes in K. olearia in comparison to the M. prima genome. Overrepresentation is defined as a ratio of the proportion of transferred genes in two genomes.
One notable family of genes missing in both the M. prima and the K. olearia, but abundantly represented in other Thermotogales genomes (5–15 genes per genome), is a family encoding proteins with similarity to COG840 (methyl-accepting chemotaxis proteins involved in signal transduction). This absence suggests that the Mesotoga and Kosmotoga lineages have different proteins for interactions with their environments than other Thermotogales.
Evolution of Threonine Synthase Gene Family
An example of an M. prima gene-family-expansion through lateral gene transfer (LGT) is threonine synthase (TS) (Supplementary fig. S3). Mesotoga prima has seven genes from this family, including one pseudogene (Theba_0615), whereas other sequenced Thermotogales genomes have at most two. Phylogenetic analyses of this gene family show that two of these genes (Theba_0160 and Theba_0167) originated from a recent duplication of a “native” Thermotogales gene. Theba_1253, a divergent homolog of the other five genes in this family, is found in most Thermotogales genomes. The remaining three genes appear to have been acquired from different bacterial lineages: Theba_0936 clusters with Kosmotoga and Thermosipho within a Synergistetes clade, Theba_2070 is found in a clade containing Clostridia and Synergistetes, and Theba_0634 is found in a cluster containing a δ-proteobacterium and a Chloroflexi (Supplementary fig. S2). It is unclear why M. prima encodes so many homologs of TS. In the methanogen Methanosacina acetivorans, one of its two TS genes has evolved a cysteate synthase function and is involved in coenzyme M synthesis (Graham et al. 2009), whereas in mammals a TS homolog has acquired a catabolic phosphor-lyase function (Donini et al. 2006). Hence, it is plausible that some of the TS genes in M. prima have been recruited for new functions.
Transferred Genes Are Clustered in the M. prima Genome
Among the genes predicted by DarkHorse as transferred, 591 are found in clusters in the genome: we discerned 160 clusters of two or more adjacent genes (3.7 genes on average, Supplementary table S2). One of the gene clusters (table 1) contains a putative reductive dehalogenase (Theba_2470), the key enzyme in halo-respiration that is primarily found in halo-respiring microorganisms such as Dehalococcoides ethenogenes (Smidt and de Vos 2004), suggesting that M. prima might contribute to the dehalogenation of polychlorinated biphenyls (PCBs) observed in the enrichment culture from which this bacterium was isolated. On a phylogenetic tree Theba_2470 groups with sequences from Clostridia, but none of the closest matches have been characterized and confirmed as reductive dehalogenases (Supplementary fig. S3). Therefore, from sequence data alone it is impossible to infer if this is indeed a reductive dehalogenase, and its biochemical characterization is underway (Edwards E, personal communication). Another gene in this cluster shows significant similarity to mercuric reductase, which is involved in mercury resistance by reducing ionic mercury [Hg(II)] to the elemental, less toxic form. This cluster appears to be a mosaic composed of genes originating from several different lineages, and may represent an environmental island targeting toxic compounds.
Table 1.
Reductive dehalogenase-containing cluster of putatively transferred genes
| Gene | Function | Taxonomic classification of the top Darkhorse hit |
|---|---|---|
| Theba_2462 | Zn-dependent protease with chaperone function | Bacteriovorax marinus SJ |
| Theba_2463 | hypothetical protein | Capnocytophaga ochracea F0287 |
| Theba_2464 | small-conductance mechanosensitive channel | Solibacillus silvestris StLB046 |
| Theba_2465 | ABC-type proline/glycine betaine transport systems, permease component | Rahnella sp. Y9602 |
| Theba_2466 | glycine betaine/L-proline transport ATP binding subunit | Streptomyces sp. SirexAA-E |
| Theba_2467 | ABC-type proline/glycine betaine transport systems, permease component | Dehalogenimonas lykanthroporepellens BL-DC-9 |
| Theba_2468 | periplasmic glycine betaine/choline-binding (lipo)protein of an ABC-type transport system (osmoprotectant binding protein) | Escherichia coli PCN033 |
| Theba_2469 | mercuric reductase/Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) component, and related enzymes | Ktedonobacter racemifer DSM 44963 |
| Theba_2470 | putative reductive dehalogenase | Clostridium difficile QCD-23m63 |
Amino Acid Composition of M. prima Proteins
Two compositional features of protein sequences have been suggested to correlate with optimal growth temperature of an organism: overrepresentation of charged amino acid residues versus polar ones (CvP bias) and overrepresentation of the IVYWREL group of amino acids (Suhre and Claverie 2003; Zeldovich et al. 2007). Previous analysis of five Thermotogales genomes (Zhaxybayeva et al. 2009) has shown that distributions of CvP values are unimodal for each of the genomes examined, with the mean CvP values > 10.62, a cutoff value suggested to indicate thermophily (Suhre and Claverie 2003). The distribution of CvP values for M. prima’s protein coding genes is also unimodal (data not shown) and has a mean value of 8.96, suggesting that compositionally M. prima’s proteins are on average not suitable for a thermophilic lifestyle. A subset of 766 putatively transferred genes was not enriched in proteins with low CvP values (data not shown). This evidence hints that M. prima’s adaptation to a lower temperature environment is not a recent evolutionary event.
Relationship of M. prima to Other Thermotogales
Traditional 16S rRNA classification places M. prima as a sister group to K. olearia (fig. 1). Detected gene flux within M. prima and other Thermotogales genomes (Zhaxybayeva et al. 2009) and skewed amino acid composition of M. prima’s proteins complicate pinpointing of the specific phylogenetic position of M. prima within Thermotogales. Pairwise comparisons of M. prima with other Thermotogales genomes reveal that the average amino acid identity (AAI) of the shared genes, a measure of relatedness complementary to the traditional 16S rRNA classification (Konstantinidis and Tiedje 2005), places the K. olearia genome as the closest to M. prima (Supplementary table S4). Functional comparisons, however, produce a different pattern. COG profile comparisons suggest that the M. prima genome is functionally more similar to that of Thermotoga lettingae (Pearson correlation coefficient [Pcc] = 0.83), than to K. olearia (Pcc = 0.78). Mesotoga prima and T. lettingae inhabit similar types of environments, and both are frequently recovered from mesothermic bioreactors and fermenters (Nesbø et al. 2010), which may explain the observed similarities between their two genomes.
Is M. prima’s Larger Genome Size Linked to Its Different Lifestyle?
To what extent are the differences in genome size within Thermotogales due to adaptive or neutral evolutionary processes? Chaffron et al. (2010) observed that genomes from similar environments tend to be similar in size, suggesting that environmental parameters determining an ecological niche can influence genome size. Although it appears that there is a correlation between optimal growth temperature and genome size in the Thermotogales (Supplemental fig. S4), this correlation fades when phylogeny is taken into account using Felsenstein’s (1985; data not shown) contrasts method. Whether this correlation, which can be dubbed “Bergmann’s Rule for genome size” in analogy to the “Bergmann’s Rule for body size” in animals (Meiri 2010), can be disentangled from the effects of phylogeny, is to be resolved by sequencing of additional Thermotogales genomes from different environments.
The direct influence of genetic drift on genome size has been recently hypothesized (Lynch 2006; Kuo et al. 2009). Lynch’s (2006) mutational-burden hypothesis postulates that due to larger effective population size (Ne), the power of genetic drift is diminished in prokaryotes, and therefore their genomes are refractory to accumulation of extraneous DNA and remain streamlined, in comparison to eukaryotes. Within prokaryotes, this hypothesis is exemplified by an extreme genome streamlining in an abundant marine bacterium Pelagibacter ubique (Giovannoni et al. 2005). Kuo et al. (2009), on the other hand, propose that lower Ne, with the associated genetic drift, can lead to an excess of non-synonymous substitutions, resulting in gene inactivation, subsequent gene loss due to deletion bias, and a more streamlined genome. How does the genome size distribution in the Thermotogales fit into these hypotheses? According to Lynch (2006), the less-streamlined M. prima genome could simply reflect a lower Ne of M. prima populations versus those of other Thermotogales. The hypothesis of Kuo et al. (2009) would predict the opposite: higher Ne, higher gene density, and lower rates of non-synonymous to synonymous substitutions in a larger genome of M. prima. In the M. prima genome we observed lower gene density than in other Thermotogales genomes. Thus, at first glance the Thermotogales genomes appear to follow the prediction of Lynch (2006). However, more extensive population-level data for Mesotoga spp. and thermophilic Thermotogales is needed to decipher the relative roles of drift and selection in genome evolution within these lineages.
The physiological characterization of M. prima under a wide range of substrates and growth conditions showed that it has a slower growth rate than other Thermotogales, with an optimal doubling time of 16.5 h compared with 3 h for its closest characterized relative K. olearia (Dipippo et al. 2009; Nesbø et al. 2012). Thus, it is tempting to speculate that M. prima may experience different selection pressures than other, faster growing Thermotogales, and therefore, from an ecological perspective, may employ a different life strategy. Slow growth rate alone does not allow us to distinguish whether M. prima predominantly encounters K-type- (i.e., maintenance of stable population close to carrying capacity K) or L-type selection (i.e., selection for stable populations close to minimal level L; Whittaker 1975). Mesotoga is often part of the “rare biosphere” (Sogin et al. 2006) in hydrocarbon-impacted environments (Nesbø CL, Foght J, and Zhaxybayeva O, unpublished), indicating that L-type selection and low Ne could be the major factors affecting its evolution. Future studies of Mesotoga populations “in the wild” are needed to elucidate other traits of its preferred life strategy, and to test if its populations are adapted to a combination of r-, K-, and L-type selection under different conditions (Whittaker 1975). It is likely that in comparison to their thermophilic relatives, Mesotoga spp. encounter more competition for resources in their mesothermic, more diverse, and species-rich environments (Kemp and Aller 2004). Perhaps M. prima has adapted to increased competition, as well as to its new thermal niche, by adjusting its growth- and life-strategies, and by acquiring new genes and functions from its new bacterial and archaeal “neighbors”.
Materials and Methods
Genome Sequencing, Assembly, Annotation, and Data Availability
Genomic DNA was isolated from an M. prima MesG1.Ag4.2 culture by using the protocol of Charbonnier and Forterre (1994). Genome sequencing, assembly and annotation were carried out by the U.S. Department of Energy Joint Genome Institute (JGI), and technical details can be found in Supplementary Material online. Pseudogenes were predicted by the JGI GenePRIMP pipeline (http://geneprimp.jgi-psf.org/). The genome and its annotation are available through the Integrated Microbial Genomes (IMG) portal (Markowitz et al. 2012) at https://img.jgi.doe.gov/, along with auxiliary data such as clustering of genes into paralogous groups and their assignment into functional categories (based on matches to COG, Pfam, TIGRfam, and InterPro databases). The genome is also available in GenBank under accession numbers CP003532 and CP003533. Pearson correlation coefficients of COG profiles were calculated in the IMG system.
Identification of M. prima-Specific Genes and Detection of Putative LGT Events
M. prima protein-coding genes were used as queries in BLASTP searches against the database of 16 completed and draft Thermotogales genomes: Kosmotoga olearia (NC_012785), Thermosipho africanus (NC_011653), Thermotoga neapolitana (NC_011978), Thermotoga maritima (NC_000853), Thermotoga sp RQ2 (NC_010483), Petrotoga mobilis (NC_010003), Thermotoga petrophila (NC_009486), Fervidobacterium nodosum (NC_009718), Thermosipho melanesiensis (NC_009616), Thermotoga lettingae (NC_009828), Thermotoga thermarum (NC_015707), Thermotoga naphthophila (NC_013642), Thermotoga maritima 2812B (CP003408), Thermotoga sp. cell2 (CP003409), Thermosipho africanus H17ap60334 (AJIP01000000), and Thermotoga maritima EMP (AJII01000000). Mesotoga prima genes with no significant matches were classified as Mesotoga-specific (E-value < 10−4 with database size set to the size of nr database). Predicted protein-coding genes of the M. prima genome were also used as queries for BLASTP searches against the nr database (E-value < 10−10). The BLAST search results were used to detect atypical ORFs using the DarkHorse program (Podell and Gaasterland 2007), excluding from consideration M. prima sequences present in nr database. Functional categories of all ORFs were assigned based on BLASTP searches against the COG database (Tatusov et al. 2001). Using the same procedure, atypical ORFs were detected and classified into functional categories for K. olearia protein-coding genes. Commonality of the distributions of putative transferred genes across functional categories in M. prima and K. olearia genomes were tested using a χ2 two-sample test.
Amino Acid Composition of Proteins
Absolute differences between charged (KRDE) and polar (NQST) amino acid residues (CvP bias) of predicted proteins in each of five genomes were calculated using in-house Perl scripts. Only proteins with fewer than two predicted transmembrane helices (1836 out of 2511) were used, as determined using the TMAP program of the EMBOSS package (Rice et al. 2000).
Supplementary Material
Supplementary figures S1–S4 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dr Kira Makarova for help with CRISPR classification, and Lauren Bradford for technical assistance. This work is supported by West Virginia University start-up funds to O.Z., by a Norwegian Research Council award (project no. 180444/V40) to C.L.N., and by National Science Foundation (DEB 0830024) and NASA Exobiology Program (NNX08AQ10G) grants to K.M.N. Mesotoga prima genome sequencing and annotation were conducted by the U.S. Department of Energy Joint Genome Institute, under contract No. DE-AC02-05CH11231 from the U.S. Department of Energy Office of Science. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Literature Cited
- Ben Hania W, et al. Cultivation of the first mesophilic representative (“mesotoga”) within the order Thermotogales. System Appl Microbiol. 2011;34:581–585. doi: 10.1016/j.syapm.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Chaffron S, Rehrauer H, Pernthaler J, Mering von C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20:947–959. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier F, Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J Bacteriol. 1994;176:1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantis TZ, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipippo JL, et al. Kosmotoga olearia gen. nov., sp. nov., a thermophilic, anaerobic heterotroph isolated from an oil production fluid. Int J System Evol Microbiol. 2009;59:2991–3000. doi: 10.1099/ijs.0.008045-0. [DOI] [PubMed] [Google Scholar]
- Donini S, et al. A threonine synthase homolog from a mammalian genome. Biochem Biophys Res Commun. 2006;350:922–928. doi: 10.1016/j.bbrc.2006.09.112. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Graham DE, Taylor SM, Wolf RZ, Namboori SC. Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase. Biochem J. 2009;424:467–478. doi: 10.1042/BJ20090999. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harriott O, Huber R, Stetter K, Betts P, Noll K. A cryptic miniplasmid from the hyperthermophilic bacterium Thermotoga sp. strain RQ7. J Bacteriol. 1994;176:2759–2762. doi: 10.1128/jb.176.9.2759-2762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoman TR, Elberson MA, Cutter LA, May HD, Sowers KR. Characterization of a defined 2,3,5, 6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl Environ Microbiol. 1998;64:3359–3367. doi: 10.1128/aem.64.9.3359-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp PF, Aller JY. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol. 2004;47:161–177. doi: 10.1016/S0168-6496(03)00257-5. [DOI] [PubMed] [Google Scholar]
- Klotz MG. The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol Biol Evol. 2003;20:1098–1112. doi: 10.1093/molbev/msg129. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-H, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19:1450–1454. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol. 2006;60:327–349. doi: 10.1146/annurev.micro.60.080805.142300. [DOI] [PubMed] [Google Scholar]
- Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri S. Bergmann’s Rule—what’s in a name? Global Ecol Biogeogr. 2010;20:203–207. [Google Scholar]
- Nelson KE, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Nesbø CL, et al. The genome of Thermosipho africanus TCF52B: lateral genetic connections to the Firmicutes and Archaea. J Bacteriol. 2009;191:1974–1978. doi: 10.1128/JB.01448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbø CL, et al. Mesotoga prima gen. nov., sp. nov., the first described mesophilic species of the Thermotogales. Extremophiles. 2012;16:387–93. doi: 10.1007/s00792-012-0437-0. [DOI] [PubMed] [Google Scholar]
- Nesbø CL, Dlutek M, Zhaxybayeva O, Doolittle WF. Evidence for existence of “mesotogas,” members of the order Thermotogales adapted to low-temperature environments. Appl Environ Microbiol. 2006;72:5061–5068. doi: 10.1128/AEM.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbø CL, Kumaraswamy R, Dlutek M, Doolittle WF, Foght JM. Searching for mesophilic Thermotogales bacteria: “mesotogas” in the wild. Appl Environ Microbiol. 2010;76:4896–4900. doi: 10.1128/AEM.02846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8:R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Saillard C, et al. The abundant extrachromosomal DNA content of the Spiroplasma citri GII3-3X genome. BMC Genomics. 2008;9:195. doi: 10.1186/1471-2164-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt H, de Vos WM. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- Smillie CS, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc Natl Acad Sci U S A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu Rev Microbiol. 2009;63:99–118. doi: 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- Suhre K, Claverie J-M. Genomic correlates of hyperthermostability, an update. J Biol Chem. 2003;278:17198–17202. doi: 10.1074/jbc.M301327200. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker RH. Communities and ecosystems. 2nd ed. New York: Macmillan Co; 1975. [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 2007;3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, et al. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci. 2009;106:5865–5870. doi: 10.1073/pnas.0901260106. [DOI] [PMC free article] [PubMed] [Google Scholar]