Abstract
Archived formalin-fixed paraffin-embedded (FFPE) tissue collections represent a valuable informational resource for proteomic studies. Multiple FFPE core biopsies can be assembled in a single block to form tissue microarrays (TMAs). We describe a protocol for analyzing protein in FFPE -TMAs using matrix-assisted laser desorption/ionization (MAL DI) imaging mass spectrometry (IMS). The workflow incorporates an antigen retrieval step following deparaffinization, in situ trypsin digestion, matrix application and then mass spectrometry signal acquisition. The direct analysis of FFPE -TMA tissue using IMS allows direct analysis of multiple tissue samples in a single experiment without extraction and purification of proteins. The advantages of high speed and throughput, easy sample handling and excellent reproducibility make this technology a favorable approach for the proteomic analysis of clinical research cohorts with large sample numbers. For example, TMA analysis of 300 FFPE cores would typically require 6 h of total time through data acquisition, not including data analysis.
INTRODUCTION
The ability to conduct mass spectrometry (MS)-based proteomic analyses of FFPE tissue opens new opportunities for clinical studies and biomarker discovery using hospital biopsy libraries. The majority of human tumor biopsies are currently stored as FFPE samples, so methods and technologies that permit analysis of large numbers of such samples are essential. When tissues are treated with formalin, an initial rapid reaction of formaldehyde principally with amino groups of basic amino acid residues such as arginine or lysine occurs, resulting in the formation of methylene bridges between amino residues with the formation of inter- and intra-molecular cross-linking of proteins1–3. Such insoluble cross-linkages maintain histomorphological information and allow preservation of the tissue integrity at the ultracellular level over time, but they also lead to difficulties with efficient protein mining. The process of antigen retrieval has been utilized for the partial reversal of such cross-links that sterically interfere in the binding of antibodies to linear protein epitopes in the tissue section4. We have established a protocol for the analysis of proteins from FFPE sections by adapting the heat-induced antigen retrieval method and following it with enzymatic hydrolysis, which provides tissue level peptide profiles and images directly from FFPE-TMA specimens using MALDI IMS5.
Advances in IMS technology over the past decade have enabled development of high-throughput proteomic approaches to identify and simultaneously localize biomolecules in tissue sections6–9. IMS is unique because it directly measures and images the analyte of interest as a consequence of the laser-induced desorption process. Thus, hundreds of analytes can be mapped simultaneously without the need for targeted reagents such as antibodies. Other imaging modalities such as fluorescence microscopy and immunohistochemistry (IHC)10–12 are generally more sensitive than IMS and use less complex instrumentation. However, IHC, for example, is a targeted imaging technology as it requires an analyte-specific antibody and where the accuracy and fidelity of the image analysis depends on the specificity of the antibody. Overall, these are complementary technologies, with each having particular advantages and limitations. IHC of FFPE tissues is still primarily used to measure a single protein at a time, and although fluorescence-imaging assays have the potential to be applied to large-scale analyses, the required hardware and software to do so are still being developed13. The reader is referred to several excellent reviews for further discussion of IHC methods14–16. Modern commercial IMS instruments can achieve spatial resolutions of 20–50 µm. However, for FFPE tissues in which trypsin must first be spotted to achieve a microdigest, these digest spots vary from 150 to 200 µm in diameter. Thus, if one assumes that analytes may migrate within this digest area, then the image spatial resolution achieved will be governed by the diameter/spacing of these digest spots. The most effective analyses are obtained with soluble proteins (2,000–50,000 Da) of moderate to high abundance. Hydrophobic proteins17, as well as proteins of higher molecular weight (MW), need special sample preparation protocols for analysis. For example, normal acquisition parameters for the mass spectrometer achieve the highest sensitivity for small molecules and proteins up to approximately 30–50 kDa. For proteins of higher MW, the instrument must be retuned and recalibrated specifically for the high MW region. Also, analysis of hydrophobic proteins requires the use of detergents, but these should be used at the lowest concentration possible to avoid signal suppression that may accompany detergent use. Nevertheless, many hundreds of specific molecular images can be generated from a single raster of a tissue section.
The application of IMS technology is enormously broad across biology and has been used in studies of animal and human tissue in both health and disease, in studies of the microbiome and of plants; it has also been used in drug development, in the study of distribution in tissue and in evaluations of drug efficacy18–21. There is substantial interest in the field of cancer biology in using IMS for the investigation of a large cohort of samples stored as TMAs, for diagnostic and prognostic purposes22, 23. At the protein level, distinct changes occur during the transformation of a healthy cell into a neoplastic cell, and these include altered expression levels, differential protein modification, changes in specific activity and aberrant localization, all of which can affect cellular function. Identifying and understanding these changes are the underlying themes in cancer proteomics. The combinatorial origin of malignant cells and the variability of the host background create molecularly distinct subgroups of tumors endowed with different phenotypes and clinical outcomes. This heterogeneity is only partially discerned by current immunohistochemical methods used by pathologists, which in many cases hinders the selection of the most appropriate diagnostic or therapeutic strategy for a given patient. Immunohistochemical techniques can be quite sensitive and selective, but they require analyte preselection, making it difficult to determine many compounds simultaneously and/or to identify unknown substances. We consider IMS to be an important new tool to distinguish different cancer histologies and subclassify individual cancer types5.
This protocol describes steps for analyzing FFPE tissue samples by IMS (Fig. 1). Under the proper sample preparation conditions for removal of the embedding medium and subsequent digestion, tryptic peptides are released from fixed proteins, and their sequence analysis and subsequent protein database searches enable identification of the nominal antecedent protein directly from tissue while preserving its spatial integrity within the tissue sample. Sequence determination of the tryptic fragments is performed using MALDI MS/MS analysis directly from the individual digest spots. Although the mechanism of antigen retrieval is still poorly understood24, old archival samples can also be analyzed using our protocol25. For example, we were able to identify proteins from a biopsy that had been stored in fixative for more than 110 years26. Further, we have investigated peptide profiling from different normal and tumor FFPE samples and across different organ sites. The analyses have proven to be robust and reproducible, even when comparing serial sections analyzed on 4 different days27. Although this general protocol is applicable to many tissue types, to achieve optimal results, the antigen retrieval parameters for each different tissue type should be optimized.
Figure 1.
Workflow for investigation of peptide content in FFPE-TMA. Flowchart showing the procedure for in situ proteomics on FFPE-TMA samples by imaging mass spectrometry.
Overall, this protocol allows protein identification from FFPE tissue sections with high accuracy and high throughput, and does not require complex chemistry or prior knowledge of the target molecules, as is the case with complementary technologies such as immunochemistry and fluorescence microscopy16.
Experimental design
TMA preparation and sectioning
Tissue biopsies are first analyzed and classified/diagnosed based on their pathology, often using standard H&E-stained sections. Cylindrical tissue cores (typically 0.6–1.0 mm in diameter, 4–6 mm in height) are removed from a specific area of interest within individual ‘donor’ paraffin blocks, and relocated in an array-like format into preformed holes, equally spaced 0.5 mm apart, in an empty recipient paraffin block (45 × 20 mm)28. The block is then sectioned using a microtome and transferred onto a slide for further analysis. A typical TMA section (15 × 15 mm, 4–8 µm thick) is shown in Figure 1. Each dot/circle corresponds to a cylindrical tissue core. The cores in the array may represent only a small portion of the original biopsy and so the choice of where to take the core in the biopsy is very important. Using a 0.6-mm core allows arraying at higher density, thereby maximizing the number of specimens for analysis (more than 500 tissue cores can be arranged on a standard glass microscope slide), while at the same time allowing inclusion of adequate internal experimental control samples, such as non-neoplastic control tissue, and tumor replicate needle cores from either the same or an unpaired biopsy. Inclusion of such controls assures standardization of parameters, such as section thickness, slide age, temperature and composition of reagents, incubation time and antigen retrieval, so that intra-assay variations are minimized. There are no imperative regulations for designing TMAs. However, the layout of the array should facilitate the analysis, for example, dividing large TMA into subsections. Distribution of the samples randomly across the array (and not sorted, e.g., by tumor stage or histological grade) helps to avoid unintended bias in the interpretation of the data.
The sections made from the FFPE-TMA are placed onto conductive slides (Indium tin oxide (ITO)-coated slides) to provide a tissue support for MALDI MS analysis29, as described in Steps 1–7 of the PROCEDURE. After drying, they can then be stored at room temperature (18–24 °C) almost indefinitely until needed for analysis.
Deparaffinization, H&E staining and antigen retrieval
Deparaffinization involves washing a wax-embedded specimen in xylene, and then rehydrating in a graded ethanol series before drying at room temperature. We have optimized the deparaffinization procedure for FFPE tissue, including TMA slides, by decreasing incubation time in xylene from 20 to 3 min with no deleterious effect on protein analysis (see http://www.abcam.com/index.html?pageconfig=resource&rid=11487 and Step 10 of the PROCEDURE).
To allow the selection of various histological regions for subsequent mass spectrometric analysis, tissue sections are typically stained with H&E for histological examination. One can then correlate a distinct histological area to the corresponding mass spectral data to obtain specific molecular information. Precise correlation of IMS images and histochemical images can be achieved by co-registering the results obtained using one tissue section for MALDI IMS analysis (Step 7, Steps 9–21 and 25–57), and a consecutive serial section for H&E staining (Steps 22–24), as described in the PROCEDURE. This co-registration is limited to the comparison of relatively large tissue features (± 50 µm) or groups of cells because of the uncertainty in alignment accuracy of the sections. Alternatively, methods have been reported in which H&E staining was performed on the same tissue section that previously underwent MALDI image acquisition29, 30.
Two common methods of antigen retrieval use either enzymatic or heat-mediated processes31–34. For IMS studies, proteolytic enzymes can be used to cleave proteins to peptides that can then subsequently be desorbed and analyzed in the MS. However, the degree of cross-linking can vary depending on the formalin fixation protocol used and, if it is not effective, this process can lead to the loss of certain epitopes35. The use of a heating step can be advantageous, because it denatures the proteins to the extent possible and allows more efficient proteolysis. This antigen-retrieval mechanism has become accepted and used by pathologists in clinical laboratories worldwide and functions as a simple and effective method to achieve satisfactory immunostaining on archival tissue sections36. Most methods use temperatures near 100 °C that can be achieved using a microwave oven, pressure cooker, autoclave or steamer. Although the temperature achieved by these methods appears to be the critical variable, studies comparing these techniques have shown that different heating methods provide similar levels of antigen retrieval, on the basis of immunostaining, if the heating times are adjusted appropriately37. We suggest using a pressure cooker as an effective and reliable method; it is very easy to use, time efficient and it uses inexpensive equipment.
pH is an important factor for recovering the formalin-modified antigenicity. Most antigen-retrieval protocols are performed at pH range 8–10. We incubate the slide in Tris-HCl buffer at pH 9.0, at 95 °C for 20 min5, 38 (Steps 11–15). After heating, we recommend leaving the slides standing in the hot buffer solution for 10–15 min before exchanging the buffer with water (Step 18). The main purpose of the cooling step is to prevent detachment of the tissue section from the slide.
In situ digestion and application of the matrix
In this procedure, trypsin solution followed by matrix solution is automatically spotted onto the section that has been previously deparaffinized and antigen retrieved. Some of the approaches used for the analysis of proteins from FFPE tissues38, 39 require considerable amounts of time and effort and have the limitation of inadequate spatial information. We previously described an on-tissue digestion technique by spotting the entire FFPE tissue section with enzyme solution in well-defined microspotted arrays5. We optimized different parameters, including the buffer used, the concentration of the enzyme, and the time and temperature. The procedure uses a reagent microspotter (Portrait 630) to apply a small drop (~160 pl per drop) of trypsin solution onto the tissue section (Step 34), creating an array of spots with 250 µm center-to-center spacing between each spot, as shown in Figure 2. A total of ~4.8 nl of trypsin solution at each spot is deposited over a series of 30 cycles at 1 drop per cycle. The digestion is carried out at room temperature with no major differences observed when compared with a digestion at 37 °C (R.C., unpublished data). With respect to time, an FFPE-TMA section with an area of ~15 × 15 mm, printed with an array of 4,000 spots, takes ~2.5 h for digestion to complete. Instrument calibration is essential for obtaining excellent drop-to-drop precision. This should be done before each run and also if the reagent has been in the reservoir for > 30 min. To optimize the droplet ejection settings, the calibration process automatically ejects droplets in a fixed pattern onto an appropriate metal calibration slide (25 × 75 mm). The shape of the calibration pattern should appear as an inverted triangular grid of spots with a size of about 7.5 mm2. After digestion, the tissue array is then robotically spotted with matrix solution (Step 38) for subsequent MALDI MS and MALDI MS/MS analyses to obtain sequence information for the tryptic peptides and thereby provide protein identification. The same amount of matrix solution (30 droplets per spot) is deposited directly on top of the tryptic spots. The time for which the droplets stay wet affects the quality of the spectra, with longer times allowing more efficient analyte extraction and better mass spectra. However, if too much liquid is deposited at one time, the resulting matrix spots are larger and may run into each other, potentially causing delocalization.
Figure 2.
Trypsin and matrix application. (a) Representative acoustic droplet ejection of picoliter volumes of reagent solution from the reagent source reservoir to a MALDI target plate holder. (b) TMA section scanned after 30 cycles of trypsin (top) and matrix (bottom), respectively.
Although we describe the use of a specific robotic instrument for reagent application, other devices may also be used. However, to achieve similar assay performance, spotting robots that provide drop deposition in the range of ≤200 pl are preferable so as to achieve an image resolution of not less than ~250 µm. Other reagent application protocols include spray deposition, accomplished using either a manual apparatus such as a thin-layer chromatography reagent sprayer or automated devices that use vibrational or pneumatic spray40–42. However the reagents are applied, it is important to achieve an even coating of reagent43.
Recently, a protocol was published describing a method for direct analysis of frozen and FFPE tissue samples44. This application requires special matrices (solid ionic matrices) for use with specific robotic spotters for the analysis of tissue. Our protocol uses one of the most commonly used matrices (α-cyano-4-hydroxycinnamic acid (CHCA)) for analysis of FFPE tissues, as well as being amenable to the use of other common matrices (see refs. 45 and 46).
Imaging mass spectrometry
The printed array is analyzed using a MALDI-time of flight (TOF)/TOF mass spectrometer operated in reflectron mode for increased mass resolution. Spectra are acquired in the range of m/z 700–5,000. In a typical procedure, the sample plate is placed in the mass spectrometer, and the laser is automatically targeted to specific spots on the plate through the use of selected fiducials. Images of the molecular species of the tryptic peptides are produced by moving the sample plate under the laser and acquiring averaged mass spectra at each spot in a raster of the target surface. A typical data array could contain thousands of spots, depending on the desired image resolution and the specified MW range. In our TMA study, each matrix spot is analyzed by averaging 1,600 laser shots acquired in 8 series of 200 shots per spot. Each series is acquired using a ‘random walk’ pattern to sample eight different locations within the matrix spot.
An important consideration for users of this protocol is the external calibration of the mass spectrometer using standards with molecular masses that cover the mass range of interest for identification. We used a custom calibration mixture consisting of four peptides: human angiotensin II (m/z 1,046.542), human [Glu1]-fibrinopeptide B (m/z 1,570.6774), bovine insulin chain A oxidized (m/z 2,530.921) and bovine insulin chain B oxidized (m/z 3,494.651). These peptides were chosen as calibrants because they fall within the 700–5,000 mass range and are relatively inexpensive and easy to obtain. The external calibration process will ensure high mass accuracy and increase statistical confidence during peptide identification, scoring, data processing and statistical analysis.
Data processing and statistical analysis
Data processing and interpretation is of paramount importance for accurate, reliable results. MALDI MS spectra require preprocessing for baseline correction, noise removal, realignment of the m/z scale, and peak selection and matching47 to reduce experimental variance within the data set. From the image data set, mass spectra from multiple regions in the sections can be exported for subsequent statistical analysis.
Data analysis software is typically provided by the manufacturer of the instruments, such as ClinProTools (Bruker Daltonics), Bioworks (Thermo Fisher) and MassLynx (Waters). For example, ClinProTools automatically picks peaks in the mass spectrum either on the basis of the calculated total average spectrum or on a single spectrum chosen by the user, and a peak list is then generated. Selection of the monoisotopic peak for each tryptic peptide requires manual editing of the peaks. In addition, spectral modification and selection filters may be used to reduce data and exclude spectra of lower quality from further processing. A subset of the statistically significant discriminator peptides between regions of interest are then selected and evaluated, on the basis of P values from the Wilcoxon/Kruskal-Wallis test and principal component values from principal component analysis, for further protein identification.
MS/MS peptides sequencing
Tandem MS/MS provides amino acid sequence information on peptide fragments from a specific peptide. Briefly, the peptide precursor of interest is selected by an ion gate (timed ion selector) that allows passage of the selected peptide of interest into a second analyzer where it is fragmented. Fragmentation may occur through metastable decay from internal energy of the ion, or it can be induced in a collision cell48.
We use the UltrafleXtreme MALDI-TOF/TOF mass spectrometer to acquire and fragment the selected peptides directly from a digested TMA section. The MALDI MS/MS spectra generated are then submitted into a MASCOT database search engine to match tryptic peptide sequences to their respective intact proteins. Together, peptide mass information derived from MALDI-TOF MS analysis, in conjunction with peptide sequence information obtained from MS/MS analysis, provides a strong basis for protein identification. It is noted that such peptide-centric methods for protein identification will identify the nominal protein with high confidence only if two or three (or more) peptides are sequenced from that protein.
Although sequencing of peptides by MS/MS is rather straightforward today, the process is not easily automated and consumes considerable time. Whereas search algorithms assist in the identification of the peptide sequence, manual interpretation of the fragmented spectrum may be needed to make an accurate assignment47.
Protein identification
A typical approach to protein identification in our laboratory is outlined in Figure 3. The resulting MS/MS spectra fragmentation patterns are searched against MASCOT database for corresponding sequence pattern. MALDI MS/MS analysis directly from an FFPE-TMA section allowed the identification of ~100 proteins.
Figure 3.
Protein identification workflow. Representative workflow for protein identification by MS/MS. (a) Tissue section is microspotted with trypsin and matrix for MALDI MS/MS analysis. (b) The mass spectrum produced is acquired, and a parent ion of m/z 1,460.9 is selected for fragmentation. (c) Magnification of the parent ion peak at m/z 1,460.9 shown in panel b. The peak at m/z 1,460.9 is the monoisotopic peak for the intact protonated peptide. The three peaks at higher mass, each separated by 1 Da, result from incorporation of one or more of the less abundant isotopes into the molecule, with 13C at a natural abundance of 1.11% being the main contributor. (d) MS/MS spectrum of the 1,460.9 parent ion and analysis of the fragment ions produced. This figure is reproduced, with permission, from reference 6.
If the database search is not fruitful, either because the protein has not been cataloged or is previously uncharacterized, or because the data are not accurate or comprehensive enough to distinguish between several entries in the database, then further information is required. It has to be considered that peptide/protein databases do not generally include the sometimes-extensive post-translational modifications of most proteins; indeed, such modifications are still unknown in most cases. However, for an analysis of post-translational modification, it is often an advantage to combine and/or compare the obtained MS-based results with in silico results from computational tools optimized for the prediction of sites likely to be modified. Database searching is essential to proteomics, but care must be taken to achieve identifications having high reliability. Generally, a minimum of two, and preferably three or more high-quality peptide identifications are necessary to establish the presence of a protein with high confidence48, 49.
The following PROCEDURE is typically used for the analysis of FFPE-TMA in our laboratory. However, for best results, some steps of the procedure (e.g., trypsin and matrix deposition) may require further optimization, depending on tissue density.
MATERIALS
REAGENTS
dH2O (Milli-Q purified water, Millipore)
Ethyl alcohol, 200 proof, absolute anhydrous ACS/USP grade (Pharmco-AAPR, cat. no. 111000200)
 It is a flammable liquid and vapor. Use in a fume hood. Wear gloves and safety goggles while handling.
It is a flammable liquid and vapor. Use in a fume hood. Wear gloves and safety goggles while handling.Xylene, ACS reagent (Acros Organics, cat. no. 42268-0040)
 It is harmful or fatal if swallowed. Causes severe eye, skin and respiratory tract irritation. Chronic exposure can cause adverse liver, kidney and blood effects. Xylene is a flammable liquid and harmful vapor. Use in a fume hood. Wear gloves and safety goggles while handling. Dispose of waste using approved disposal protocol.
It is harmful or fatal if swallowed. Causes severe eye, skin and respiratory tract irritation. Chronic exposure can cause adverse liver, kidney and blood effects. Xylene is a flammable liquid and harmful vapor. Use in a fume hood. Wear gloves and safety goggles while handling. Dispose of waste using approved disposal protocol.Trizma Base, minimum 99.9% titration (Sigma, cat. no. T1503).
 Causes eye, skin irritation. May be harmful if inhaled. Causes respiratory tract irritation. Wear chemical safety goggles protective gloves and use only in a well-ventilated area.
Causes eye, skin irritation. May be harmful if inhaled. Causes respiratory tract irritation. Wear chemical safety goggles protective gloves and use only in a well-ventilated area.Hydrochloric acid (min 35%–max 38% (vol/vol); Fisher Scientific, cat. no. A508-500)
 It is corrosive. Causes burns by all exposure routes. Thermal decomposition can lead to release of irritating gases and vapors. Wear chemical safety goggles and appropriate protective gloves and clothing to prevent skin exposure.
It is corrosive. Causes burns by all exposure routes. Thermal decomposition can lead to release of irritating gases and vapors. Wear chemical safety goggles and appropriate protective gloves and clothing to prevent skin exposure.Sodium hydroxide solution (50% (wt/wt), Fisher Scientific, cat. no. SS254-500)
 It causes severe burns if in contact with eyes and skin. May be harmful if inhaled or swallowed. Use only under a chemical fume hood. Wear personal protective equipment.
It causes severe burns if in contact with eyes and skin. May be harmful if inhaled or swallowed. Use only under a chemical fume hood. Wear personal protective equipment.Trypsin Gold, MS grade (100 µg; Promega, cat. no. V5280)
Acetic acid glacial (99.99 + % (vol/vol), Sigma-Aldrich, cat. no. 338826)
 It is flammable. May be harmful if inhaled. Causes severe skin burns. Material is extremely destructive to the tissue of the mucous membranes and upper respiratory tract. Handle with gloves and wear safety goggles.
It is flammable. May be harmful if inhaled. Causes severe skin burns. Material is extremely destructive to the tissue of the mucous membranes and upper respiratory tract. Handle with gloves and wear safety goggles.Ammonium bicarbonate (Sigma, cat. no. A-6141)
Acetonitrile (Fisher Scientific, cat. no. A998-4)
 It is toxic. Wear gloves when handling and operate in a ventilated chemical hood.
It is toxic. Wear gloves when handling and operate in a ventilated chemical hood.Trifluoroacetic acid (TFA, Fluka, cat. no. 73605)
 It is highly corrosive. Causes severe burns to contact areas of the body, including eyes, inhalation pathways and skin. Always work in the fume hood. Dilutions of TFA are created by adding TFA to water. Never add water to concentrated TFA. Use glass pipettes with bulbs to transfer TFA.
It is highly corrosive. Causes severe burns to contact areas of the body, including eyes, inhalation pathways and skin. Always work in the fume hood. Dilutions of TFA are created by adding TFA to water. Never add water to concentrated TFA. Use glass pipettes with bulbs to transfer TFA.α-Cyano-4-hydroxycinnamic acid (CHCA, Fluka, cat. no. 28480)
 It causes irritation to the eyes, skin and the respiratory system. Wear gloves and safety goggles.
It causes irritation to the eyes, skin and the respiratory system. Wear gloves and safety goggles.Ammonium hydroxide (28–30% (wt/wt), Sigma-Aldrich, cat. no. 320145)
 Inhalation of vapors causes respiratory tract irritation to pulmonary edema. Contact with liquid may cause severe pain, damage to skin, eye damage, and permanent blindness. Use glass pipettes with bulbs to transfer ammonium hydroxide. Wear chemical safety goggles and appropriate protective gloves. Handle in the fume hood.
Inhalation of vapors causes respiratory tract irritation to pulmonary edema. Contact with liquid may cause severe pain, damage to skin, eye damage, and permanent blindness. Use glass pipettes with bulbs to transfer ammonium hydroxide. Wear chemical safety goggles and appropriate protective gloves. Handle in the fume hood.Insulin chain B, oxidized from bovine pancreas (Sigma, cat. no. I-6383)
Insulin chain A, oxidized (Sigma, cat. no. I-1633)
[Glu1]-Fibrinopeptide B, human (Sigma, cat. no. F-3261)
Angiotensin II, human (Sigma, cat. no. A-9525)
Formic acid (88%, J.T. Baker, cat. no. 0128-01)
 Danger. Corrosive, causes severe burns to all body tissue. May be fatal if swallowed. Harmful if inhaled. Wear safety goggles, proper gloves and use under fume hood.
Danger. Corrosive, causes severe burns to all body tissue. May be fatal if swallowed. Harmful if inhaled. Wear safety goggles, proper gloves and use under fume hood.Eosin Y solution, intensified, histology grade (Fisher Scientific, cat. no. 314-630)
 It is an irritant and causes skin, severe eye and respiratory tract irritation. It is a flammable liquid and vapor. This substance has caused adverse reproductive and teratogenic effects in humans. May cause CNS depression. May cause liver, kidney and heart damage. Causes moderate skin irritation. Handle with gloves and wear safety goggles. Do not keep Eosin Y solution container open for long periods of time.
It is an irritant and causes skin, severe eye and respiratory tract irritation. It is a flammable liquid and vapor. This substance has caused adverse reproductive and teratogenic effects in humans. May cause CNS depression. May cause liver, kidney and heart damage. Causes moderate skin irritation. Handle with gloves and wear safety goggles. Do not keep Eosin Y solution container open for long periods of time.Phloxine B (Fisher Scientific, cat. no. 18472-87-2)
 It is an irritant and causes skin and eye irritation. Harmful if inhaled or ingested. Wear safety goggles and gloves.
It is an irritant and causes skin and eye irritation. Harmful if inhaled or ingested. Wear safety goggles and gloves.Hematoxylin (Sigma-Aldrich, cat. no. H-3136)
 It is an irritant. Do not keep the hematoxylin container open for long periods of time. Causes skin and eye irritation. Harmful if swallowed. Wear safety goggles and gloves.
It is an irritant. Do not keep the hematoxylin container open for long periods of time. Causes skin and eye irritation. Harmful if swallowed. Wear safety goggles and gloves.Glycerol (Acros Organics, cat. no. 15892-0010)
 It is an irritant. Glycerol may cause irritation to skin, eyes and respiratory tract. Wear appropriate protective gloves and safety goggles.
It is an irritant. Glycerol may cause irritation to skin, eyes and respiratory tract. Wear appropriate protective gloves and safety goggles.Sodium iodate (Alfa Aesar, cat. no. 40196)
 It is an oxidizer. May be flammable. Harmful if swallowed or inhaled. Causes irritation to skin, eyes and respiratory tract.
It is an oxidizer. May be flammable. Harmful if swallowed or inhaled. Causes irritation to skin, eyes and respiratory tract.Aluminum potassium sulfate × 12 H2O (dodecahydrate; Acros Organics, cat. no. 7784-24-9)
 It is an irritant and can be harmful if swallowed. It causes eye and skin irritation, and may cause respiratory tract irritation. Wear appropriate protective gloves and safety goggles.
It is an irritant and can be harmful if swallowed. It causes eye and skin irritation, and may cause respiratory tract irritation. Wear appropriate protective gloves and safety goggles.
EQUIPMENT
Gloves, safety goggles and kimwipes absorbent wipes paper (Fisher Scientific, cat. nos 19-130-1597, 19-034-794, 06-666A)
Indium tin oxide (ITO)-coated conductive slides (Delta Technologies, cat. no. CG-81IN-S115)
SuperFrost/Plus microscope slides (Fisher Scientific, cat. no. 12-550-15)
Plastic Coplin staining jar (Fisher Scientific, cat. no. 19-4)
Glass Coplin staining jar (Fisher Scientific, cat. no. 08-813E)
Paper filters (Whatman-Schleider & Schuell, cat. no. 1003-917)
Digital decloaking chamber (Biocare Medical, cat. no. DC2002)
Hot-hand protector (Fisher Scientific, cat. nos 08-647-729, 08-647-730)
Vacuum desiccator (Fisher Scientific, cat. no. 08-594-15B)
Milli-Q Advantage A10 water system production unit (Millipore, cat. no. Z00Q0V0WW)
Ultrasonic cleaner 5200 (Branson) Sonic water bath
MTP Slide-Adapter II (Bruker Daltonics, cat. no. 235380)
Minicentrifuge (VWR, cat. no. 93000-196)
Instant-recall memory timer (Fisher Scientific, cat. no. 02-401-7)
Slide preparation mounting medium, xylene based (Cytoseal XYL, Richard-Allan Scientific, cat. no. 8312-4)
Microscope cover slides (Fisher Scientific, cat. no. 12-548-C, 25 × 25 mm and cat. no. 12-548-5M, 24 × 50 mm)
Microtome knives (C.L. Sturkey, cat. no. DT315RD)
Rotary microtome HM 325 (MICROM International)
Epson Scan Perfection 4990 PHOTO flatbed scanner (Epson)
Portrait 630 spotter software (Labcyte) or similar robotic reagent deposition
Mass spectrometer (e.g., UltrafleXtreme MALDI-TOF/TOF, Bruker Daltonics)
Mass spectrometric software: FlexControl 3.3, FlexAnalysis 3.3, FlexImaging 2.1, ClinProTools 2.2, BioTools 3.2 software (Bruker Daltonics)
MASCOT database search engine (Matrix Science; http://www.matrixscience.com/)
Kimwipes (Kimberly-Clark)
REAGENT SETUP
TMA sections Cut at 5 µm and mount on (ITO)-coated conductive slides. Paraffin-embedded sections can be stored at 18–24 °C for several months.
Ethanol Dilute 100% ethanol with Milli-Q–purified water to 70% and 95% (vol/vol). Freshly prepare on the day of the experiment.
Tris buffer (10 mM Tris base, pH 9.0) To prepare 1 liter of buffer, completely dissolve 1.21 g Tris Base in 1,000 ml of Milli-Q–purified water. Adjust pH to 9.0 with NaOH or HCl. Can be stored at room temperature for approximately 6 months.
Acetic acid (50 mM) Prepare 500 ml of 50 mM acetic acid by adding 1.4 ml of acetic acid glacial (99.99 + %) to 498.6 ml of Milli-Q–purified water. Can be stored at 4 °C for several months.
Trypsin (0.5 µg µl−1) Reconstitute the trypsin powder (100 µg) with 200 µl of 50 mM acetic acid to obtain a stock solution with a concentration of 0.5 µg µl−1.  For long-term storage the stock solution can be divided into 100-µl aliquots and stored at or below −80 °C. Thaw the reconstituted trypsin at room temperature, placing on ice immediately after thawing. To maintain maximum product activity, we strongly suggest avoiding freeze-thaw cycles.
For long-term storage the stock solution can be divided into 100-µl aliquots and stored at or below −80 °C. Thaw the reconstituted trypsin at room temperature, placing on ice immediately after thawing. To maintain maximum product activity, we strongly suggest avoiding freeze-thaw cycles.
Ammonium bicarbonate (100 mM) To prepare 500 ml of ammonium bicarbonate, dissolve 3.9 g in 500 ml of Milli-Q–purified water. Can be stored at 4 °C for approximately 3 months.
Trypsin solution (0.075 µg µl−1) Add an aliquot (100 µl) of trypsin 0.5 µg µl−1 to 500 µl of 100 mM ammonium bicarbonate. Add 60 µl of acetonitrile in water and vortex to mix. Freshly prepare immediately before use.
MALDI matrix solution Prepare a 2 ml solution of CHCA (10 mg ml−1) in 50% (vol/vol) acetonitrile and 0.5% (vol/vol) TFA in water. Sonicate in a sonic water bath for 10 min. Freshly prepare on the day of the experiment.
Ammonium hydroxide (5%, vol/vol) Dilute 1 volume of 30% (vol/vol) ammonium hydroxide to 5 volumes of dH2O.  Use glass pipettes with bulbs to transfer ammonium hydroxide. Work under the fume hood. Can be stored at room temperature for several months if stored tightly sealed in a vented cabinet away from acids, heat and sunlight.
Use glass pipettes with bulbs to transfer ammonium hydroxide. Work under the fume hood. Can be stored at room temperature for several months if stored tightly sealed in a vented cabinet away from acids, heat and sunlight.
Peptide calibration standard mix Prepare a stock solution of peptide standards containing 4 pmol µl−1 of insulin chain B oxidized from bovine pancreas; 2 pmol µl−1 of insulin chain A oxidized; 1 pmol µl−1 of [Glu1]-fibrinopeptide B human; 1 pmol µl−1 angiotensin II human in acetonitrile 50% (vol/vol); and 0.5% (vol/vol) formic acid in water. Aliquot the stock solution in 5-µl aliquots and store at or below − 80 °C for several months.
Peptide calibration standard mix + matrix Add 5 µl of peptide calibration standard to 5 µl of MALDI matrix solution. Vortex to mix, then centrifuge for a few seconds to collect solution in the bottom of the tube. Freshly prepare before use.
Hematoxylin solution Dissolve 1 g hematoxylin in 100 ml glycerol. Dissolve 45.93 g aluminum potassium sulfate dodecahydrate in 354 ml dH2O. Dissolve 0.1 g sodium iodate in 25 ml water. Add the aluminum potassium sulfate solution to the hematoxylin solution slowly while mixing well. Add the sodium iodate solution, mix well (Carazzi method of preparation of hematoxylin solution stain; see http://stainsfile.info/StainsFile/stain/hematoxylin/aluminum/carazzi.htm).  Light sensitive, store in dark at room temperature. Solution is good for ~6 months. Gravity-filter before use.
Light sensitive, store in dark at room temperature. Solution is good for ~6 months. Gravity-filter before use.
Eosin solution Dissolve 1 g of phloxine B in 100 ml dH2O to make 1% (wt/vol) phloxine B stock solution. Add 0.57 ml of phloxine B stock solution to 50 ml Eosin Y solution. Mix well. The solution is good for ~6 months and can be stored at room temperature.
EQUIPMENT SETUP
Setup to create a sample spotting run The setup consists of creating an experiment protocol in the Portrait software; see Table 1.
TABLE 1.
Setup trypsin and matrix spotting method for TMA sample.
| Area sample (mm) |
Spatial resolution (µm) |
Cycle repeat Mode |
Minimum droplet timing (ms) |
Minimum repeat timing (s) |
Number of cycles |
Number of droplets |
Method mode |
|---|---|---|---|---|---|---|---|
| ~15 | 250 | Loop pattern | 100 | 1 | 10 | 1 | Fly By |
Mass spectrometer The UltrafleXtreme and Autoflex Speed MALDI TOF/TOF mass spectrometers are equipped with a SmartBeam laser50 and controlled by the FlexControl software package. The Ultraflex II MALDI TOF/TOF mass spectrometer can also be used, as it produces high quality of spectra, but in a longer amount of time, as it is equipped with a laser beam tunable at low frequencies (from 1 to 200 Hz); in contrast, the laser beam of the UltrafleXtreme and the Autoflex Speed may run at up to 1,000 Hz. MS and MS/MS analysis can also be performed with other instruments, including MALDI quadrupole-TOF, MALDI quadrupole-ion mobility separation-TOF, MALDI quadrupole ion trap-TOF and MALDI Fourier transform mass spectrometers. Below, we describe the setting of the UltrafleXtreme MALDI TOF/TOF mass spectrometer.
Operation of the MALDI TOF/TOF mass spectrometer A FlexControl method needs to be created for peptide analysis. Instrument settings (i.e., the mass range, detector voltage) are optimized in order to obtain a good quality spectrum either for MS or MS/MS analysis.
For MS analysis we operate the mass spectrometer in positive ion reflectron mode, optimized for higher sensitivity, acquiring spectra in a range of m/z 700–5,000.
For IMS analysis, an AutoXecute method needs to be created using the FlexControl software package provided. This method provides a variety of functions to customize automatic data acquisition, and contains all the information on how the spectra are automatically acquired, including FlexControl method, FlexAnalysis method for pre-processing, number of laser shots and movement of laser. A default AutoXecute method is used as a template to create new user-defined method name. We load the FlexControl method previously created for the MS analysis (see above). We set an acquisition of 1,600 laser shots to be summed up in 200 shots step at each spot position, with the laser performing a ‘random walk’ during the acquisition.
For MS/MS analysis we operate the mass spectrometer in Lift mode, optimized to acquire the precursor ions first and the fragmented masses after.
The parameter settings to operate the mass spectrometer for MS and MS/MS analyses are described in Table 2.
TABLE 2.
Mass spectrometer operating parameters for the MS (reflector) and MS/MS (LIFT) modes.
| Operation Mode |
Pulsed ion extraction delay (ns) |
Ion source voltage 1 (kV) |
Ion source voltage 2 (kV) |
Reflector voltage 1 (kV) |
Reflector voltage 2 (kV) |
Reflector detection voltage 2 (kV) |
Sample rate (ns) |
|---|---|---|---|---|---|---|---|
| MS | 130 | 25 | 22.65 | 26.5 | 13.45 | 2.493 | 0.5 |
| MS/MS | 70 | 19 | 3.2 | 29.5 | 13.85 | 2.696 | 0.5 |
Image acquisition setup A MALDI imaging experiment is setup as a series of measurements at defined x-y positions of the sample tissue using the FlexImaging software. A new sequence in the FlexImaging software needs to be created. The command launches the New Sequence Wizard that is used as guideline during the setup of the new FlexImaging sequence. For the automated acquisition using FlexImaging, the optical image of the spotted sample must be aligned with the sample carrier inside the instrument. Thus, three teach points of the matrix spot microarray, including the upper left, the upper right and the lower right spot positions are defined. The spectra are acquired using the AutoXecute method previously created (see operation of the MALDI TOF/TOF mass spectrometer).
Software The Bruker Daltonics software packages incorporate all the modules that are essential for the control of the instrument: acquisition, processing, display and storage of the data.
Preprocessing of the image MS data is performed using FlexAnalysis 3.3. Smoothing is carried out with 20 cycles of a Savitzky-Golay smoothing algorithm with a width of 0.1 kDa. Baseline subtraction is performed using a median algorithm with flatness and median level value of 0.5. Other programs such as MATLAB may also be used for this step.
We use ClinPro Tools 2.2 software to process MS data for statistical analysis. Processing uses a baseline subtraction with a convex hull algorithm at 0.8% flatness of the baseline and smoothing with 10 cycles of a Savitzky-Golay smoothing algorithm with a width of 0.1 m/z. ClinProTools automatically normalizes spectra with respect to the total ion current.
For MS/MS analysis, each MS/MS spectrum is preprocessed with FlexAnalysis 3.3. Monoisotopic peaks are selected using the SNAP peak picking algorithm with a signal/noise threshold of 5. The MASCOT database search engine is used for peptide mass–mapping identification of proteins. Freely available algorithms such as X!Tandem and OMSSA can also be used for database identification. SwissProt database is used for matching peptide sequences to their respective intact proteins (http://ca.expasy.org/sprot/).
PROCEDURE
FFPE-TMA sectioning  40 min to heat water bath; 15 min for sectioning; 8 h for drying
40 min to heat water bath; 15 min for sectioning; 8 h for drying
-
1|
Fill a clean glass dish with dH2O and bring it to a temperature of 47–50 °C; the water bath temperature should be 5–10 °C below the paraffin melting point. In the meantime keep the FFPE-TMA paraffin block (see EXPERIMENTAL DESIGN for further details of TMA preparation) in iced water to avoid the block (and the specimen) to crack.
-
2|
Test the conductivity of the ITO-coated conductive slides with a multimeter, and mark the conductive side.
-
3|
Insert the blade into the disposable blade carrier, and select section thickness (5 µm) on the microtome.
-
4|
Trim the paraffin block to expose the entire sample surface; cut 5 µm slices. When sections begin to appear, gently pick up the end of the ribbon with a brush and hold it up, away from the knife side.
-
5|
Place the paraffin ribbon in a glass dish filled with room-temperature dH2O. This intermediate step between the cutting and the warmer water bath is used to prevent tissue wrinkling on the warmer water bath.

-
6|
Transfer the paraffin ribbon into a 47–50 °C water bath with a wet brush. Separate the ribbon into individual serial sections using a needle.
-
7|
Mount one floating section onto an ITO-coated glass slide by immersing the ITO slide under the section at an angle of ~45° and slowly removing the slide with the section on top from the water bath. The section should adhere to the conductive surface of the slide with no wrinkling or distortion of the section. This slide will be used for MALDI IMS analysis.
-
8|
Mount a consecutive serial section onto a SuperFrost/Plus microscope slide, following the same procedure described in Step 7. This slide will be used for histological staining.
-
9|
Dry sections by placing them on a warming surface at 37 °C for at least 8 h.
 Paraffin-embedded sections can be stored for several months.
Paraffin-embedded sections can be stored for several months.
FFPE tissue deparaffinization  20 min
20 min
-
10|
Dewax FFPE tissue sections, including both ITO and SuperFrost/Plus microscope slides (from Steps 7 to 9). Box 1 outlines our preferred method for doing this.
Box1 | FFPE-TMA DEPARAFFINIZATION.
Set up Coplin jars filled with xylene, 70% (vol/vol), 95% (vol/vol) and 100% (vol/vol) ethanol, and Milli-Q–purified water (use the glass Coplin jar for xylene, and the plastic Coplin jar for Milli-Q–purified water and ethanol).
Immerse the slide in xylene for 3 min. Repeat once in fresh xylene for 3 min.
Immerse the slide in 100% (vol/vol) ethanol twice, 95% (vol/vol) and 70% (vol/vol) ethanol one time, for 1 min duration each to rehydrate tissue sections.
-
Wash the slide in Milli-Q–purified water twice, for 3 min each.
 At no time from this point onwards should the slides be allowed to dry out. All the above solutions should be changed regularly, i.e., after three or four deparaffinization procedures.
At no time from this point onwards should the slides be allowed to dry out. All the above solutions should be changed regularly, i.e., after three or four deparaffinization procedures.
Antigen retrieval
 1 h
1 h
-
11|
Place an ITO slide from Step 9 directly into Tris buffer (see REAGENT SETUP).
-
12|
Add 500 ml of dH2O to the decloaking chamber pan and place the plastic Coplin staining jar, containing the ITO slide completely submerged in 10 mM Tris buffer (pH 9.0), into the decloaker slightly on the edge of the grate, so that the jar is not directly on the source of heat.
-
13|
Set the program as described in Table 3.
-
14|
Place the lid on top of staining jar, but do not tighten it. Make sure the chamber gasket is in place. The gasket ensures an airtight chamber. Lock the decloaker lid and make sure the vent switch located on the lid handler is in the closed position. The dot on the handle must match with the ‘closed’ wording on the lid.
-
15|
Press the ‘start’ button to begin the programmed run. The timer will start to count down when the correct pressure and temperature are reached.
 The time required to reach the programmed temperature (95 °C) and pressure are variable, depending on the number of slides in the chamber and the temperature at which the instrument is operated. At this point, record the temperature and pressure. It should take ~5 min for the chamber to reach 95 °C and then 20 min to maintain that temperature. The pressure should remain below 5 psi (typically, 2 psi is the pressure reported on the pressure gauge).
The time required to reach the programmed temperature (95 °C) and pressure are variable, depending on the number of slides in the chamber and the temperature at which the instrument is operated. At this point, record the temperature and pressure. It should take ~5 min for the chamber to reach 95 °C and then 20 min to maintain that temperature. The pressure should remain below 5 psi (typically, 2 psi is the pressure reported on the pressure gauge).
-
16|
Once the first part of the program has completed, the decloaker will repeatedly beep. Press the ‘start’ button to continue to the next part of the program.
-
17|
The decloaker chamber will now drop in temperature and pressure to 90 °C and 0 psi, respectively. Once it reaches these two points (~10 min), the chamber will repeatedly beep again. Press the ‘start’ button again. The decloaking chamber can now be turned off.
-
18|
Toggle the valve to make sure that pressure has been released so the lid may be safely removed. Open the lid in such a way that it is pointing away from yourself to avoid steam contact burns. By using a hot-hand protector, carefully remove the staining jar and place on the counter. Allow the jar to cool on the bench top for 10 min in the buffer.
 High temperature and pressure are involved in this process. Make sure the lid is completely closed and that pressure is evacuated before trying to open the lid. Wear gloves and safety goggles when opening. To prevent burns, use a hot-hand protector to open the chamber and remove items.
High temperature and pressure are involved in this process. Make sure the lid is completely closed and that pressure is evacuated before trying to open the lid. Wear gloves and safety goggles when opening. To prevent burns, use a hot-hand protector to open the chamber and remove items. -
19|
Exchange the Tris buffer with Milli-Q H2O five times, pouring out half of the liquid each time, with the final rinse being entirely water. Stand the ITO slide vertically on a paper filter until it is completely dry.
-
20|
Acquire an optical image of the ITO slide. This picture can be useful to define a measurement region in the image acquisition setup, as the matrix sometimes masks the tissue border (see EQUIPMENT SETUP).
-
21|
Place the ITO slide in a bench vacuum desiccator (vacuum of approximately − 12 psi) until needed.
 For short-term antigen retrieval, sections can be stored in vacuum desiccators for up to 2 d. For the long term, dried sections can be stored at − 80 °C for at least 3 months.
For short-term antigen retrieval, sections can be stored in vacuum desiccators for up to 2 d. For the long term, dried sections can be stored at − 80 °C for at least 3 months.
TABLE 3.
Decloaking chamber setting program for antigen retrieval.
| Setting | Temperature (°C) | Time | Operation |
|---|---|---|---|
| SP (set point) 1 | 95 | 20 min | Chamber will heat the sample at the desired temperature (95 °C) |
| SP (set point) 2 | 90 | ~10 s | Temperature cools down to 90 °C Chamber holds steadily at 90 °C |
H&E staining  30 min
30 min
-
22|
Stain the microscope slide from Step 9 using the standard H&E staining method (Box 2).
-
23|
Cover slipping: dry the back of the microscope slide and wrap the tissue section with a Kimwipe to get rid of excess xylene. Put 1–3 drops of Cytoseal on the tissue section and then cover it with a cover slide. Make sure no air bubbles are left under the slide.
-
24|
Let the slide dry until the Cytoseal has hardened for ~15 min.
Box 2 | H&E STAINING.
Set up Coplin jars filled with 70%, 95% (vol/vol) and 100% (vol/vol) ethanol as well as Milli-Q–purified water, hematoxylin solution, eosin solution and xylene (use the glass Coplin jar for xylene, H&E solution, and the plastic Coplin jar for Milli-Q–purified water and ethanol).
Immerse the slide sequentially through the ethanol series of 95% (vol/vol), 70% (vol/vol) and Milli-Q–purified water for 30 s duration each.
Immerse the slide in hematoxylin solution for 2 min.
Wash the slide in Milli-Q–purified water for 20 s.
Dehydrate tissue sections gradually by passing the slides through 70% (vol/vol) and 95% (vol/vol) ethanol for 30 s duration each.
Immerse the slide in eosin solution for 1 min.
Immerse the slide in 95% (vol/vol) and 100% (vol/vol) ethanol for 30 s each.
-
Immerse the slide in xylene for 2–2.5 min.
 The same solvents can be used for multiple tissue sections, but they should be changed daily or more often if they become too contaminated (i.e., strongly colored by the stains).
The same solvents can be used for multiple tissue sections, but they should be changed daily or more often if they become too contaminated (i.e., strongly colored by the stains).
In situ digestion and matrix application  3 h
3 h
-
25|
Prepare the trypsin solution as described in the REAGENT SETUP.
-
26|
Fill the source reservoir with trypsin solution by slowly pipetting 500–600 µl into one of the margin spaces between the baffle and the reservoir wall, taking care not to introduce bubbles (see filling reagent solution details in Fig. 4a). Cover the source reservoir with the lid.
 Do not allow any liquid to sit on top or sides of the baffle. This will produce wicking and contact of the liquid with the lid resulting in a fluid height error that prevents subsequent spotting.
Do not allow any liquid to sit on top or sides of the baffle. This will produce wicking and contact of the liquid with the lid resulting in a fluid height error that prevents subsequent spotting. -
27|
Launch the Portrait software and load the source reagent reservoir into the Portrait 630 instrument. By selecting the specific source type reagent defined by the manufacturer, ejection parameters are optimized for trypsin deposition.
-
28|
Insert the ITO slide from Step 21 and the calibration slide into the appropriate spaces of the target plate holder (MTP Slide-Adapter II) as shown in Figure 4b. These specific positions in the target plate holder are defined. Load the target plate holder into the instrument.
-
29|
Calibrate ejection parameters by clicking the Calibrate icon from the toolbar of the Portrait software window, or clicking ‘YES’ to the calibration prompt after loading the target plate holder. The ‘Calibration-Ejection’ dialog box opens and indicates the calibration parameter (transducer focus position and transducer power) values. Click the ‘Next’ button. The system will spot a calibration pattern using the specified default setting. A good calibration shows a triangular array of spots, as illustrated in Figure 4c. This process involves optimizing the droplet ejection settings.
 If the size and shape of the calibration pattern is distorted, the process will need to be repeated with automatic or manual adjustment of the power and focus of the transducer. The triangular array image (see Fig. 4c) shows two appropriate markers for automatically adjusting the parameters. Move the upper marker to the upper leftmost spot and the lower marker to the apex spot of the triangle and click ‘ReCal Eject’; the software will use these marker positions to estimate better values for the power and focus settings, respectively. The manual adjustment of the ejection parameters can be done by clicking the ‘Back’ button to return to the ‘Calibration-Ejection’ dialog box. Select the ‘Override Defaults’ box and increase or decrease by 10% the focus and power values. Click ‘Next’ to rerun the calibration with the new parameters. Repeat until a good calibration image is generated. The calibration process is a very important step in the spotting procedure, as it allows a very precise drop-to-drop spotting. Calibration must be done prior to each run, and again if the reagent has been sitting in the source for more than 30 min.
If the size and shape of the calibration pattern is distorted, the process will need to be repeated with automatic or manual adjustment of the power and focus of the transducer. The triangular array image (see Fig. 4c) shows two appropriate markers for automatically adjusting the parameters. Move the upper marker to the upper leftmost spot and the lower marker to the apex spot of the triangle and click ‘ReCal Eject’; the software will use these marker positions to estimate better values for the power and focus settings, respectively. The manual adjustment of the ejection parameters can be done by clicking the ‘Back’ button to return to the ‘Calibration-Ejection’ dialog box. Select the ‘Override Defaults’ box and increase or decrease by 10% the focus and power values. Click ‘Next’ to rerun the calibration with the new parameters. Repeat until a good calibration image is generated. The calibration process is a very important step in the spotting procedure, as it allows a very precise drop-to-drop spotting. Calibration must be done prior to each run, and again if the reagent has been sitting in the source for more than 30 min. -
30|
Select the option ‘Perform X/Y Calibration’ to adjust for slight X/Y offset. A calibration grid with a vertical and horizontal calibration bar will be scanned. Use the mouse to move the calibration bars through the center of a column and row. This additional calibration will increase the accuracy of array spotting.
-
31|
Click ‘Finish’ to apply the calibration setting.

-
32|
Scan the image of the sample to identify the region to be spotted and draw a spotting array block on the sample image. Each spot in the array is specified by X and Y coordinates from the number of the columns and the rows, and it represents the precise location where the reagent will be deposited.
-
33|
Create an experiment method by setting spotting parameters. Our optimized parameter setting to spot 10 cycles of 1 drop each of either trypsin or matrix solution on TMA sample tissue (size ~15 × 15 mm) is described in Table 1. For optimum spotting of small areas (less than 10 × 10 mm), we recommend including a ‘minimum repeat timing’ of ~50 s in the setup spotting method that specifies the delay before revisiting the first spot between repeat cycles. This will allow enough time for spots to dry and avoid formation of large droplets on the same tissue that may merge and thus affect lateral image resolution. The ‘Minimum repeat timing’ option can be modified from the ‘Properties’ window listed in the ‘View’ menu of the Portrait software.
-
34|
Select the appropriate reagent method for trypsin solution. Run 1 cycle of trypsin, and scan the image of the sample to check that trypsin was deposited onto preformed spotting array block (see Step 32, and Fig. 2b). Run 3 series of 10 cycles of trypsin, each spaced by the calibration process (see Steps 29–31 for calibration).

-
35|
After trypsin spotting, remove and clean the source reservoir and fill it with the matrix solution (see REAGENT SETUP) following the same directions described for trypsin solution filling (Step 26).
-
36|
Load the source reagent reservoir into the Portrait 630 instrument. By selecting the specific source type defined by the manufacturer, ejection parameters are optimized for matrix deposition.
-
37|
Calibrate the instrument according to the directions in Steps 29–31.
-
38|
Select the appropriate reagent method for matrix solution. Run 3 series of 10 cycles of matrix solution spotting using the same spotting setup created for the tryptic solution. Matrix will be then spotted onto the same spots where the enzymatic solution was previously placed.
-
39|
At the end of the matrix spotting, remove the MALDI plate and check, under a light microscope, the resulting matrix surface for efficient crystallization. Typically, 30 repeated cycles yield a good crystal formation, thereby allowing the acquisition of good-quality spectra (see Figs. 2b and 4d).
-
40|
Prepare peptide standard calibration mix + matrix as described in REAGENT SETUP. Immediately spot 1 µl of this solution on the MALDI target plate. Let the drop dry at room temperature.
 The matrix is light sensitive; we recommend protecting the slide from direct light.
The matrix is light sensitive; we recommend protecting the slide from direct light. Spotted samples can be stored in vacuum desiccators overnight.
Spotted samples can be stored in vacuum desiccators overnight.
Figure 4.
Portrait spotting setup. (a) Description of loading the source reagent reservoir of Portrait 630 instrument. Reagent solution, trypsin or matrix, is slowly pipetted into the source reservoir between the baffle and the reservoir wall. The source is filled with a volume ranging from 500 to 600 µl. (b) Target plate holder view for calibration and sample slide insertion. With the round corner at the holder on the right, place the calibration slide below the ITO slide. (c) Enlarged view of calibration patterns for ejector parameter and X/Y offset calibration, respectively. (d) Enlarged view of the TMA section after trypsin and matrix spotting.
IMS analysis  ~1 h
~1 h
-
41|
Before starting an imaging experiment an optical image of the spotted sample must be acquired. Place the target plate holder with the tissue section facing down onto the scanner surface and acquire the image. Use a resolution of at least 1,200 dpi. This optical image is used to align the sample in the sample carrier inside the instrument for imaging data acquisition.
-
42|
Load the sample into the mass spectrometer and operate the MALDI TOF/TOF mass spectrometer for MS analysis as described in Table 2. To operate the instrument, create a FlexControl method as described in the EQUIPMENT SETUP section.
-
43|
Calibrate the instrument using the peptide standard mix. The target holder is recognized by the instrument with the corresponding ‘Geometry’ file and appears in the spot array window displayed in the ‘Carrier’ dialog box of the FlexControl software. Go to the position on the plate where the peptide standard calibration was deposited. The MALDI plate/stage is moved by selecting one of the spots in the array window. For fine adjustment, the plate can be moved by clicking the mouse in the receptacle window until the peptide standard spot is found. Adjust the sliding intensity scale bar, initially at a minimum energy setting, and click the ‘Start’ button in the receptacle window to fire the laser on the peptide standard spot until an acceptable data signal is acquired. You can increase the laser intensity in order to maximize resolution and signal intensity while also minimizing noise. A center panel window should now display a mass spectrum with peaks at m/z of approximately m/z 1,046.542 (human angiotensin II), m/z 1,570.6774 (human [Glu1]-fibrinopeptide B), m/z 2,530.921 (bovine insulin chain A oxidized) and m/z 3,494.651 (bovine insulin chain B oxidized).
-
44|
Click the ‘Add’ button in the receptacle window.
-
45|
Repeat Steps 43 and 44 until you have added a total of 500–1,000 shots. Change the spectrum viewer from ‘single spectrum’ to ‘summed spectrum’ by selecting the corresponding icons on the toolbar of the FlexControl software.
-
46|
Click the ‘Calibration’ tab at the bottom-center section of the spectrum panel window.
-
47|
Select ‘Peptide calibration standard’ from the ‘Mass control List’.
-
48|
Select ‘Human angiotensin II’ in the compound mass list. The view of the summed mass spectrum should zoom into this peak. Click below and to the left of the apex of the human angiotensin II peak. An arrow will appear above the apex signifying the peak has been calibrated. Calibration will also be confirmed by a check mark beside human angiotensin II peak in the compound mass list window.
-
49|
Repeat Step 46 for the remaining peptide standards. Click the ‘Apply’ button.
-
50|
On the ‘File’ menu, select ‘Save Method as’ and save the calibrated method in your own user folder for upcoming experiment.
-
51|
Revert back to the single spectrum view in the toolbar and click ‘Clear Sum’ button in the laser receptacle window.
-
52|
Locate some spots on the tissue and optimize the laser intensity on those spots to obtain the best resolution and signal-to-noise ratios for the peptide peaks observed. High laser power can initiate scissions in the molecular structure; even at lower incident laser power it is very difficult to define what constitutes a representative spectrum. We suggest initially setting the laser intensity at a minimum energy setting and then increasing it until an acceptable data signal is acquired.
-
53|
Save the FlexControl method.
-
54|
Create an AutoXecute method as described in EQUIPMENT SETUP.
-
55|
Setup an imaging run as described in EQUIPMENT SETUP for image acquisition setup.
-
56|
Start the automatic acquisition.
-
57|
Preprocess spectra with proper software. This step reduces experimental variance within the data set. Depending on the quality of the spectra, the use of different parameter settings, such as removal of noise and baseline subtraction, may be carried out. The result is an array of spots or ‘pixels’ covering the tissue section, with a mass spectrum linked to each individual pixel. Individual peptide images are then visualized with FlexImaging software. For details, see the EQUIPMENT SETUP section.
 For further MS/MS analysis, sections can be stored in vacuum desiccators for up to several weeks.
For further MS/MS analysis, sections can be stored in vacuum desiccators for up to several weeks.
Statistical analysis  ~1 h
~1 h
-
58|
Co-register the IMS image with the H&E-stained section following directions described in the ‘co-register image’ dialog box of the FlexImaging software.
-
59|
Mark regions of interest by drawing a defined area in the displayed image in the FlexImaging software.
-
60|
Copy all spectra belonging to the defined region of interest into a newly created directory and combine them into separate folders for each group of study.
-
61|
Load the folders in this newly created directory into ClinProTools software and process them using the routine setting ‘data preparation’ in the program (see EQUIPMENT SETUP for details).
-
62|
Manually select the monoisotopic peak for each tryptic peptide and run ‘peak calculation’ to generate a table of average intensity and standard deviations of all peaks picked from the spectra in each folder.
-
63|
Evaluate and analyze peaks that are significantly different between each group using a minimum twofold intensity difference threshold and the Wilcoxon/Kruskal-Wallis test in the software, or using any other suitable process listed in the software.
MS/MS analysis and protein identification  10–15 min (protein identification via database searching may take several days)
10–15 min (protein identification via database searching may take several days)
-
64|
After MS acquisition (Step 56) and data pre-processing and analysis (Steps 57–63), a specific ion of interest is submitted to LIFT TOF/TOF acquisition. The specific instrument operation details are described in Table 2. Acquisition conditions must be modified to generate high fragment ion yields. This is done by increasing laser fluence to optimize a larger number of precursor ions per shot.
-
65|
Process data with FlexAnalysis software (our software setting is described in EQUIPMENT SETUP).
-
66|
Load the peptide sequence into BioTools to convert the spectrum to a MASCOT generic format file (.mgf).
-
67|
Submit the .mgf file into Mascot search engine and specify your search criteria. Our criteria search is typically performed with a precursor ion tolerance of 200 p.p.m. and a fragment ion tolerance of ± 0.4 kDa. We also include up to three missed cleavages and variable modifications of methionine oxidation, N terminus acetylation and histidine/tryptophan oxidation.
-
68|
Perform homology searches by using the SwissProt database.

Troubleshooting advice can be found in Table 4.
TABLE 4.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | The first few sections are often unusable | The surface of the sample is uneven | Remove the first few sections with a small brush. When the surface of the sample evens, an intact tissue section or ribbon will form. Cut several sections to ensure that the region of interest has been sampled |
| 15 | If the pressure is higher than 5 psi, an error signal will be activated (The LED panel will display ‘ERR’) | The decloaking chamber pan is not filled with enough water, temperature and pressure rise behind normal limits | Ensure that there is enough water added to the system |
| 31 | Error messages due to the fluid height change in the source reservoir may occur and the previous focus position will no longer be correct | Depending on the surface tension of the liquid and the surface interactions with the plastic, the meniscus of some fluids (more likely an enzyme solution, water, but not matrix solutions) may shift in the reservoir so that it is suddenly at a different height | This may be prevented by keeping the reservoir and baffles clean and replacing them when the surfaces have become scratched or pitted. We recommend cleaning the baffle with 5% (vol/vol) ammonium hydroxide (see REAGENT SETUP) |
| 34 | Trypsin spotting cannot be visualized after scanning the image of the sample | Dried trypsin droplets are not visible on the tissue | Scan the sample image immediately after one run. By zooming into the spot array, translucent droplets of moisture should be visible where the tryptic solution was spotted (see Fig. 2b) |
| Box 1 | Some paraffin residue can still be observed on the slides | The xylene solution is diluted | Pour out diluted xylene into an appropriate chemical waste disposal container and refill with fresh xylene |
![]()
Step 1, Heating water bath to appropriate temperature: 40 min
Steps 2–8, FFPE-TMA sectioning: 15 min
Step 9, Section drying: 8 h
Step 10, FFPE tissue deparaffinization: 20 min
Steps 11–21, Antigen retrieval: 1 h
Steps 22–24, H&E staining: 30 min
Steps 25–40, In situ digestion and matrix application: 3 h
Steps 41–57, IMS analysis: ~1 h
Steps 58–63, Statistical analysis: ~1 h
Steps 64 and 65, MS/MS analysis: 15 min
ANTICIPATED RESULTS
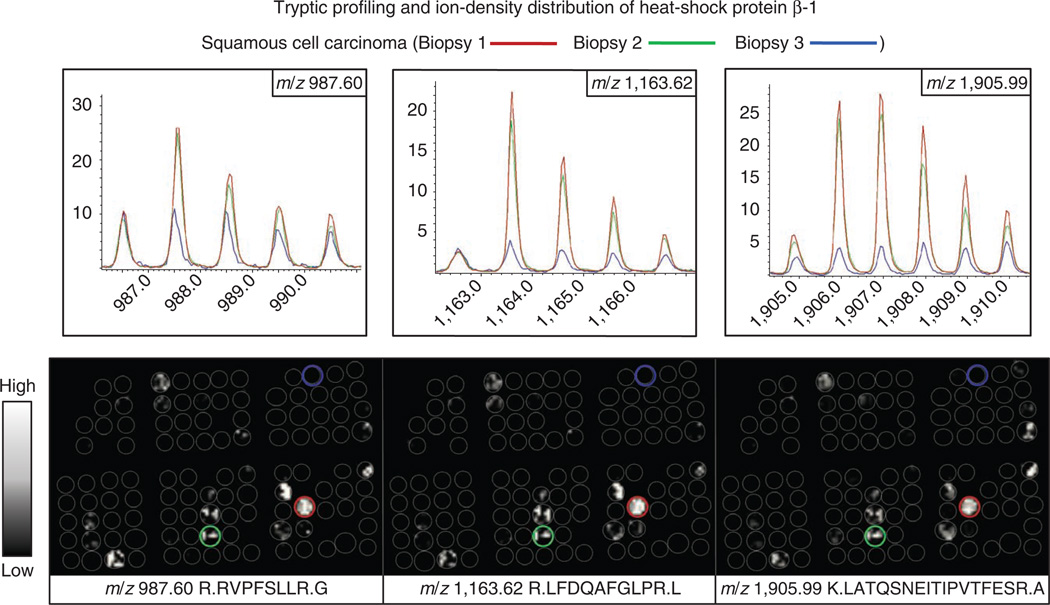
Our protocol demonstrates that under the proper conditions for removal of the embedding medium and digestion, tryptic peptides are released from discrete histological areas of fixed tissue sections and are amenable to analysis by IMS. Figure 5 demonstrates the possibility of classification of tumor samples in a TMA at the peptide level. Spectra from four biopsies were compared through statistical analysis for classification. Two were classified as squamous cell carcinoma and the other two biopsies as adenocarcinoma, in agreement with the pathology diagnosis. These findings suggest that MALDI IMS is complementary to histochemical stains in identifying molecular signatures and may help to distinguish different lung cancer histologies and subclassify individual cancer types. The advantage of IMS to directly visualize the molecular content of a sample to maintain spatial integrity becomes critical for the analysis of highly heterogeneous tissue samples. Figure 6 shows an example of incongruence ion-density distributions of three different tryptic peptides, corresponding to heat shock protein β-1 (m/z 987.60, 1,163.62 and 1,905.99)5. Heat shock protein β-1 was found to be localized almost exclusively to the cancerous regions of a subset of squamous cell carcinoma biopsies of a TMA. When three different squamous cell carcinoma biopsies were analyzed, only two showed a similar peak intensity and spatial distribution of the tryptic peptides for this protein. This demonstrates the protein heterogeneity that can be measured in biopsies of the same nominal disease classification.
Figure 5.
IMS correlation with pathological diagnosis. (a–d) Representation of the statistical classification of four TMA cores correlating with the pathological diagnosis. Pathologist diagnosis is shown in panels a–d; four TMA cores have undergone H&E staining, and pathological analysis using light microscopy. Adenocarcinoma (a,c) and squamous cell carcinoma (b,d) were marked in each biopsy for the cancerous region (red line) and the noncancerous region (green line). (e–h) The marked cancer regions were then co-registered with serial TMA sections analyzed by MALDI IMS and ClinProTools for classification, enabling the spot position of each spectrum to be linked to that same precise histological region in the TMA. Adenocarcinoma is shown in red and squamous cell carcinoma in green. For the statistical classification, mass spectra from the cancer regions of each biopsy were first grouped to create an average spectrum representative of both the adenocarcinoma and squamous cell carcinoma. The peaks present in these average spectra were compared through statistical analysis to identify a subset of peaks that were significantly different between each group to be used as the class identifiers. A support vector machine (SVM) algorithm was used to build a classification model. The SVM model was run against all spectra in the data set and the outcome of each classification was visualized using the class imaging function in FlexImaging software. This model classified two biopsies as squamous cell carcinoma (f,h, peptide images in green) and the other two biopsies as adenocarcinoma (e,g, peptide images in red), in agreement with the pathology diagnosis. This figure is reproduced, with permission, from reference 5.
Figure 6.
MALDI images of three tryptic peptides originating from heat shock protein β-1 detected in two of three squamous cell carcinoma core biopsies analyzed in a TMA. This demonstrates the protein heterogeneity that can be measured in biopsies of the same nominal disease classification. This figure is reproduced, with permission, from reference 5.
Acknowledgments
We thank P.M. Angel, M.L. Reyzer and E.H. Seeley for helpful discussions and critical reviewing of the manuscript. We also thank J.L. Allen, E.H. Seeley and K. Schwamborn for development and optimization of the deparaffinization and antigen retrieval methods, and M. Reid Groseclose for providing some TMA images and the figures in the Anticipated Results section. This work was supported by the US National Institutes of Health/National Institute of General Medical Sciences grants 5RO1GM058008-11 and DOD W81XWH-05-1-0179.
Footnotes
AUTHOR CONTRIBUTIONS Both authors contributed equally to the development of the protocol and the writing of the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Metz B, et al. Identification of formaldehyde-induced modifications in proteins: Reactions with insulin. Bioconjugate Chem. 2006;17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- 2.Rahimi F, Shepherd CE, Halliday GM, Geczy CL, Raftery MJ. Antigen-epitope retrieval to facilitate proteomic analysis of formalin-fixed archival brain tissue. Anal. Chem. 2006;78:7216–7221. doi: 10.1021/ac060294s. [DOI] [PubMed] [Google Scholar]
- 3.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohistochemistry. Am. J. Surg. Pathol. 2000;24:1016–1019. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 4.Shi SR, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 5.Groseclose MR, Massion PP, Chaurand P, Caprioli RM. Highthroughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics. 2008;8:3715–3724. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 8.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J. Proteome Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 9.Lemaire R, et al. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 10.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol. Cell Proteomics. 2005;4:384–393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Wessels JT, et al. In vivo imaging in experimental preclinical tumor research-a review. Cytometry A. 2007;71:542–549. doi: 10.1002/cyto.a.20419. [DOI] [PubMed] [Google Scholar]
- 12.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat. Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 13.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat. Rev. Mol. Cell. Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 14.Becker KF, Taylor CR. ‘Liquid morphology’: immunochemical analysis of proteins extracted from formalin-fixed paraffin-embedded tissues: combining proteomics with immunohistochemistry. Appl. Immunohistochem. Mol. Morphol. 2011;19:1–9. doi: 10.1097/PAI.0b013e3181f50883. [DOI] [PubMed] [Google Scholar]
- 15.Teruya-Feldstein J. The immunohistochemistry laboratory: looking at molecules and preparing for tomorrow. Arch. Pathol. Lab. Med. 2010;134:1659–1665. doi: 10.5858/2009-0582-RAR1.1. [DOI] [PubMed] [Google Scholar]
- 16.Leong TY, Cooper K, Leong AS. Immunohistology-past, present, and future. Adv. Anat. Pathol. 2010;17:404–418. doi: 10.1097/PAP.0b013e3181f8957c. [DOI] [PubMed] [Google Scholar]
- 17.Grey AC, Chaurand P, Caprioli RM, Schey KL. MALDI imaging mass spectrometry of integral membrane proteins from ocular lens and retinal tissue. J. Proteome Res. 2009;8:3278–3283. doi: 10.1021/pr800956y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang YL, et al. Connecting chemotypes and phenotypes of cultured marine microbial assemblages by imaging mass spectrometry. Angew. Chem. Int. Ed. Engl. 2011;50:5839–5842. doi: 10.1002/anie.201101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iijima M. Visualization of lateral water transport pathways in soybean by a time of flight-secondary ion mass spectrometry cryo-system. J. Exp. Bot. 2011;62:2179–2188. doi: 10.1093/jxb/erq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyzer ML, Caprioli RM. MALDI-MS-based imaging of small molecules and proteins in tissues. Curr. Opin. Chem. Biol. 2007;11:29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 21.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 2008;80:5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djidja MC, et al. Novel molecular tumour classification using MALDI-mass spectrometry imaging of tissue micro-array. Anal. Bioanal. Chem. 2010;397:587–601. doi: 10.1007/s00216-010-3554-6. [DOI] [PubMed] [Google Scholar]
- 23.Caprioli RM. Deciphering protein molecular signatures in cancer tissues to aid in diagnosis, prognosis, and therapy. Cancer Res. 2005;65:10642–10645. doi: 10.1158/0008-5472.CAN-04-3581. [DOI] [PubMed] [Google Scholar]
- 24.Shi S-R, Shi Y, Taylor CR. Antigen retrieval immunohistochemistry. J. Histochem. Cytochem. 2011;59:13–32. doi: 10.1369/jhc.2010.957191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proc. Natl. Acad. Sci. USA. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westermark P, Nilsson GT. Demonstration of amyloid protein Aa in old museum specimens. Arch. Pathol. Lab. Med. 1984;108:217–219. [PubMed] [Google Scholar]
- 27.Schwamborn K, Caprioli RM. MALDI imaging mass spectrometry— painting molecular pictures. Mol. Oncol. 2010;4:529–538. doi: 10.1016/j.molonc.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kononen J, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 29.Chaurand P, et al. Integrating histology and imaging mass spectrometry. Anal. Chem. 2004;76:1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 30.Schwamborn K, et al. Identifying prostate carcinoma by MALDI-Imaging. Int. J. Mol. Med. 2007;20:155–159. [PubMed] [Google Scholar]
- 31.Evers P, Uylings HBM, Suurmeijer AJH. Antigen retrieval in formaldehyde-fixed human brain tissue. Methods. 1998;15:133–140. doi: 10.1006/meth.1998.0616. [DOI] [PubMed] [Google Scholar]
- 32.Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J. Histochem. Cytochem. 2006;54:739–743. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- 33.Sato TA, Aoki Y, Toyama A, Shimada T. A novel method for analyzing formalin-fixed paraffin-embedded (FFPE) tissue sections by mass spectrometry imaging. Neurosci. Res. 2008;61:S19. doi: 10.2183/pjab/83.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi SR, Cote RJ, Taylor CR. Antigen retrieval techniques: current perspectives. J. Histochem. Cytochem. 2001;49:931–937. doi: 10.1177/002215540104900801. [DOI] [PubMed] [Google Scholar]
- 35.Taylor CR, et al. Comparative study of antigen retrieval heating methods: microwave, microwave and pressure cooker, autoclave, and steamer. Biotech. Histochem. 1996;71:263–270. doi: 10.3109/10520299609117171. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita S. Heat-induced antigen retrieval: Mechanisms and application to histochemistry. Prog. Histochem. Cytochem. 2007;41:141–200. doi: 10.1016/j.proghi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Hoetelmans RWM, van Slooten HJ, Keijzer R, van de Velde CJH, van Dierendonck JH. Comparison of the effects of microwave heating and high pressure cooking for antigen retrieval of human and rat Bcl-2 protein in formaldehyde-fixed, paraffin-embedded sections. Biotech. Histochem. 2002;77:137–144. [PubMed] [Google Scholar]
- 38.Aerni HR, Cornett DS, Caprioli RM. High-throughput profiling of formalin-fixed paraffin embedded tissue using parallel electrophoresis and matrix assisted laser desorption ionization mass spectrometry. Anal. Chem. 2009;81:7490–7495. doi: 10.1021/ac900974j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Mining the archival formalin-fixed paraffin-embedded tissue proteome: opportunities and challenges. Mol. Biosyst. 2008;4:712–720. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- 40.Reyzer ML, Chaurand P, Angel PM, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by MALDI imaging mass spectrometry. Methods Mol. Biol. 2010;656:285–301. doi: 10.1007/978-1-60761-746-4_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuerenberg M, Luebbert C, Deininger SO, Ketterlinus R, Suckau D. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat. Methods. 2007;4:iii–iv. [Google Scholar]
- 42.Nilsson A, et al. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS ONE. 2010;5:e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal. Chem. 2007;79:6862–6867. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- 44.Wisztorski M, Franck J, Salzet M, Fournier I. MALDI direct analysis and imaging of frozen versus FFPE tissues: what strategy for which sample? Methods Mol. Biol. 2010;656:303–322. doi: 10.1007/978-1-60761-746-4_18. [DOI] [PubMed] [Google Scholar]
- 45.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 2006;78:827–834. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 46.Hardesty WM. Ph.D. Dissertation. Vanderbilt University; 2010. Proteomic Analysis and Classification of Metastatic Melanoma by MALDI Imaging Mass Spectrometry. http://etd.library.vanderbilt.edu/available/etd-08252010-081820/unrestricted/Hardesty.pdf. [Google Scholar]
- 47.Norris JL, et al. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int. J. Mass Spectrom. 2007;260:212–221. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [DOI] [PubMed] [Google Scholar]
- 49.Tabb DL, Friedman DB, Ham AJL. Verification of automated peptide identifications from proteomic tandem mass spectra. Nat. Protoc. 2006;1:2213–2222. doi: 10.1038/nprot.2006.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holle A, Haase A, Kayser M, Hohndorf J. Optimizing UV laser focus profiles for improved MALDI performance. J. Mass Spectrom. 2006;41:705–716. doi: 10.1002/jms.1041. [DOI] [PubMed] [Google Scholar]