Abstract
Marine algal toxins such as brevetoxins, okadaic acid, yessotoxin, and ciguatoxin are polyether compounds. The fate of polyether toxins in the aqueous phase, particularly bacterial biotransformation of the toxins, is poorly understood. An inexpensive and easily available polyether structural analog salinomycin was used for enrichment and isolation of potential polyether toxin degrading aquatic marine bacteria from Florida bay area, and from red tide endemic sites in the South Florida Gulf coast. Bacterial growth on salinomycin was observed in most of the enrichment cultures from both regions with colony forming units ranging from 0 to 6 × 107 per mL. The salinomycin biodegradation efficiency of bacterial isolates determined using LC-MS ranged from 22% to 94%. Selected bacterial isolates were grown in media with brevetoxin as the sole carbon source to screen for brevetoxin biodegradation capability using ELISA. Out of the two efficient salinomycin biodegrading isolates MB-2 and MB-4, maximum brevetoxin biodegradation efficiency of 45% was observed with MB-4, while MB-2 was unable to biodegrade brevetoxin. Based on 16S rRNA sequence similarity MB-4 was found have a match with Chromohalobacter sp.
Keywords: Polyether, salinomycin, brevetoxin, biodegradation, Chromohalobacter sp., red tide, Karenia brevis, dinoflagellate
Introduction
Marine shorelines are important public and ecological resources that serve as a home to a variety of wildlife and provide public recreation. Harmful algal blooms with discolored water, known as `red tide', occur worldwide and appear to be increasing in both frequency and duration.[1,2] Large-scale occurrences of red tide are a serious threat to marine animals and to the public health. A marine dinoflagellate, Karenia brevis, is associated with the Florida red tide and it produces a suite of highly potent neurotoxins known as the brevetoxins. Blooms of this organism are most often located in the Gulf of Mexico off the southwest coast and, occasionally, the Atlantic coast of Florida.
Brevetoxins are highly stable polyketide compounds and belong to a family of heat-stable, lipid-soluble, highly oxygenated, cyclic polyether molecules. The neurotoxic effects of brevetoxin are due to its agonistic activity on voltage sensitive sodium channels. K. brevis blooms have been implicated in massive fish kills, bird deaths, and marine mammal mortalities. Fish and seagrass can accumulate high concentrations of brevetoxin and serve as vector for toxin transfer to upper trophic levels.[3] If shellfish derived from the brevetoxin-contaminated sea is consumed by humans, a life threatening neurotoxic shellfish poisoning results. Even the aerosols from the red tide contain brevetoxins and can cause respiratory illness in the exposed person or animal.[4]
Brevetoxin has been shown to undergo photochemical degradation when exposed to natural sunlight.[5] The ether linkage is known to confer a high degree of resistance to biological degradation. On the other hand, the C-O-C molecular signature is common in various natural compounds such as syringic acid, vanillic acid and other lignin precursors, and in cellulose. Synthetic ether-containing compounds include agrochemicals (herbicides), polyethers (PEG, PPG, and PTMG), and detergents. Under aerobic conditions, enzymes with various ranges of substrate specificity (peroxidase, monoxygenases, dioxygenases, P450) can cleave the ether bond. Such reactions result in the incorporation of oxygen and subsequent production of a hemiacetal, which spontaneously dismutates in aqueous solution to the corresponding alcohol and aldehyde.[6,7] Three enzymes, PEG dehydrogenase, PEG-aldehyde dehydrogenase and PEG-carboxylate dehydrogenase (ether-cleaving) have been found to be involved in the metabolism of PEG by Flavobacterium sp.[8] In PEG-utilizing sphingomonads[9] membrane-bound PEG dehydrogenase (PEG-DH) was found to be highly active towards PEG 6,000 and 20,000.
Bacteria that are associated with dinoflagellates include strains of Pseudomonas (most common), Nocardia, Vibrio, Aeromonas, Flavobacterium and Moraxella.[10] Most studies have found that two bacterial phyla, Alphaproteobacteria and CFB (Cytophaga-Flavobacter-Bacteroides), are more commonly found in association with dinoflagellates.[11,12] Algal and bacterial interactions play an important role in the ecology of harmful algal bloom. Reports have demonstrated that marine bacteria are capable of inter-converting dinoflagellate paralytic shell-fish toxins (tetrahydropurines) and possess the capacity to carry out side chain modifications.[13] The ability to transform paralytic shellfish toxins was predominantly a trait of bacteria within the Roseobacter clade. Roseobacter clade isolates from coastal and oceanic environments are known to exhibit diverse modes of metabolism and can transform lignin.[14] Although no metabolites have been detected, uptake of brevetoxin PbTx-2 but not PbTx-3 by phytoplanktons has been reported.[15] Some of the native bacteria possess algicidal properties and can affect K. brevis populations.[16,17] However, the susceptibility of toxin molecules to marine bacterial enzymatic activity has not been explored.
The brevetoxins have been naturally released to the marine environment by K. brevis for many years. At the two study sites in the Florida Gulf Coast, the brevetoxin (PbTx-2 and PbTx-3) concentration in the seawater was found to be in the range of 1.5 μg L−1 during low K. brevis population to 23.7 μg L^#x2212;1 during high K. brevis population.[4] The heterotrophic bacterial biodegradation of an algal toxin, microcystin, has been reported in fresh water ecosystems.[18,19] There is valid ground for the assumption that, the frequent occurrence of brevetoxins in marine water from red tide events will act as a strong selective force on certain marine bacteria to adapt and utilize the carbon in the toxin molecule for growth.
Some of the marine algal toxins such as brevetoxins, ciguatoxin, okadaic acid, yessotoxin etc., have polyether structures. Significant amounts of toxin material will be needed for conducting repeated enrichment cultures for isolating polyether toxin degrading marine bacteria. However, polyether algal toxins are not easily available, very expensive, and thus limiting the amount of toxin that could be used in the enrichment media and the number of enrichment cultures that could be performed for the isolation of marine bacteria. This serious problem might be overcome by using a less-expensive structural analog for initial screening. After extensive literature search we chose the polyether antibiotic salinomycin as the closest structural analog, which has 83% structural similarity with polyether toxin okadaic acid. Salinomycin produced by Streptomyces albus, belong to the polyether ionophore group of antibiotics, which comprises more than one hundred different compounds. Several members of the group have found commercial application as anticoccidials in poultry farming and as growth promoters for cattle, pigs and chickens. Gram-negative group of bacteria are generally ionophore resistant but the gram-positive group are not.
The objectives of this study were to isolate potential polyether algal toxin degrading marine bacteria from red tide endemic and non-red tide areas of Florida using salinomycin enrichment cultures and to further evaluate the brevetoxin biodegradation capability of salinomycin utilizing marine bacterial isolates.
Materials and methods
Enrichment and isolation of bacteria
Whole-water samples were collected within the Florida Gulf coast area, and the Florida Bay. The sampling locations within the Florida Gulf coast area were chosen to be representative of red tide endemic sites with varying population of Karenia brevis, the dinoflagellate responsible for red tide. Water samples were collected within the Florida gulf coast area by the Karenia monitoring program at the Fish and Wildlife Research Institute in St. Petersburg Florida. The water samples from K. brevis free areas were obtained from Florida Bay area through SERC (Southeast Environmental Research Center, FIU) water quality management project. All the samples after collection were transported and stored at 4°C until use. The water samples were collected and used for starting enrichment cultures during red tide season (August–December).
The enrichment cultures were prepared by inoculating one mL of each marine water sample into 125 mL Erlenmeyer flasks containing 20 mL of nitrogen and phosphorus amended synthetic seawater media (SSW+).[20] The enrichment process was repeated using 0.1 mL of inoculum and an incubation period of 4 weeks each. At the initial stages of enrichment a 5 μg mL−1 concentration of salinomycin (Sigma-Aldrich) was maintained, after four enrichment transfers, the concentrations was increased to 20 μg mL−1. The samples that produced turbid culture in salinomycin-supplemented SSW+ after the enrichment process were selected for further study. Salinomycin and no-salinomycin controls were included in each of the enrichment. Flasks were shaken at 200 rpm on a reciprocal shaker in an incubator set at 27°C for 4 weeks. The bacterial population in the enrichment cultures was estimated by dilution plating on marine agar.
Quantitative salinomycin biodegradation study
Based on the results from colony forming unit counts, six marine bacterial isolates were selected and a quantitative salinomycin biodegradation study was initiated. The cultures of salinomycin degrading bacteria were grown in 250 mL Erlenmeyer flasks containing 50 mL of medium (SSW+ with glucose to increase cell yield) on a rotary shaker (200 rpm) at 27°C. Bacterial cells were harvested during exponential growth phase by centrifugation (10,000 × g, 10 min, 4°C), washed once with sterile SSW+ solution and resuspended in SSW+ medium and used immediately. Individual bacterial isolates were grown in 25 mL of SSW+ containing 20 μg mL−1 of salinomycin inoculated with 1 × 106 CFU mL−1 and incubated at 27°C for 4 weeks. A salinomycin amended control SSW+ medium with and without the bacterial cell inoculum were maintained for comparison. There were three replicate flasks for each bacterial isolate treatment including control flasks with and without salinomycin. After 4 weeks of incubation salinomycin was extracted from enrichment flasks via a liquid-liquid partition with chloroform. The chloroform was evaporated under nitrogen and the salinomycin was resuspended in 100% methanol. The salinomycin content was determined using a modified version of the LC-MS procedure described by Cha et al.[21]
The LC system was a Thermo Spectra system equipped with an autosampler and a quaternary pump. The detector employed was an LCQ Finnigan Mat used on the ESI mode. Separation was carried out using a Whatman Partisil ODS-3, 4.6 × 250 mm column with 10μm particle size. The operating conditions were: mobile phase used were 15% of 0.1% formic acid with 85% of methanol in isocratic conditions with a 0.5mL/minute flow rate; run time was 14 minutes and injection volume was 100 μL. Calibration plot was prepared from a 20 μg mL−1 standard salinomycin. For each of the isolates the salinomycin biodegradation efficiency percentage was calculated relative to the sterile salinomycin control. Statistical analysis of the data was done using the SPSS statistical software package version 13.0 (SPSS Inc. 233 S. Wacker Dr. Chicago, Illinois).
Dinoflagellate cultures and brevetoxin production
Brevetoxin production from K. brevis was carried out in the toxic algae culture facility, Department of Chemistry and Biochemistry at FIU. K. brevis was grown in 4L carboys at 19–21°C, in a fully defined artificial seawater media at 28–32 ppt, with illumination from GE Cool White lights on a 12 hr/ 12 hr light-dark cycle. Cultures were maintained in Wilson's NH-15 media, f/2 media, and L1 media.[20] Brevetoxins purified from laboratory cultures by a combination of chloroform extraction preparative TLC and finally preparative HPLC.[22] PbTx-2 and –3 are subjected to a final purification by HPLC (85% isocratic methanol) using a Microsorb-MV, C-18 column (5 M, 25 cm bed) and monitored by UV at 215 nm.
Biodegradation of brevetoxin
The marine bacterial isolates showing efficient salinomycin degrading ability were selected and a replicated brevetoxin biodegradation experiment was set up using selected four marine bacterial isolates from salinomycin biodegradation study and one bacterial isolate (WS+) from brevetoxin producing K. brevis culture. The WS+ was isolated through multiple cycles of enrichment using brevetoxin as the sole carbon source. The individual bacterial cultures were grown in 250 mL Erlenmeyer flasks containing 50 mL of medium (SSW+ with glucose to increase cell yield) on a rotary shaker (200 rpm) at 27°C.
Bacterial cells were harvested during exponential growth phase by centrifugation (10,000 × g, 10 min, 4°C), washed once with sterile SSW+ solution and resuspended in SSW+ medium and used immediately. Bacterial isolates were grown in 20 mL of SSW+ containing 500 ng mL−1 of brevetoxin (PbTx-3) as the sole carbon source and were inoculated with 1 × 106 CFU mL−1 and incubated at 27°C. A brevetoxin amended control SSW+ medium with and without the bacterial cell inoculum were maintained for comparison. There were 3 replicate flasks for each bacterial isolate treatment including control flasks with and without brevetoxin. After 8 weeks of incubation brevetoxin was extracted from enrichment flasks via a liquid-liquid partition with chloroform. The chloroform was evaporated under nitrogen and the brevetoxin was resuspended in 100% methanol. The brevetoxin content was determined using ELISA.[23] For each of the isolates the brevetoxin biodegradation efficiency percentage was calculated relative to the sterile brevetoxin control. Statistical analysis of the data was done using the SPSS statistical software package version 13.0 (SPSS Inc. 233 S. Wacker Dr. Chicago, Illinois).
MB-4 isolate 16S rRNA sequence analysis
The 16S rRNA gene was PCR amplified from genomic DNA isolated from pure MB-4 bacterial colonies. Primers used are universal 16S primers that correspond to positions 0005F and 0531R for a 500bp sequence, and 0005F and 1513R for the 1500bp sequence. Amplification products were purified from excess primers and dNTPs and checked for quality and quantity by running a portion of the products on an agarose gel. Cycle sequencing of the 16S rRNA amplification products was carried out using DNA polymerase and dye terminator chemistry. Excess dye-labeled terminators were then removed from the sequencing reactions. The samples were electrophoresed on ABI 3130 Genetic Analyzer. Sequence analysis was preformed using Sherlock® DNA microbial analysis software and database.
Results and discussion
Enrichment on salinomycin
The K. brevis population in the Florida Gulf Coast water samples ranged from 0 to 54,000 cells per liter as shown in Table 1. Initial observable bacterial growth (turbidity) in the salinomycin enrichment flasks took more than 10 days to develop. In enrichment studies, when salinomycin was used as a carbon source there was an increase in bacterial population in 4 out of 6 samples in each location. The highest bacterial population obtained in the salinomycin enrichment was 6 × 107 colony forming units per mL in enrichment flasks with Florida Bay isolate MB-2. Because of the endemic exposure to brevetoxin, polyether degrading bacteria were expected to be present in waters with K. brevis population. However, no such relationship was observed, the MB-2 isolate from Florida Bay showed increased population growth and isolates MB-9 and MB-10 both from Gulf Coast were unable to utilize salinomycin as a carbon source.
Table 1.
Bacterial population (cfu) in salinomycin enrichment cultures.
| Isolate | Site | CFU* mL−1 | K. brevis (cells L−1)a |
|---|---|---|---|
| MB-1 | Florida Bay | 2 (±0.11) × 105 | |
| MB-2 | Florida Bay | 6 (±0.88) × 107 | |
| MB-3 | Florida Gulf Coast | 6 (±0.55) × 106 | 23,000 |
| MB-4 | Florida Gulf Coast | 1 (±0.07) × 105 | 23,000 |
| MB-5 | Florida Bay | 1 (±0.06) × 105 | |
| MB-6 | Florida Gulf Coast | 1 (±0.09) × 106 | 23,000 |
| MB-7 | Florida Gulf Coast | 3 (±0.35) × 105 | 667 |
| MB-8 | Florida Bay | 1 (±0.08) × 105 | |
| MB-9 | Florida Gulf Coast | 0 | 23,000 |
| MB-10 | Florida Gulf Coast | 0 | 54,300 |
| MB-11 | Florida Bay | 0 | |
| MB-12 | Florida Bay | 0 |
CFU = Colony forming units, mean of three replications, values in parenthesis are SE.
data provided by Fish and Wildlife Research Institute, St. Petersburg, FL.
Salinomycin biodegradation
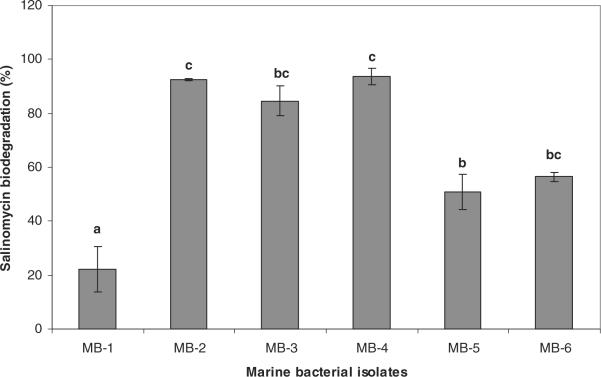
In the quantitative salinomycin biodegradation study (Fig. 1), significant (P < 0.05) variations among the marine bacterial isolates in terms of their salinomycin biodegradation efficiency (%) were observed. One isolate from Florida Bay (MB-2) and two isolates from Gulf Coast (MB-3 and MB-4) showed significantly (P < 0.05) higher salinomycin degradation efficiency than the rest of the 3 isolates. The MB-1 isolate from Florida Bay showed the least with 22% salinomycin biodegradation.
Fig. 1.
Salinomycin biodegradation efficiency (%) of marine bacterial isolates. Bars with different alphabets indicate significant difference (P < 0.05).
There is little information about the persistence and degradation of antibiotics in marine water ecosystem. In aquatic system such as in river water ionophore antibiotics were found to be only in trace levels.[21] In soil salinomycin has shown to be least persistent with the half-life in the range of 5 days.[24] Cell free enzymes from soil bacteria Pseudomonas stutzeri and Enterobacter agglomerans found to degrade salinomycin in vitro.[25] Microbial activity during composting of manure contaminated with salinomycin was shown to be effective in reducing salinomycin.[26] In the present study the lack of clear difference between two regions regarding the marine bacterial capability to degrade salinomycin indicates that possibly bacteria have been encountering and metabolizing diverse groups of naturally occurring organic compounds with ether bonds in the marine water.
Brevetoxin biodegradation
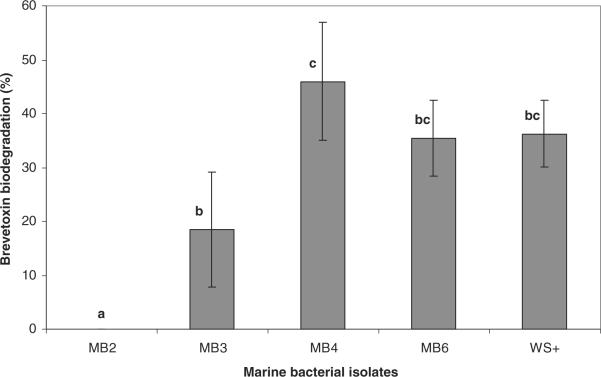
In the brevetoxin biodegradation study (Fig. 3), among the marine bacterial isolates tested MB-4 showed the maximum brevetoxin biodegradation efficiency of 45% based on ELISA, whereas MB-2 showed no reduction in brevetoxin content. Although isolates MB-2, MB-3 and MB-4 were highly capable of degrading salinomycin, there was a significant (P < 0.05) difference in their biodegradation activity in the presence of brevetoxin as the sole carbon source. The isolate WS+ that was isolated through enrichment on brevetoxin and isolate MB-6 showed close to 36% biodegradation of brevetoxin.
Fig. 3.

Partial 16S rRNA gene sequences of marine bacterial isolate MB-4.
The molecular conformation of brevetoxins is the major determinant of its toxicity. A 30Å long “cigar-shaped” molecular segment of brevetoxin is considered to be the bioactive site[27] of the toxin molecule. Even minor transformations in the brevetoxin structure due to enzymatic oxidation of terminal aldehyde, such as oxidation and esterification of ring D, have been found to detoxify the molecule.[28]
The potential biochemical interactions between marine bacteria and the brevetoxins in the ecological context merit further study. If brevetoxins that are released undergo biotransformation or biodetoxification by marine bacteria in the bloom, the toxicity of the derivatives and by-products needs to be studied to better understand the distribution and efficiency of toxin entry into the food web. Such information will help in efforts aimed at modeling and predicting brevetoxin trophic transfer in a given system.
Identification of isolate MB-4
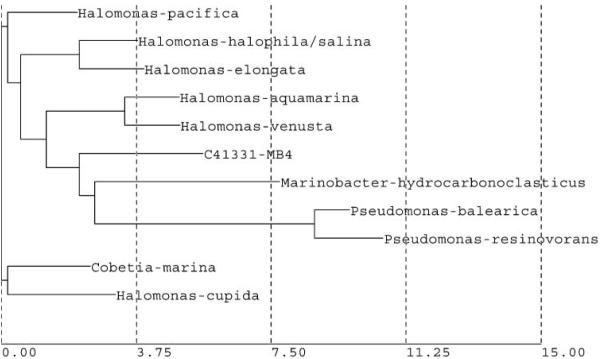
The partial 16S rRNA gene sequence of marine bacterial isolate MB-4 (Fig. 3) was used for database search and construction of phylogenetic tree using neighbor joining method (Fig. 4). Based on the results the isolate MB-4 belongs phylogenetically to a branch that also contains genus Halomonas, Cobetia, Marinobacter and Pseudomonas. The closest match was with Halomonas pacifica, but the partial 16S rRNA gene sequence match report showed a 92.42% similarity between MB-4 and Halomonas pacifica. Since the difference was more than 5% the match was considered not significant. However, based on the on-line homology BLAST search (GenBank) showed that the 16S rDNA sequence of MB-4 was found to be most similar to the sequence of Chromohalobacter sp. EF198248 with 99% match. The bacterial strains presenting 16S rRNA gene similarities lower than 99.5% and higher than 97% may belong to different species but full 16S rDNA sequence analysis and other tests are needed for definitive taxonomic identity.[29]
Fig. 4.
Phylogenetic tree of isolate MB-4 constructed using neighbor joining method.
The genus Chromohalobacter belongs to the family Halomonadaceae under the class Gamma Proteobacteria. The genus Chromohalobacter was established by Ventosa and others in 1989, with the reclassification of Chromobacterium marismortui as Chromohalobacter marismortui.[30] Chromohalobacter sp. are gram negative, aerobic, motile, non-spore forming rod shaped bacteria. Phylogenetic groups closely related to Chromohalobacter are Halomonas canadensis and Halomonas israelensis, which were included as members of the genus Chromohalobacter.[31] The Chromohalobacter species show a broad range of tolerance to salinity (0.5–25% NaCl) and temperature (5–42°C).[32]
Osmoprotection is achieved by the accumulation of compatible solutes. Chromohalobacter sp. bacterial habitat includes Dead Sea, oceans, saline subterranean lakes, marine salterns and salt preserved foods. Several strains of chromohalobacter sp. have been shown to be resistant to a range of antibiotics.[32] A member of Halomonadaceae, Alcanivorax borkumensis has been shown to biodegrade marine alkane hydrocarbons.[33] The Chromohalobacter species have been reported to be resistant to heavy metals and degrade aromatic hydrocarbons[34], organophosphorus pesticides[35], and aminomethane sulfonate.[36]
Conclusions
Marine bacteria from Florida red tide endemic region as well as from non-red tide region were able to utilize the polyether compound salinomycin as a substrate for growth. Most of the salinomycin enrichment culture isolates obtained from water samples showed bacterial growth and the maximum population obtained was 6 ×107 colony forming units per mL. The salinomycin and brevetoxin biodegradation efficiency of bacterial isolates varied significantly (P < 0.05). Among the marine bacterial isolates tested, MB-4 (Chromohalobacter sp.) showed maximum efficiency of biodegradation of salinomycin (94%) and of brevetoxin (45%). Although no direct relationship was observed between salinomycin and brevetoxin biodegradation capability, the use of structural analog salinomycin was found to be effective in isolation of potential polyether toxin degrading marine bacteria. Further research is needed to characterize the enzymes, metabolites and other factors that are involved in the bacterial brevetoxin biodegradation process.
Fig. 2.
Brevetoxin biodegradation efficiency (%) of marine bacterial isolates. Bars with different alphabets indicate significant difference (P < 0.05).
Acknowledgments
This work was supported by funding from NIH-FIU-ARCH-PILOT-S11ES011181. We thank Earnest Truby (Fish and wildlife research institute, St. Petersburg, FL) and Pete Lorenzo (Southeast Environmental Research Center, FIU) for providing water samples. We also thank Dr. Yang Cai and Damaris Hernandez (Department of Chemistry and Biochemistry, FIU) for help with LC-MS analysis.
References
- [1].Chretiennot-Dinet MJ. Global increase of algal blooms, toxic events, casual species introductions and biodiversity. Oceanis. 2001;24:223. [Google Scholar]
- [2].Knap A, Dewailly E, Furgal C, Galvin J, Baden D, Bowen B, Depledge M, Duguay L, Fleming LE, Ford T, Moser M, Owen R, Suk WA, Unluata U. Indicators of ocean health and human health: Developing a research and monitoring framework. Environ. Health Perspect. 2002;110:839. doi: 10.1289/ehp.02110839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Flewelling LJ, Naar JP, Abbot JP, Baden DG, Barros NB, Bossart GD, Bottein MD, Hammond DG, Haubold EM, Heil CA, Henry MS, Jacocks HM, Leighfield TA, Pierce RH, Pitchford TD, Rommel SA, Scott PS, Steidinger KA, Truby EA, Van Dolah FM, Landsberg JH. Red tides and marine mammal mortalities. Nature. 2005;435:755–756. doi: 10.1038/nature435755a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Backer LC, Fleming LE, Rowan A, Cheng YS, Benson J, Pierce RH, Zaias J, Bean J, Bossart GD, Johnson D, Quimbo R, Baden DG. Recreational exposure to aerosolized brevetoxins during Florida red tide events. Harmful Algae. 2003;2:19–28. [Google Scholar]
- [5].Hardman RC, Cooper WJ, Bourdelais AJ, Gardinali P, Baden DG. Brevetoxin degradation and by-product formation via natural sunlight. The Xth International Conference on Harmful Algae; St. Petersburg Beach, Florida, USA. October 21–25, 2002; Steidinger, K.A.; Landsberg, J.H.; Tomas, C.R.; Vargo, G.A. Eds. Florida Fish and Wildlife Conservation Commission, Florida Institute of Oceanography, and Intergovernmental Oceanographic Commission of UNESCO, St. Petersburg, Florida, USA; 2004, 153-154. [PMC free article] [PubMed] [Google Scholar]
- [6].White GF, Russell NJ, Tidswell EC. Bacterial scission of ether bonds. Microbiol. Rev. 1996;60:216–232. doi: 10.1128/mr.60.1.216-232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kawai F. Microbial degradation of polyethers. Appl. Microbiol. Biotechnol. 2002;58:30–38. doi: 10.1007/s00253-001-0850-2. [DOI] [PubMed] [Google Scholar]
- [8].Kawai F, Yamanaka H. Biodegradation of polyethylene glycol by symbiotic mixed culture (obligate mutualism) Arch. Microbiol. 1986;146:125–129. doi: 10.1007/BF00402338. [DOI] [PubMed] [Google Scholar]
- [9].Sugimoto M, Tanabe M, Hataya M, Enokibara S, Duine JA, Kawai F. The first step in polyethylene glycol degradation by Sphingomonads proceeds via a flavoprotein alcohol dehydrogenase containing flavin adenine dinucleotide. J. Bacteriol. 2001;183:6694–6698. doi: 10.1128/JB.183.22.6694-6698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tosteson TR, Ballantine DL, Tosteson CG, Hensley V, Bardales A. Associated bacterial flora, culture growth and toxicity of the benthic dinoflagellates, Ostreopsis lenticularis and Gambierdiscus toxicus. Appl. Environ. Microbiol. 1989;55:137–141. doi: 10.1128/aem.55.1.137-141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hold GL, Smith EA, Rappe MS, Maas EW. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 2001;37:161–173. [Google Scholar]
- [12].Green DE, Llewellyn LE, Negri AP, Blackburn SI, Bolch CJS. Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol. Ecol. 2004;47:345–357. doi: 10.1016/S0168-6496(03)00298-8. [DOI] [PubMed] [Google Scholar]
- [13].Smith EA, Grant F, Ferguson CMJ, Gallacher S. Bio-transformations of paralytic shellfish toxins by bacteria isolated from bivalve mollusks. Appl. Environ. Microbiol. 2001;67:2345–2353. doi: 10.1128/AEM.67.5.2345-2353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gonzalez JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedros-Alio C, Moran MA. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tracey LM, Prince EK, Naar J, Kubanek J. Loss of waterborne brevetoxins from exposure to phytoplankton competitors. Harmful Algae. 2008;7:762–771. [Google Scholar]
- [16].Mayali X, Doucette GJ. Microbial community interactions and population dynamics of an algicidal bacterium active against Karenia brevis (Dinophyceae) Harmful Algae. 2002;1:277–293. [Google Scholar]
- [17].Roth PB, Twiner MJ, Wang Z, Bottein Dechraoui MY, Doucette GJ. Fate and distribution of brevetoxin (PbTx) following lysis of Karenia brevis by algicidal bacteria, including analysis of open A-ring derivatives. Toxicon. 2007;50:1175–1191. doi: 10.1016/j.toxicon.2007.08.003. [DOI] [PubMed] [Google Scholar]
- [18].Rapala J, Lahti L, Sivonen K, Niemela SI. Biodegradability and adsorption on lake sediments of cyanobacterial hepatotoxins and anatoxin-A. Lett. Appl. Microbiol. 1994;19:423–428. doi: 10.1111/j.1472-765x.1994.tb00972.x. [DOI] [PubMed] [Google Scholar]
- [19].Christoffersen K, Lyck S, Winding A. Microbial activity and bacterial community structure during degradation of microcystins. Aquat. Microb. Ecol. 2002;27:125–136. [Google Scholar]
- [20].Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals. Plenum Publishing Corp.; New York: 1975. pp. 29–60. [Google Scholar]
- [21].Cha JM, Yang S, Carlson KH. Rapid analysis of trace levels of antibiotic polyether ionophores in surface water by solid-phase extraction and liquid chromatography with ion trap tandem mass spectrometric detection. J. Chromatogr. A. 2005;1065:187–198. doi: 10.1016/j.chroma.2004.12.091. [DOI] [PubMed] [Google Scholar]
- [22].Poli MA, Mende TJ, Baden DG. Brevetoxins, unique activators of voltage-sensitive sodium channels bind to specific sites in rat brain synaptosomes. Mol. Pharmacol. 1986;30:129–135. [PubMed] [Google Scholar]
- [23].Naar J, Bourdelais A, Tomas C, Kubanek J, Whitney P, Fleweling L, Steindinger KA, Lancaster J, Baden DG. Competitive ELISA to detect brevetoxins from Karenia brevis (Formerly Gymnodinium breve) in seawater, shellfish, and mammalian body fluid. Environ. Health Perspect. 2002;110:179–185. doi: 10.1289/ehp.02110179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schlüsenera MP, Bester K. Persistence of antibiotics such as macrolides, tiamulin and salinomycin in soil. Environ. Pollut. 2006;143:565–571. doi: 10.1016/j.envpol.2005.10.049. [DOI] [PubMed] [Google Scholar]
- [25].Vertesy L, Heil K, Fehlhaber HW, Ziegler W. Microbial decomposition of salinomycin. J. Antibiot. 1987;40:388–390. doi: 10.7164/antibiotics.40.388. [DOI] [PubMed] [Google Scholar]
- [26].Ramaswamy J, Prasher SO, Patel RM, Hussain SA, Barrington SF. The effect of composting on the degradation of a veterinary pharmaceutical. Bioresour. Technol. 2010;101:2294–2299. doi: 10.1016/j.biortech.2009.10.089. [DOI] [PubMed] [Google Scholar]
- [27].Rein KS, Baden DG, Gawley RF. Conformational analysis of the sodium channel modulator brevetoxin A, comparison with brevetoxin B conformations, and a hypothesis about the common pharmacophore of the site 5′ toxins. J. Org. Chem. 1994;59:2101–2106. [Google Scholar]
- [28].Morohashi A, Satake M, Murata K, Naoki H, Kaspar HF, Yasumoto T. Brevetoxin B3, a New Brevetoxin Analog Isolated from the Greenshell Mussel Perna canaliculus Involved in Neurotoxic Shellfish Poisoning in New Zealand. Tetrahedron Lett. 1995;36:8995–8998. [Google Scholar]
- [29].Tindall BJ, Rossello`-Mo' ra R, Busse HJ, Ludwig W, Kampfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- [30].Ventosa A, Gutierrez MC, Garcia MT, Ruiz-Berraquero F. Classification of Chromobacterium marismortui in a new genus, Chromohalobacter gen. nov., as Chromohalobacter marismortui comb. nov., nom. rev. Int. J. Syst. Bacteriol. 1989;39:382–386. [Google Scholar]
- [31].Arahal DR, Garćıa MT, Ludwig W, Schleifer KH, Ventosa A. Transfer of Halomonas canadensis and Halomonas israelensis to the genus Chromohalobacter as Chromohalobacter canadensis comb. nov. and Chromohalobacter israelensis comb. nov. Int. J. Syst. Evol. Microbiol. 2001;51:1443–1448. doi: 10.1099/00207713-51-4-1443. [DOI] [PubMed] [Google Scholar]
- [32].Beutling DM, Peconek J, Stan-Lotter H. Chromohalobacter beijerinckii: A psychrophilic, extremely halotolerant and enzymatically active microbe from salted food with the capacity for biogenic amine production. Eur. Food Res. Technol. 2009;229:725–730. [Google Scholar]
- [33].Van Beilen JB, Marín MM, Smits TH, Röthlisberger M, Franchini AG, Witholt B, Rojo F. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 2004;6:264–273. doi: 10.1111/j.1462-2920.2004.00567.x. [DOI] [PubMed] [Google Scholar]
- [34]. [(accessed July 2010)];Chromohalobacter salexigens DSM 3043. http;//genome.jgi-psf.org/chrsa/chrsa.home.html.
- [35].Sabdono A, Radjasa OK. Phylogenetic diversity of organophosphorus pesticide-degrading coral bacteria from mid-west coast of Indonesia. Biotechnology. 2008;7:694–701. [Google Scholar]
- [36].Ternan NG, McMullan G. Utilization of aminomethane sulfonate by Chromohalobacter marismortui VH1. FEMS Microbiol. Lett. 2002;207:49–53. doi: 10.1111/j.1574-6968.2002.tb11027.x. [DOI] [PubMed] [Google Scholar]