Abstract
Background:
CyclinD1 is an essential sensor and activator of cell cycle initiation and progression; overexpression of cyclinD1 is linked to various human cancers, including breast cancer. The elevated cyclinD1 in some types of cancers is believed to be associated with tumor progression and response to systemic treatments.
Aims:
In this study, we anticipate to address the questions in human breast cancer; the function of cyclinD1 in mediating chemoresponses; and the signaling pathway cooperating with cyclinD1 to interfere with the drug functions.
Materials and Methods:
Using the cell clones, concurrent ectopic expression of the wild-type or K112E-mutated human cyclinD1 protein in the MCF7 and MDA-MB231 (MB231) breast cancer cells to study the function of cyclinD1 in responses to the chemotherapeutic treatments. Three drugs, cisplatin (CDDP), 5-fluorouracil (5-FU), and Gemzar were used in this study; the 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay, cell cycle and cell death analysis, clonogenic survival assay, acridine orange (AO)/ethidium bromide (EB) staining, and Western blot assay were conducted to evaluate the drugs’ effects in the cell clones.
Results:
The cell clones expressing the D1 protein in MCF7 and MB231 cells result in distinct effects on the responses to chemotherapeutic treatments. Particularly with Gemzar, ectopic expression of cyclinD1 protein in MCF7 cells results in a potentiated effect, which is CDK4 kinase activity dependent, whereas in MB231 cells, an opposite effect was observed. Moreover, our results suggested that the distinct chemosensitivities among those cell clones were not resulted from accelerated cell cycle, cell proliferation driven by the cyclinD1CDK4/6-Rb-E2F signaling chain, rather, they were results of the cell cycle-independent functions led by cyclinD1 alone or in complex with CDK4.
Conclusions:
Our results suggest that the functions of cyclinD1 protein in modulating chemoresponses in the MCF7 and MB231 cells are independent to its function as cell cycle initiator through activation of CDK4/6. Furthermore, the signals modulated by cyclinD1 upon treatment are determined by the drug and the cellular network.
Keywords: Breast cancer, CDK4/6 kinase, chemosensitivity, cyclinD1, NF-kB, TGF-β
INTRODUCTION
CyclinD1 is an initiator and activator of cell cycle and its level is under strict control in normal cells, whereas it is overexpressed in a wide range of human tumors, including breast cancer as being reported in about 15% of human primary breast cancers and in up to 50% of invasive breast cancers.[1] In cooperation with CDK4/6 kinase, cyclinD1 regulates cell cycle progression, angiogenesis, lipogenesis, and mitochondrial function.[2,3] Additionally, cyclinD1 also modulates the activity of various cellular transcription factors directly or indirectly, including estrogen receptor (ER),[4] androgen receptor (AR),[5] DMP1,[6] BETA2/NeuroD,[7] STAT3,[8] and C/EBPβ[9] without the participation of the CDKs. Overall, overexpression of cyclinD1 has been linked to tumorigenesis, tumor progression, metastasis, invasion, and chemosensitivity. In contrast to promotion of cell cycle, overexpression of cyclinD1 could suppress cell growth and induce apoptosis[10–12] in some circumstances.
The function of cyclinD1 displaying in cancer treatment seems ambiguous and complicated due to contradictory results from different studies in different cancer types. Increased protein level or ectopic overexpression of cyclinD1 has been shown to increase the sensitivity to chemotherapeutic treatments or radiation treatment in multiple myeloma,[13] in mammary epithelial cells and in breast tumor cell lines.[10,14,15] But, in human oral squamous cell carcinoma,[16] inhibition of cyclinD1 expression increases sensitivity of response to CDDP. Furthermore, using different approaches, including DNA replication inhibitor,[17] topoisomerase II inhibitors,[18,19] and protein kinase inhibitor,[20] has demonstrated a link between overexpression of cyclinD1 and a reduced response to anticancer treatments. A previous study in our laboratory has reported that ectopic expression of cyclinD1 in an Ela-myc transgenic mouse tumor–derived cell line imposed resistance to CDDP.[21] The discrepancies stated above, and more, may be due to different models and different drugs selected for the study or may be due to the different cell types and organs of origin chosen with, under one circumstance, cyclinD1 functioning as a prosurvival factor but as a proapoptotic factor under another circumstance. The molecular mechanisms underlying the selective effects of cyclinD1 on cytotoxic drug-induced signaling remain ambiguous.
The objectives of this study were to understand the function of the cyclinD1 protein in modulating the chemoresponses in human breast cancer cells, whether its functions in the regulation of chemoresponse require the CDK4 kinase activity and whether the function of cyclinD1 in the chemotherapeutic signaling is linked to its functions in the cell cycle promotion.
MATERIALS AND METHODS
Cell lines, retroviral vectors, and stable cell clones
MCF15 cell line was obtained from Karmanos Cancer Institute.[22] MCF7, MB231 cell lines were originally obtained from the American Type Culture Collection (ATCC). The full-length coding region of wild-type or K112E-mutated human cyclinD1 cDNA was cloned into the mammalian retroviral expression vector, pMSCV-IRES-GFP (Addgene, Cambridge, MA, USA) and was used to establish the overexpression clones. The pCL-Ampho vector was used as helper to produce the infective viral particles in the HEK293T packaging cells. The viral particles were then infected into MCF7 and MB231 cells further sorted by fluorescence-activated cell sorting (FACs) using GFP as a selective marker to purify the overexpression clones.
Reagents
The chemical synthetic CDK4 inhibitor, naphtho[2,1-a] pyrrolo [3,4-c] carbazole-5,7(6H, 12H)-dione (NPCD)[23] was dissolved with dimethyl sulfoxide (DMSO) and kept at −20°C until use. The chemotherapeutic agent, 5-FU, was purchased from Sigma Aldrich (St Louis, MO, USA) prepared in DMSO and stored at −20°C. CDDP was purchased from American Pharmaceutical Partners, Inc. (Schaumburg, IL, USA), dissolved in phosphate-buffered saline (PBS) and stored at −20°C. Gemzar (clinical injection formula of gemcitabine) was obtained from Eli Lilly (Indianapolis, IN, USA) and prepared in PBS stored at −20°C. Unless specified separately, primary antibodies used in this study were purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA), including mouse monoclonal anti-β-actin (sc-47778) and rabbit polyclonal antibodies, anti-cyclinD1 (sc-718), anti-cyclinD3 (sc-182), anti-CDK4 (sc-260), anti-CDK6 (sc-7180), anti-CDK2 (sc-163), anti-cyclin E (sc-481), anti-Rb (sc-50), anti-pRb (Ser807/811; sc-16670), anti-pRb (Ser780; sc-12901), anti-p27 (sc-528), anti-p21 (sc-397), anti-Myc (sc-788), and anti-p53 (sc-6243 and sc-126). The anticleavage poly-(ADP-ribose) polymerase (PARP) (Cat#9541) rabbit polyclonal antibody was purchased from Cell Signaling Technology, Inc (Danvers, MA, USA). Mouse monoclonal anti-BCL-2 antibody was purchased from Millipore (Billerica, MA, USA). Peroxidase-conjugated antimouse (NA931) and antirabbit (NA934) secondary antibodies were purchased from Amersham Biosciences (Piscataway, NJ, USA).
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assay
Cells were seeded in a 96-well microplate at 4000 cells per well in multiple repetitions to ensure 6 wells per dose for each time point in the subsequent treatments, and then the cells were incubated at 37°C with 5% CO2. To measure the cell proliferation, the culture medium in each 96-well plate was changed into serum-free or 10% fetal bovine serum (FBS) condition after 24 h of seeding, and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was performed after 4, 24, 48, 72, and 96 h of conditional culture. To evaluate the responses of cell clones to anticancer treatments, after 24 h of seeding, the culture media were changed into fresh media containing the selected drugs at a specified concentration with DMSO or PBS as vehicle control, and the MTT assay was performed at the end of each treatment following routine procedure.[23] The experiment was repeated at least 3 times to ensure data reproducibility.
Clonogenic survival assay
Cells were seeded into 24-well culture plates at 1 × 105 cells per well and allowed to adhere overnight, and then were treated with the selected drug or a vehicle control for 24 h. The cells were trypansized and evenly reseeded at a lower density (1, 2, and 3 × 102 cells per well) in triplicate in a 6-well culture plate. Cells were cultured for 10–15 days, with medium change every 3 days, to let the viable cells propagate to form visible colonies. The colonies were fixed with methanol–acetic acid (3:1), stained with 1% crystal violet at room temperature; then the number of colonies in each well was counted and photographed. The experiment was repeated 3 times and the presented data are the means after statistical analysis of collected results.
Acridine orange and ethidium bromide staining
Cells were seeded into 96-well plates at 3000 cells per well overnight and then were exposed to selected drugs at the specified concentration with 6 repetitions per treatment. AO/EB staining and photo processing were performed after 24 h as described earlier.[23] Multiple photos were taken at randomly selected areas of the wells to avoid prejudicial data selection.
Cell cycle analysis
Cells were cultured in 6-cm dishes to 70%–80% confluence. The culture media were then changed into either serum-free or 5% FBS without or with Gemzar at a final concentration of 1 μM. After 48 h incubation, both floating and adherent cells were collected, combined, washed with cold PBS, and then fixed overnight with 70% ethanol in PBS at −20°C. The cell cycle analysis and data processing were performed as described before.[23]
Protein extraction and Western blot assay
Total proteins were extracted and quantified using a Bradford assay (Bio-Rad Laboratories Inc. Hercules, CA, USA). An equal amount of protein from each sample was fractioned in SDS-PAGE and then transferred onto an Immobilon-P Nylon membrane (Millipore, Bedford, MA, USA). After blocking with 5% milk, the membrane was incubated with specific primary antibody overnight at 4°C. The membrane was washed 3 times with PBS-Tween20, incubated with horseradish peroxidase-conjugated secondary antibody for 2 h, followed by 3 washes. The signal was captured with ECL chemiluminescent substrates (Pierce, Rockford, IL, USA) visualized on X-ray film (ISC BioExpress, Kaysville, UT, USA).
Soft-agar colony formation assay
There were about 1000 cells per well from each cell clone seeded with the soft-agar (0.35% agar in 10% FBS DMEM) medium into the 6-well culture plate, which was pre-coated with a layer of basal agar (0.5% agar in 10% FBS DMEM) in triplicate. The cell containing soft-agar medium was covered with 10% FBS DMEM culture medium and then maintained at 37°C, 5% CO2 for 14–20 days with the top medium refreshed every 4 days. At the end of the culturing, the top medium was discarded and the colonies were counted under the Leica DM IRB inverted fluorescence microscope (Leica Microsystems Inc., Bannockburn, IL, USA) using an automatic colony counting system.
Statistical analyses
All MTT, FACs, clonogenic survival and soft-agar colony formation assays were performed in multiple repetitions in each experiment, and the experiments were repeated at least 3 times. The data were presented as mean ± SE. Statistical comparisons between groups were made with the Student's t test. A P value < 0.05 is considered as significant.
RESULTS
The basal levels of cyclinD1 and CDK4 do not show direct correlation with chemosensitivities of breast cancer cells
We first evaluated the response of MCF7, MB231, and MCF15 cells to the therapeutic drugs 5-FU, CDDP, and Gemzar using MTT assay as shown in Figure 1a; the basal levels of G1 phase cell cycle proteins in these cells are shown in Figure 1b. Among the 3 cell lines, the MCF15 is the most sensitive one in response to all the drugs, as reported characteristic of this cell line.[22] The MCF7 and MB231 cells showed similar sensitivities to the CDDP and Gemzar, whereas the MB231 cells were more resistant to 5-FU. The levels of D1 and CDK4 were not significantly different among the 3 cell lines; therefore, the levels of these proteins are not determinants of the chemosensitivities of these breast cancer cells.
Figure 1.
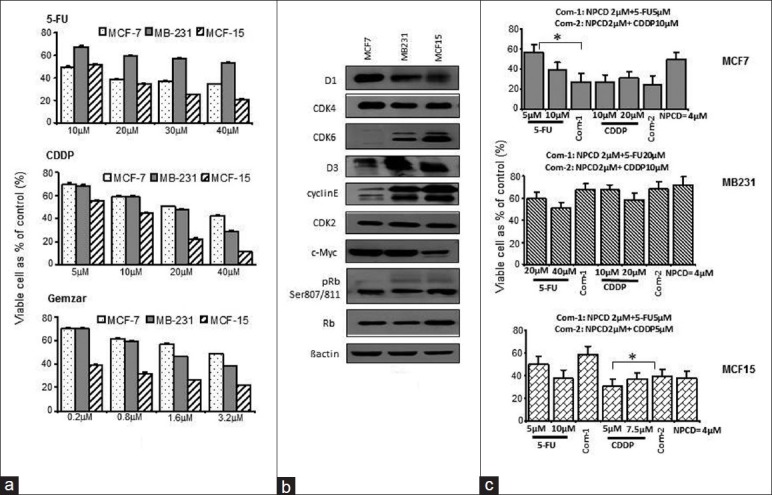
Protein expression patterns and the chemoresponses of human breast cancer cell lines. (a) Diphenyltetrazolium bromide (MTT) detection of the chemosensitivities of MCF7, MB231, and MCF15 cells to 5-FU, cisplatin (CDDP), and Gemzar. (b) The basal levels of cell cycle regulatory proteins in 3 cell lines determined by western blot assay. (c) Combination of naphtho [2, 1-a] pyrrolo [3, 4-c] carbazole-5, 7 (6H, 12H)-dione (NPCD) is synergistic with the effects of 5-FU in the MCF7 cell and the attenuate effects of CDDP in MCF15 cells. *P < 0.05
We then treated the 3 cell lines with 5-FU and CDDP alone or with the combination of 5-FU or CDDP with NPCD. Through inhibiting the Rb phosphorylation, blocking cell cycle progression, NPCD has been shown to have therapeutic effects as a single agent in these breast cancer cells.[23] As shown in Figure 1c, NPCD synergizes 5-FU but has no effect on CDDP in the MCF7 cells (top) and has no effects on either drug in the MB231 cells (middle), while NPCD attenuated the effects of 5-FU and CDDP in the MCF15 cells (bottom). This result indicated that the cyclinD1 or the cyclinD1CDK4/6 kinase is engaged in the signaling response to treatments in human breast cancer, whereas the functions may be cell-type specific and drug specific.
To pursue the role of cyclinD1 and the cyclinD1CDK4/6 kinase in the regulation of chemoresponsivity in human breast cancer, we ectopically expressed the wild type of cyclinD1 (D1) or the K112E mutant cyclinD1 (D1KE) protein in the MCF7 and MB231 cells. The GFP-positive clones were maintained, enriched, confirmed by RT-PCR and Western blot assay and then subjected to downstream studies. It has been proved that the K112E mutated cyclinD1 protein could not activate the CDK4 kinase although the complex is still formed.[24]
Expression of ectopic cyclinD1 protein sensitizes the MCF7 cells’ response to CDDP, Gemzar, and CDK4 inhibitor, which may require the CDK4 kinase activity
MCF7, a widely studied ER-positive breast cancer cell line, ectopic expression of the D1 or D1KE protein resulted in distinct effects on responses to anticancer treatments.
The responses to anticancer treatments detected by MTT assay after 72 h of treatments are shown in Figure 2a. The viable cells were 5% fewer with CDDP treatment and 10%–13% fewer with Gemzar treatment in the D1 clones compared with the vector and D1KE clones. In response to NPCD, the D1KE clones showed 20% more viable cells than the vector and the D1 clones and 5% fewer viable cells in the D1 clones compared with the vector clones. Overall, the D1 clones were more sensitive to the chemotherapeutic drug; on the other hand, the sensitivities between the vector and the D1KE clones were very similar.
Figure 2.
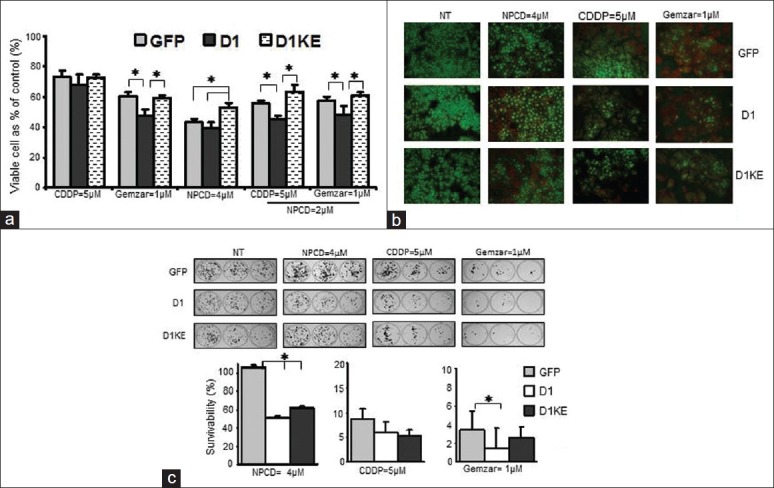
Effects of ectopic expression of D1 or D1KE protein in MCF7 cell on the responses to NPCD, CDDP, and Gemzar. (a) Diphenyltetrazolium bromide (MTT) to determine the chemosensitivities in response to CDDP, Gemzar alone or their combination with NPCD. (b) Acridine orange/ethidium bromide (AO/EB) staining to evaluate the cell death after 24-h treatment with NPCD, CDDP, or Gemzar. (c) Clonogenic survival assay assessment of the drugs’ persistent effects. The 3 wells indicated the consecutive seeding numbers of the same cell clone from same treatment. The chart represents the mean of 3 individual experiments shown as the survivability. *P < 0.05
Drug-induced cell death was evaluated by the AO/EB staining in these clones after 24 h of drug treatments as shown in Figure 2b. The ratio of dead cells (red or orange) to the living cells (green) was higher in the D1 clones than in the vector and the D1KE clones further confirming the enhanced sensitivities of the D1 clones to the treatments. The persistent effects of these anticancer treatments on the different cell clones were examined by the clonogenic survival assay and the results summarized from 3 individual experiments are shown in Figure 2c. The survivability was calculated with the average colony number from each treatment in each cell clone as the percentage of the average colony number from the same clone without treatment. The results further confirmed the higher sensitivity of the D1 clones to all the treatments among 3 clones, whereas the D1KE clones also were more sensitive than the vector clones but less significantly than the D1 clones. The difference exhibited from the clonogenic survival and MTT assay was that the survivability was much lower than the viability determined in the MTT assay when the same cell clone received the same dose of the drug, which suggested the persistent effects and the differences in response to the chemotherapeutic drugs among cell clones constantly exist. Together, the 3 techniques consistently detected the higher sensitivity of D1 clones in response to the anticancer treatments, whereas the D1KE clones respond differently to each drug compared with the D1 clones.
Simultaneously targeting the CDK4 kinase and ectopically expressing cyclinD1 protein potentiate the effects of CDDP but antagonize the effects of Gemzar
NPCD, the CDK4 inhibitor, affects the chemosensitivity of MCF7 cells as shown in Figure 1c, and to ascertain whether these effects exist during the ectopic expression of D1 or D1KE protein, we then performed the MTT assay using the combination of 2 μM NPCD with CDDP and Gemzar in the treatment of those cell clones.
Inhibition of CDK4 activity potentiates the effects of CDDP as shown in Figure 2a. The viable cells dropped from 73% to 55% in the vector clones, from 68% to 45% in the D1 clones, and from 72% to 63% in the D1KE clones. In contrast, inhibition of CDK4 activity has minor effect on Gemzar in the vector clones with the viable cell dropping from 60% to 57% but with no obvious effects in the D1 and the D1KE clones. Moreover, simultaneous inhibition of the CDK4 did not switch the patterns of the 3 cell clones in response to CDDP or Gemzar compared with the patterns in response to CDDP and Gemzar alone, which, the D1 clones are more sensitive to both the drugs.
Collectively, ectopic expression of the wild-type cyclinD1 protein sensitizes the MCF7 cell responses to anticancer agents and in contrast, the K112E mutant expression clones showed less effect, which indicated the requirement of CDK4 kinase activity in the D1 function under this circumstance.
Ectopic expression of cyclinD1 protein in MCF7 cells has no effects on the cell cycle progression; rather it promotes the anchorage-independent cell growth in a kinase activity-independent manner
Were the different sensitivities responding to anticancer treatments observed in MCF7 derivatives linked to the D1 function in promoting cell proliferation?
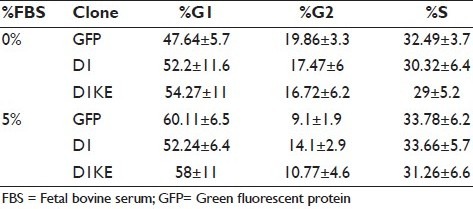
We assessed the levels of several G1–S phase cell cycle regulatory proteins under serum-free and 10% FBS culture conditions in the MCF7 derivatives [Figure 3a]. The levels of CDK4, CDK6, CDK2, cyclinE, and the endogenous D1 proteins were higher in the vector clones than in the D1 and D1KE clones, whereas the level of Myc protein was higher in the D1 and the D1KE clones than in the vector clones. The Rb protein was induced slightly by 10% FBS and its levels were similar in 3 cell clones. The CDK4/6 phosphorylated Rb proteins, detected by 2 specific antibodies, were dramatically induced by the addition of 10% FBS in the vector and D1 clones reaching similar levels among the 3 clones while under serum-free conditions; only in the D1KE clones, its level was as high as at the 10% FBS condition. It could indicate a continuously activated Rb phosphorylation in the D1KE clones. We then evaluated cell proliferation by MTT assay as shown in Figure 3b. In a 96-h period, either serum-free or in 10% FBS, the growth rate was similar among the 3 cell clones at the same condition, and there was no difference in the growth rate under 5% versus 10% FBS in all cell clones (data not shown). We further analyzed the cell cycle of the MCF7 cell clones by FACs as shown in Table 1. Compared with the 10% FBS culture condition, serum deprivation resulted in an increase of G1 phase cell population in the 3 clones that was 16% in the vector clones and 9% in both the D1 and D1KE clones. The distinct effects of D1 and D1KE proteins on cell growth in MCF7 cell were observed in soft-agar assay as summarized in Figure 3c from 3 individual experiments. The D1KE clones displayed the highest rate of colony formation and the vector clones showed the lowest rate. Obviously, the cyclinD1 protein is able to promote MCF7 cell anchorage-independent cell growth, which is CDK4 activity independent or possibly the CDK4 activity may inhibit this function.
Figure 3.
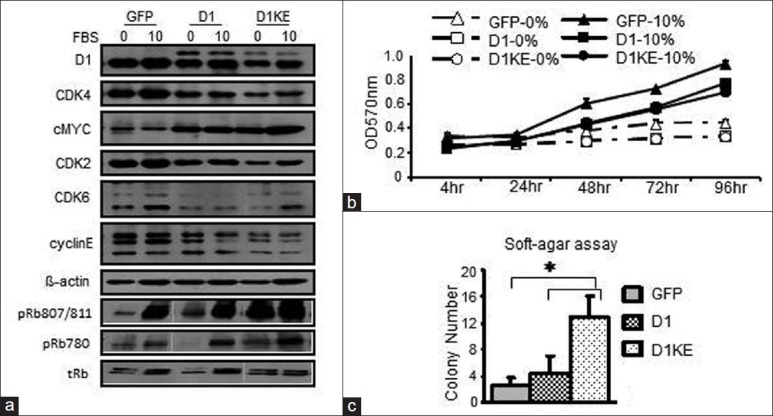
Effects of ectopic expression of D1 or D1KE protein in MCF7 cell on the cell cycle protein expression and cell growth. (a) The expression of several cell cycle regulatory proteins in MCF7 cell clones under serum free or 10% fetal bovine serum (FBS) culture conditions. (b) Cell proliferation measured by diphenyltetrazolium bromide assay in MCF7 cell clones under serum free and 10% FBS culture conditions within 96 h. (c) Soft-agar assay to assess the anchorage independent growth of the MCF7 cell clones. *P < 0.05
Table 1.
Cell cycle patterns of MCF7 cell clones
Ectopic expression of the cyclinD1 protein attenuates the chemoresponses of MB231 cells and is partially CDK4 activity dependent
In the MB231, a triple (ER, PR, and Her2) negative, more malignant basal-like breast cancer cell line, we applied the same strategy to investigate the role of cyclinD1 protein and its kinase partner in the chemoresponses.
The responses of the MB231 cell derivatives to CDDP, Gemzar, and NPCD were detected by MTT after 72 h of treatment as shown in Figure 4a. In response to the CDDP, the D1 clones showed 24% more, and the D1KE clones showed 6% more, viable cells than the vector clones; to Gemzar, there were 20% more viable cells in the D1 and 8% more in the D1KE clones than in the vector clones; to NPCD, 9% more viable cells in the D1 clones and 5% more in the D1KE clones than in the vector clones. Clearly, the D1 clones were less sensitive to these anticancer agents than the vector clones, whereas the D1KE clones also showed decreased sensitivity but less significantly than the D1 clones showed. Next, we applied treatments by combination of 2 μM NPCD with CDDP or Gemzar and the results are shown in Figure 4a. The NPCD potentiated the CDDP effects in all the MB231 derivatives as seen from the dropped viable cell ratio when compared with CDDP being used alone, from 62% to 40% in the vector clones, from 85% to 52% in the D1 clones and from 67% to 51% in the D1KE clones. NPCD showed a very minor effect on Gemzar in the 3 clones. Regardless of the effects on CDDP and Gemzar, the addition of NPCD did not change the response patterns among the 3 clones, showing that the D1 and D1KE clones were less sensitive to the treatments.
Figure 4.
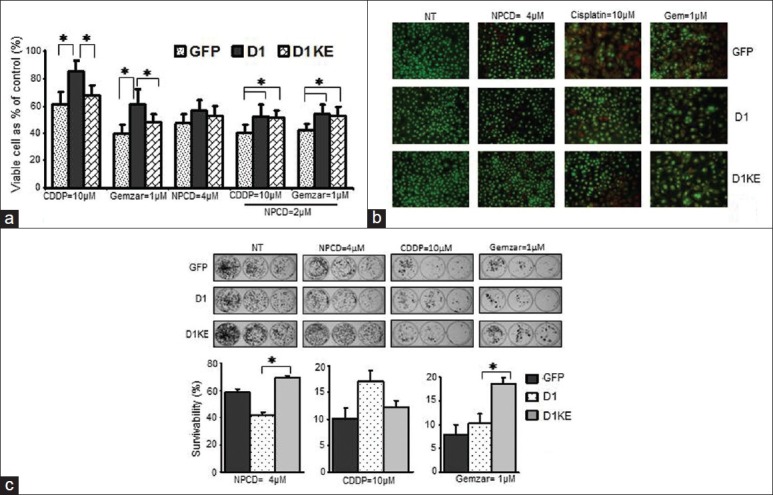
Chemoresponses of the MB231 derivatives to NPCD, CDDP and Gemzar. (a) The responses to single drugs or to combination of NPCD with CDDP or Gemzar detected by MTT assay. (b) AO/EB staining evaluation of cell death after 24-h treatment. (c) Clonogenic survival assay assessment of the persistent effects of the drugs. The 3 wells indicated three consecutive seeding numbers of the same cell clone from same treatment. The chart represents the mean of 3 individual experiments shown as the survivability. *P < 0.05
Drug-induced cell death was evaluated by AO/EB staining and clonogenic survival assay with the same treatments. As shown in Figure 4b, upon the CDDP and Gemzar treatment, the ratio of dead cells (red or orange) over the living cells (green) was higher in the vector clones than in the D1 and the D1KE clones.
The survivability determined by clonogenic survival assay summarized from 3 individual experiments is shown in Figure 4c. Compared with the vector clones, the D1 clones were more sensitive to the NPCD with 20% less survivability but resistant to CDDP with 8% more cell survival and were slightly resistant to Gemzar. The D1KE clones were more resistant to all treatments with about 10% more survivability than the vector clones and they were less resistant to CDDP than the D1 clones with 5% less survivability. The response to drug persistent effects among the 3 clones further confirmed that the D1 and D1KE clones are less sensitive to the treatments.
Ectopic expression of neither the D1 nor the D1KE protein in MB231 has any effect on cell cycle but does enhance the anchorage-independent cell growth
We next evaluated the cell cycle regulatory proteins and the cell cycle profile in the MB231 clones. The levels of several G1 phase proteins were estimated under the serum-free or 10% FBS culture conditions as shown in Figure 5a; the D1, CDK4, and p27 proteins were obviously induced by 10% FBS in the vector and the D1 clones but only with minor induction in the D1KE clones. At the serum-free condition, these proteins were more abundant in the D1KE clones than in the vector and the D1 clones. The levels of CDK2, CDK6, cyclinE, Rb, and CDK4/6 phosphorylated Rb proteins were similar in all 3 clones and without obvious change between the 2 culture conditions. The levels of Myc protein were similar among the 3 cell clones without obvious induction by 10% FBS in all the cell clones.
Figure 5.
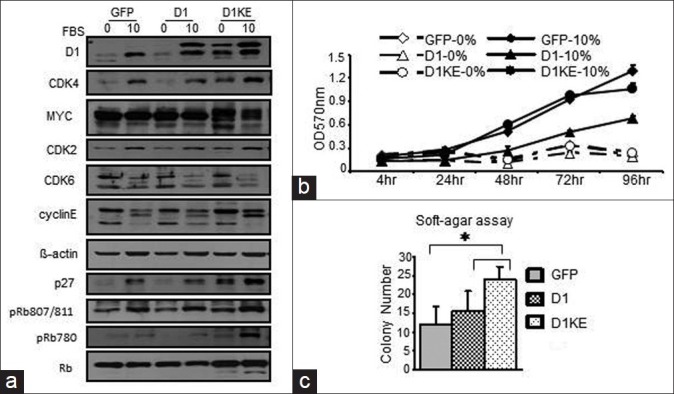
Effects of ectopic expression of D1 or D1KE protein in MB231 cell on the cell cycle protein expression and cell growth. (a) Western blot assay determination of the expression of several cell cycle regulatory proteins under serum free or 10% fetal bovine serum (FBS) culture conditions. (b) Cell proliferation measured by diphenyltetrazolium bromide assay under serum-free and 10% FBS culture conditions within 96 h. (c) Soft-agar assay to assess the anchorage-independent growth of the MB231 cell clones. *P< 0.05.
The cell proliferation was evaluated over a 96-h time period as shown in Figure 5b. Ectopic expression of the D1 protein did not promote the MB231 cell growth or change the effect of serum on cell growth. The cell cycle analysis further confirmed the MTT results, which demonstrated that neither the D1 clones nor the D1KE clones showed growth advantage under serum-free or 5% FBS culture condition [Supplementary Table]. However, the anchorage-independent cell growth was highly induced in the D1KE clones, whereas there was only minor induction observed in the D1 clones as shown in Figure 5c. Overexpression of D1 protein was reported to enhance the anchorage-independent cell growth[25,26]; and here, our results further indicated that the function of D1 in promoting the anchorage-independent cell growth is likely CDK4 activity independent.
Supplementary Table.
Cell cycle patterns of MB231 cell clones
So far, we could exclude the possibility that ectopic expressed cyclinD1 proteins modulate chemoresponses through facilitating cell cycle and cell growth in the MCF7 and MB231 cells. Besides, ectopic expression of D1 or D1KE protein in these 2 cell lines exhibits opposite effects on the cells’ response to the same treatment.
Gemzar triggers different signaling in MCF7 and MB231 cells, and in co-operation with ectopic expression of cyclinD1 protein, resulting in distinct effects
Gemcitabine (2’,2’-difluorodeoxycytidine) is a nucleoside analogue with potent activity against various solid tumors.[27,28] It has been used as first-line drug to treat the breast cancer patients with metastasis or relapse after receiving other chemotherapies. As the D1 and D1KE clones derived from MCF7 and MB231 cells showed different sensitivities to Gemzar, we then performed further studies to reveal the signaling associated with cyclinD1 in response to Gemzar treatment.
In the MCF7 cell clones, given 1 μM of Gemzar for 24 h, several cell cycle proteins were assessed as shown in Figure 6a. The levels of D1, CDK4, CDK6, CDK2, cyclinE, and p53 proteins increased upon the treatment in all 3 cell clones with the vector clones increasing most dramatically, whereas the p21 and p27 increased only in the vector clones. We did not detect obvious change of several key factors involved in apoptotic or nuclear factor kappa B (NF-kB) pathways at the RNA or protein level in response to the Gemzar treatment. Cell cycle was assessed after 48 h of treatment as shown in Figure 6b; the only obvious change induced by Gemzar in the 3 cell clones was a 12% decrease of the G1 phase in the D1KE clones. Moreover, the 3 cell clones shared very similar cell cycle patterns either with or without Gemzar treatment condition. The cell cycle patterns did not reflect the changes in the cell cycle regulatory proteins; we hypothesize that in the MCF7 cell clones, the cell cycle pathway is connected with the signaling interference by Gemzar therefore the changes of cell cycle proteins resulted from the response to Gemzar treatment.
Figure 6.
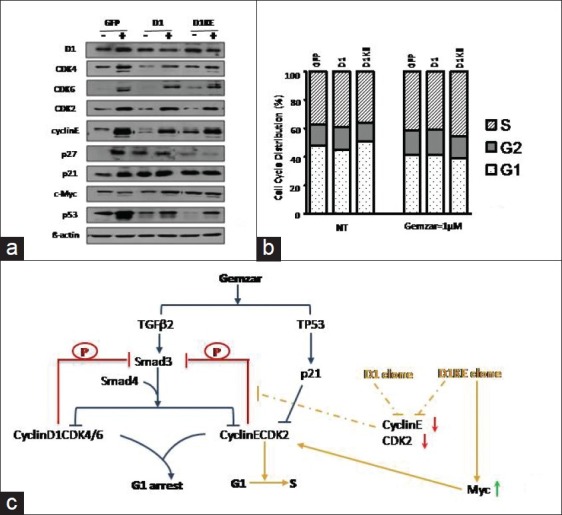
The patterns of protein expression and cell cycle distribution in the MCF7 cell derivatives responding to Gemzar treatment. (a) Western blot assay detection of the cell cycle regulatory proteins after 24-h treatment. (b) Cell cycle analysis after 24-h treatment. (c) The hypothetic model of the signaling responses upon Gemzar treatment in the 3 clones. Gemzar triggers the TGFb and TP53 in the MCF7 cells, which leads to G1 arrest, meanwhile, it also upregulates the cyclinD1, CDK4, CDK2, and cyclinE protein, which will inhibit Smad3 by phosphorylation. The major differences between the 3 clones are the levels of cyclinE and CDK2 lower in the D1 and D1KE than in the vector clones. The inhibition effects on Smad3 will be less effective in the D1 and D1KE clones, which results in the increased sensitivity to Gemzar treatment. While in the D1KE clones, the level of Myc protein is higher than other 2 clones, which will promote the G1-S phase transition so as to result the less sensitive to Gemzar than the D1 clones
In the MB231 cell clones, after 24 and 48 h of treatment with 1 μM of Gemzar, the levels of cell cycle regulatory proteins were evaluated as shown in the Figure 7b. The cyclinD1 protein dramatically diminished in both the D1 and the D1KE clones but not in vector clones while the CDK4 protein decreased in the D1KE clones only, with no changes of CDK6 protein in the 3 clones. Meanwhile, an increase of cyclinD3 protein was detected in all 3 cell clones, whose functions may compensate for the D1 protein. The increase of cyclinE, CDK2, p27, and p53 proteins after Gemzar treatment was detected in the vector clones only, whereas a decrease of cyclinE protein was detected in the D1 clones. The Myc protein level dramatically decreased in the Gemzar-treated vector and D1 clones, more obviously in the D1 clones, and with no obvious change in the D1KE clones. Gemzar treatment caused an increase of cleaved PARP protein in all cell clones with the vector clones increasing most obviously and with increase of BCL-2 protein in all cell clones as well.
Figure 7.
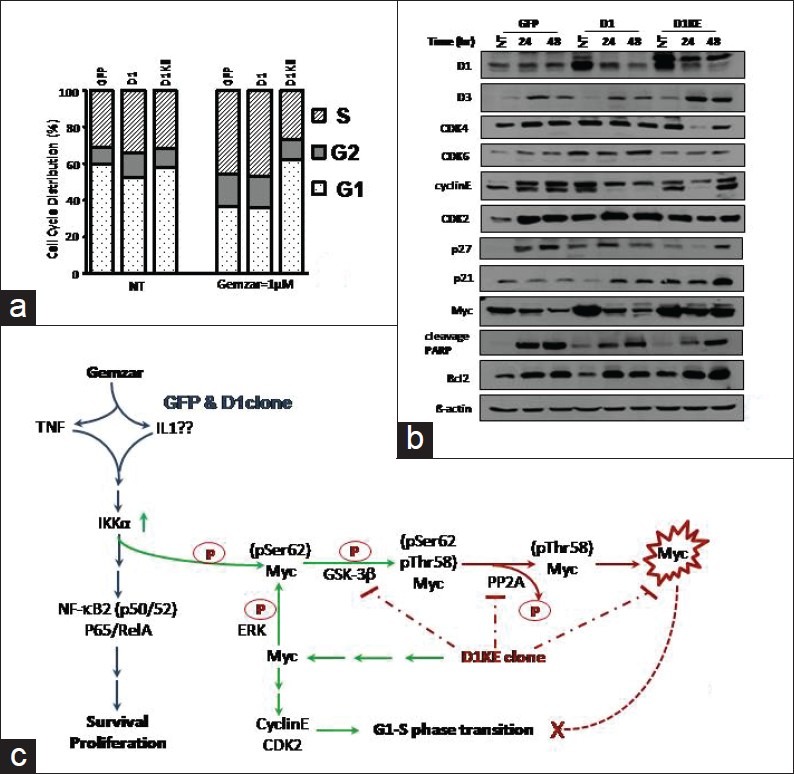
The patterns of protein expression and cell cycle distribution in the MB231 derivatives responding to Gemzar treatment. (a) Cell cycle analysis after 24-h treatment. (b) Expression of several cell cycle regulatory proteins after 24 and 48 h of the treatment. (c) The hypothetic model of the signaling responses upon Gemzar treatment. Gemzar triggers NF-kB signaling in the MB231 cells resulting resistance to the drug treatment and degrade the Myc protein. In the D1KE clones, Myc protein becomes more stable, which leads to more resistance to the Gemzar treatment and G1 arrest
Cell cycle analysis of the MB231 cell clones after 48 h of treatment with Gemzar Figure 7a showed that the D1KE clones exhibited more abundant G1 phase (25% more) and a less S-phase cell population (20% less) than the vector and the D1 clones while there was no distinguishable difference between the vector and the D1 clones. Compared with the nontreated groups, Gemzar induced diminished G1 phase, which was about 24% in the vector clones and 17% in the D1 clones, but with no obvious change in the D1KE clones. This result indicated that the CDK4 kinase activity participates in the MB231 cell response to Gemzar.
DISCUSSIONS
CyclinD1 was primarily defined as cell cycle regulator, in the past 2 decades, more functions have been discovered with some of them being noncatalytic or CDK4/6-independent functions. One rising question, the role of D1 in the human cancer responses to treatments has been taken into account, while only limited studies have clearly addressed the relationship between D1 level and sensitivity to anticancer treatment except for the endocrine resistance in breast cancer.[29–31]
In this study, ectopic expression of neither the wild-type nor the K112E-mutated cyclinD1 protein had significant impact on the Rb phosphorylation, cell cycle progression, or cell growth in the MCF7 and MB231 breast cancer cells. This result is contradictory to the classic D1 function, especially the D1KE clones do not show diminished Rb phosphorylation, delayed cell cycle, or restrained cell proliferation. The possible explanation for this result could be that the already abundant D1 protein in both cell lines limits the exogenous D1 proteins’ functions due to the availability of CDK4, CDK6, or p21 and p27. Moreover, exogenously expressed D1 protein provides a negative feedback signal leading to downregulation of endogenous D1 expression, which is the case in MCF7 cell clones as shown the RT-PCR results in supplementary Figure a. Another reason could be, in these 2 cell lines, continuously activated cell cycle progression has been governed by factors other than cyclinD1CDK4/6, such as Myc, to activate cyclinECDK2, drive cells into S phase.[32] The anchorage-independent cell growth was dramatically enhanced in the D1KE clones, and over stronger than the D1 clones in both cell lines, which suggested D1 functioning as the catalytic function-independent context.
Supplementary Figure.
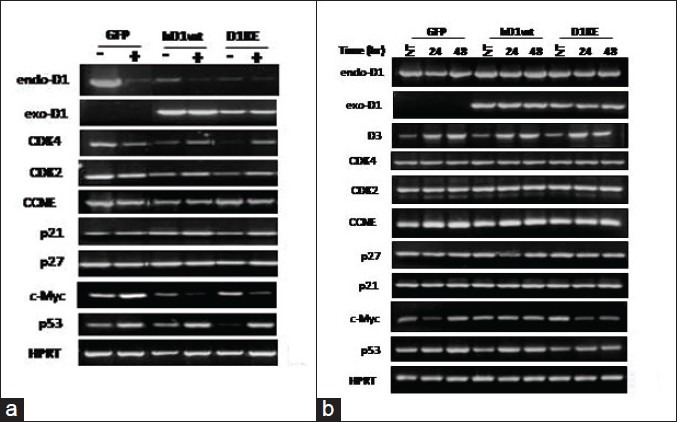
(a) RT-PCR assay detection of the genes expression in the MCF7 cell clones without or with 1 μM of Gemzar treatment for 24 h. (b) RT-PCR assay detection of the genes expression in the MB231 cell clones without or with 1 μM of Gemzar treatment for 24 and 48 h
In MCF7 cells, ectopic expression of D1 protein sensitized the cells to CDDP, Gemzar, and NPCD, as demonstrated by MTT, clonogenic survival assay, and AO/EB staining, and these effects are probably CDK4 kinase activity dependent. Gemzar treatment induced obvious increases of the CDK6, CDK2, cyclinE, and p53 proteins and the changes were more obvious in the vector clones than in the D1 and D1KE clones. As Hernandez-Vargas et al.[33] reported, gemcitabine induced expression and activation of the tumor necrosis growth factor-beta2 (TGFβ2) and TP53 signaling in the MCF7 cells and the genes involved in the cell cycle; metabolism and cell growth signaling changed more obviously. The cyclinD1CDK4 and cyclinECDK2 kinases were shown to repress small mothers against decapentaplegic (Smad3)-mediated G1 arrest by phosphorylation of the Smad3 protein in the MCF7 cells,[34,35] whereas the Smad3, as one of the important effectors in the TGFβ signaling pathway, is triggered by gemcitabine in this cell line. Combining their reports and our finding here, we think that the enhanced sensitivity to Gemzar in the D1 clones is due to the lower levels of CDK4, CDK2, and cyclinE proteins in this particular clone, which leads to impaired inhibition effect on the Smad3-mediated G1 arrest as we illustrated in Figure 6c. However, how ectopic expression of D1 proteins affects cell cycle proteins’ expression is still unrevealed. The subsequent effects in mediating the responses of different cell clones to Gemzar or to other chemotherapeutic drugs remain unclear. Future studies are needed to address these questions.
In the MB231 cells, ectopic expression of either the wild-type or the K112E-mutated D1 protein attenuated the sensitivity to CDDP and Gemzar, which did not result from accelerated cell proliferation. Gemzar treatment caused dramatically diminished Myc protein in the vector and D1 clones but not in the D1KE clones and the changes were not due to transcriptional repression [Supplementary Figure b]; meanwhile, BCL-2 protein increased in the 3 clones with more obvious in the D1KE clones. The MB231 cell displays a higher basal level of NF-kB activity, which would be further induced[36–38] and contributes to the resistance of hormone and chemotherapeutic treatments. Gemcitabine triggers NF-kB transcriptional activity through activation of the I-kappa B kinase alpha (IKKa)[33] and, more importantly, this response is gemcitabine and the MB231 cell line bispecific. The IKKa associates and phosphorylates the D1 protein at Threonine286 leading to D1 protein nuclear exportation and degradation[39] as here we detected obvious decrease of D1 protein in the Gemzar-treated D1 and D1KE clones. The oncoprotein Myc displays prominent and direct effects on invasion, migration, and metastasis besides its function on survival and proliferation.[40] Myc sensitizes tumor necrosis factor (TNF)-induced apoptosis through inhibition of NF-kB transcription activity by impaired p65/RelA (v-rel-reticuloendotheliosis viral oncogene homologue A) transactivation;[41] especially in MB231 cells, the highly expressed Myc protein is implicated as a regulator of poor prognosis. The stability of Myc protein is controlled at multiple stages, including the phosphorylation and ubiquitin proteolysis system.[42] A recent study has shown in MCF7 cells[43] that IKKa directly interacts with Myc to prolong Myc protein half-life. Here, in the MB231-derived cell clones, the different sensitivities in responding to Gemzar are likely associated with Myc protein level and its stability. Gathering results from others and ours showing here, the hypothetic signaling network in response to Gemzar treatment is illustrated in Figure 7c. The patterns of Myc protein shown on the Western blot displays differently among the 3 cell clones, that are without treatment appearing as multiple bands in the D1KE clones while there was only a dominant single band in the vector and D1 clones. After exposure to Gemzar, in the D1KE clones, the Myc protein remained abundant without obvious change and cells were arrested in G1 phase, while in the other 2 clones, the Myc protein dramatically decreased and cells were retained in S phase. The mechanisms on how the ectopic expression of D1 or D1KE protein affects the Myc protein level remain to be further studied.
Additionally, in both MCF7 and MB231 cells, attenuation effects of the CDK4 inhibitor on Gemzar were consistently detected. Hyperactivation of the cyclinD1CDK4/6 exists in different types of human tumors, and targeting the CDK4/6 has been shown as a promising anticancer approach.[44,45] Unexpectedly, inhibition of CDK6[46] or CDK4[47] could restrain NF-kB signaling, and the functions conflict with the function of Gemzar. Therefore, from a clinical perspective, combining targeted therapy with common chemotherapeutic drugs in the treatment of human cancers has to be done cautiously.
In conclusion, we report that ectopic expression of the wild-type or K112E-mutated human cyclinD1 proteins in MCF7 and MB231 breast cancer cells results in different effects on the response to CDDP and Gemzar. In the MCF7 cells, ectopic expression of D1 protein sensitizes responses to CDDP, Gemzar, and CDK4 inhibitor in a CDK4 kinase activity-dependent context. In the MB231 cells, ectopic expression of D1 protein imposes resistance to CDDP, Gemzar and CDK4 inhibitor, which is less CDK4 kinase activity dependent. The changes of the drug sensitivities in the MCF7 and MB231 cells are not resulted from D1-induced acceleration of cell cycle by activating CDK4/6 and repressing Rb. Meanwhile, the CDK4 inhibitor has a potent effect on CDDP but attenuates the effect on Gemzar in both MCF7 and MB231 cells and their derivatives. To our knowledge, this is the first study through concurrent ectopic expression of the wild type of D1 and D1KE proteins to investigate the function of cyclinD1 in modulating chemoresponses in breast cancer cells. Furthermore, inhibition of CDK4 activity diminishes the effects of Gemzar, which indicates that the combination of targeted therapy with chemotherapeutic drugs must be done cautiously in the clinical application.
The mechanisms on how ectopic expression of D1 or D1KE protein interferes with cellular protein expression and the associated signaling in mediating response to Gemzar are further to be explored.
AUTHOR'S PROFILE
Dr. Yuan Sun, Department of Pathology of Guangxi Medical University, Nanning, Guangxi, China, Hormel Institute of the University of Minnesota, USA
Dr. Dianzhong Luo, Department of Pathology of Guangxi Medical University, Nanning of Guangxi, China.
Dr. D.J. Liao, Department of Translational Cancer Research, Hormel Institute, the University of Minnesota, USA.
ACKNOWLEDGMENTS
This work was supported by a grant from National Science Foundation of China (#81060200) to Dr. D.-Z. Luo and a grant from the Department of Defense of United States (DOD Award W81XWH-11-1-0119) to D.J. Liao. We thank Fred Bogott, M.D., Ph.D., at Austin Medical Center, Austin of Minnesota, USA, for his excellent English editing of this manuscript.
Footnotes
Source of Support: This work was supported by a grant from National Science Foundation of China (#81060200) to Dr. D.-Z. Luo and a grant from the Department of Defense of United States (DOD Award W81XWH-11-1-0119) to D.J. Liao
Conflict of Interest: None declared
REFERENCES
- 1.Li Z, Wang C, Prendergast GC, Pestell RG. Cyclin D1 functions in cell migration. Cell Cycle. 2006;5:2440–2. doi: 10.4161/cc.5.21.3428. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L, et al. Cyclin D1 induction of cellular migration requires p27KIP1. Cancer Res. 2006;66:9986–94. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Wang C, Jiao X, Lu Y, Fu M, Quong AA, et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol Cell Biol. 2006;26:4240–56. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zwijsen RM, Wientjens E, Klompmaker R, Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–15. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 5.Knudsen KE, Cavenee WK, Arden KC. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59:2297–301. [PubMed] [Google Scholar]
- 6.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ratineau C, Petry MW, Mutoh H, Leiter AB. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J Biol Chem. 2002;277:8847–53. doi: 10.1074/jbc.M110747200. [DOI] [PubMed] [Google Scholar]
- 8.Bienvenu Fdr, Gascan H, Coqueret O. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J Biol Chem. 2001;276:16840–7. doi: 10.1074/jbc.M100795200. [DOI] [PubMed] [Google Scholar]
- 9.Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–34. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 10.Han EK, Begemann M, Sgambato A, Soh JW, Doki Y, Xing WQ, et al. Increased expression of cyclin D1 in a murine mammary epithelial cell line induces p27kip1, inhibits growth, and enhances apoptosis. Cell Growth Differ. 1996;7:699–710. [PubMed] [Google Scholar]
- 11.Sofer-Levi Y, Resnitzky D. Apoptosis induced by ectopic expression of cyclin D1 but not cyclin E. Oncogene. 1996;13:2431–7. [PubMed] [Google Scholar]
- 12.Pratt MA, Niu MY. Bcl-2 controls caspase activation following a p53-dependent cyclin D1-induced death signal. J Biol Chem. 2003;278:14219–29. doi: 10.1074/jbc.M209650200. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda Y, Sakai A, Tsuyama N, Katayama Y, Munemasa S, Asaoku H, et al. Ectopic cyclin D1 overexpression increases chemosensitivity but not cell proliferation in multiple myeloma. Int J Oncol. 2008;33:1201–13. [PubMed] [Google Scholar]
- 14.Coco Martin JM, Balkenende A, Verschoor T, Lallemand F, Michalides R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999;59:1134–40. [PubMed] [Google Scholar]
- 15.Niu MY, Menard M, Reed JC, Krajewski S, Pratt MA. Ectopic expression of cyclin D1 amplifies a retinoic acid-induced mitochondrial death pathway in breast cancer cells. Oncogene. 2001;20:3506–18. doi: 10.1038/sj.onc.1204453. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Zhang Z, Yang X, Chen W, Zhang P. Inhibition of cyclin D1 expression by cyclin D1 shRNAs in human oral squamous cell carcinoma cells is associated with increased cisplatin chemosensitivity. Int J Cancer. 2009;124:483–9. doi: 10.1002/ijc.23964. [DOI] [PubMed] [Google Scholar]
- 17.Hochhauser D, Schnieders B, Ercikan-Abali E, Gorlick R, Muise-Helmericks R, Li WW, et al. Effect of cyclin D1 overexpression on drug sensitivity in a human fibrosarcoma cell line. J Natl Cancer Inst. 1996;88:1269–75. doi: 10.1093/jnci/88.18.1269. [DOI] [PubMed] [Google Scholar]
- 18.Kornmann M, Danenberg KD, Arber N, Beger HG, Danenberg PV, Korc M. Inhibition of cyclin D1 expression in human pancreatic cancer cells is associated with increased chemosensitivity and decreased expression of multiple chemoresistance genes. Cancer Res. 1999;59:3505–11. [PubMed] [Google Scholar]
- 19.Kuhn DJ, Smith DM, Pross S, Whiteside TL, Dou QP. Overexpression of interleukin-2 receptor alpha in a human squamous cell carcinoma of the head and neck cell line is associated with increased proliferation, drug resistance, and transforming ability. J Cell Biochem. 2003;89:824–36. doi: 10.1002/jcb.10557. [DOI] [PubMed] [Google Scholar]
- 20.Orr MS, Reinhold W, Yu L, Schreiber-Agus N, O’Connor PM. An important role for the retinoblastoma protein in staurosporine-induced G1 arrest in murine embryonic fibroblasts. J Biol Chem. 1998;273:3803–7. doi: 10.1074/jbc.273.7.3803. [DOI] [PubMed] [Google Scholar]
- 21.Biliran H, Wang Y, Banerjee S, Xu H, Heng H, Thakur A, et al. Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin Cancer Res. 2005;11:6075–86. doi: 10.1158/1078-0432.CCR-04-2419. [DOI] [PubMed] [Google Scholar]
- 22.Shen KC, Miller F, Tait L, Santner SJ, Pauley R, Raz A, et al. Isolation and characterization of a breast progenitor epithelial cell line with robust DNA damage responses. Breast Cancer Res Treat. 2006;98:357–64. doi: 10.1007/s10549-006-9173-4. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, Li YX, Wu HJ, Wu SH, Wang YA, Luo DZ, et al. Effects of an indolocarbazole-derived CDK4 inhibitor on breast cancer cells. J Cancer. 2011;2:36–51. doi: 10.7150/jca.2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci U S A. 1994;91:709–13. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez RM, Ruiz-Miro M, Dolcet X, Aldea M, Gari E. Cyclin D1 interacts and collaborates with Ral GTPases enhancing cell detachment and motility. Oncogene. 2011;30:1936–46. doi: 10.1038/onc.2010.577. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q, Wulfkuhle J, Ouatas T, Fukushima P, Stetler-Stevenson M, Miller FR, et al. Cyclin D1 overexpression in a model of human breast premalignancy: Preferential stimulation of anchorage-independent but not anchorage-dependent growth is associated with increased cdk2 activity. Breast Cancer Res Treat. 2000;59:27–39. doi: 10.1023/a:1006370603147. [DOI] [PubMed] [Google Scholar]
- 27.Sandler A, Ettinger DS. Gemcitabine: single-agent and combination therapy in non-small cell lung cancer. Oncologist. 1999;4:241–51. [PubMed] [Google Scholar]
- 28.Storniolo AM, Enas NH, Brown CA, Voi M, Rothenberg ML, Schilsky R. An investigational new drug treatment program for patients with gemcitabine: Results for over 3000 patients with pancreatic carcinoma. Cancer. 1999;85:1261–8. [PubMed] [Google Scholar]
- 29.Ahnstrom M, Nordenskjold B, Rutqvist LE, Skoog L, Stal O. Role of cyclin D1 in ErbB2-positive breast cancer and tamoxifen resistance. Breast Cancer Res Treat. 2005;91:145–51. doi: 10.1007/s10549-004-6457-4. [DOI] [PubMed] [Google Scholar]
- 30.Rudas M, Lehnert M, Huynh A, Jakesz R, Singer C, Lax S, et al. Cyclin D1 expression in breast cancer patients receiving adjuvant tamoxifen-based therapy. Clin Cancer Res. 2008;14:1767–74. doi: 10.1158/1078-0432.CCR-07-4122. [DOI] [PubMed] [Google Scholar]
- 31.Stendahl M, Kronblad A, Ryden L, Emdin S, Bengtsson NO, Landberg G. Cyclin D1 overexpression is a negative predictive factor for tamoxifen response in postmenopausal breast cancer patients. Br J Cancer. 2004;90:1942–8. doi: 10.1038/sj.bjc.6601831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Chen Y, Olopade OI. MYC and breast cancer. Genes Cancer. 2010;1:629–40. doi: 10.1177/1947601910378691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández-Vargas H, Rodríguez-Pinilla S, Julián-Tendero M, Sánchez-Rovira P, Cuevas C, Antón A, et al. Gene expression profiling of breast cancer cells in response to gemcitabine: NF-kB pathway activation as a potential mechanism of resistance. Breast Cancer Res Treat. 2007;102:157–72. doi: 10.1007/s10549-006-9322-9. [DOI] [PubMed] [Google Scholar]
- 34.Zelivianski S, Cooley A, Kall R, Jeruss JS. Cyclin-dependent kinase 4-mediated phosphorylation inhibits Smad3 activity in cyclin d-overexpressing breast cancer cells. Mol Cancer Res. 2010;8:1375–87. doi: 10.1158/1541-7786.MCR-09-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooley A, Zelivianski S, Jeruss JS. Impact of cyclin E overexpression on Smad3 activity in breast cancer cell lines. Cell Cycle. 2010;9:4900–7. doi: 10.4161/cc.9.24.14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata A, Nagaya T, Imai T, Funahashi H, Nakao A, Seo H. Inhibition of NF-kB activity decreases the VEGF mRNA expression in MDA-MB-231 breast cancer cells. Breast Cancer Res Treat. 2002;73:237–43. doi: 10.1023/a:1015872531675. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Eppenberger-Castori S, Eppenberger U, Benz CC. The NF-kB pathway and endocrine-resistant breast cancer. Endoc Relat Cancer. 2005;12:S37–46. doi: 10.1677/erc.1.00977. [DOI] [PubMed] [Google Scholar]
- 38.Monks NR, Pardee AB. Targeting the NF-kappa B pathway in estrogen receptor negative MDA-MB-231 breast cancer cells using small inhibitory RNAs. J Cell Biochem. 2006;98:221–33. doi: 10.1002/jcb.20789. [DOI] [PubMed] [Google Scholar]
- 39.Kwak YT, Li R, Becerra CR, Tripathy D, Frenkel EP, Verma UN. IkB kinase alpha regulates subcellular distribution and turnover of cyclin D1 by phosphorylation. J Biol Chem. 2005;280:33945–52. doi: 10.1074/jbc.M506206200. [DOI] [PubMed] [Google Scholar]
- 40.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You Z, Madrid LV, Saims D, Sedivy J, Wang CY. c-Myc sensitizes cells to tumor necrosis factor-mediated apoptosis by inhibiting nuclear factor kB transactivation. J Biol Chem. 2002;277:36671–7. doi: 10.1074/jbc.M203213200. [DOI] [PubMed] [Google Scholar]
- 42.O’Hagan RC, Ohh M, David G, de Alboran IM, Alt FW, Kaelin WG, Jr, et al. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–91. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh PY, Lu YS, Ou DL, Cheng AL. IkappaB kinases increase Myc protein stability and enhance progression of breast cancer cells. Mol Cancer. 2011;10:53. doi: 10.1186/1476-4598-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N, Fang H, Li Y, Xu W. Recent research in selective cyclin-dependent kinase 4 inhibitors for anti-cancer treatment. Curr Med Chem. 2009;16:4869–88. doi: 10.2174/092986709789909611. [DOI] [PubMed] [Google Scholar]
- 45.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 46.Sung B, Pandey MK, Aggarwal BB. Fisetin, an inhibitor of cyclin-dependent kinase 6, down-regulates nuclear factor-kB-regulated cell proliferation, antiapoptotic and metastatic gene products through the suppression of TAK-1 and receptor-interacting protein-regulated IkB alpha kinase activation. Mol Pharmacol. 2007;71:1703–14. doi: 10.1124/mol.107.034512. [DOI] [PubMed] [Google Scholar]
- 47.Thoms HC, Dunlop MG, Stark LA. p38-mediated inactivation of cyclin D1/cyclin-dependent kinase 4 stimulates nucleolar translocation of RelA and apoptosis in colorectal cancer cells. Cancer Res. 2007;67:1660–9. doi: 10.1158/0008-5472.CAN-06-1038. [DOI] [PubMed] [Google Scholar]


