Abstract
Five mutations in the S-cone-opsin gene (OPN1SW) that give rise to different single amino-acid substitutions (L56P, G79R, S214P, P264S, R283Q) are known to be associated with tritan color-vision deficiency. Here we report a sixth OPN1SW mutation (T190I) and the associated color vision phenotype. S-opsin genotyping and clinical evaluation of color vision were performed on affected and unaffected family members and normal controls. Chromatic contrast was tested at different levels of retinal illuminance. Affected family members were heterozygous for a nucleotide change that substituted the amino acid isoleucine (I) in place of threonine (T) that is normally present at position 190 of the S-opsin. The mutation is in extracellular loop II (EII). The association between making tritan errors and having the T190I mutant S opsin was strong (p > 0.0001: Fisher's exact test). The performance of subjects with the T190I mutation was significantly different from that of normal trichromats along the tritan vector under all conditions tested (Mann-Whitney U: p < 0.05), but not along the protan or deutan vectors. Individuals with the T190I S-opsin mutation behaved as mild tritans at 12.3–92.3 Td, but as tritanopes at 1.2–9.2 Td, for both light-adapted and dark-adapted conditions. The results are consistent with the mutant opsin causing abnormal S-cone function.
Keywords: tritan color-vision deficiency, rod-cone interaction, S-opsin mutation
Introduction
Inherited tritan color-vision deficiency is caused by mutations in the S-cone-opsin gene, officially designated OPN1SW, located on chromosome 7 (Nathans, Thomas & Hogness, 1986). Five mutations in the S-cone-opsin gene that give rise to different single amino-acid substitutions are known to be associated with tritan color-vision deficiency. These are: replacement of leucine by proline at codon 56 [L56P (Gunther, Neitz & Neitz, 2006)]; replacement of glycine by arginine at codon 79 [G79R (Weitz, Miyake, Shinzato, Montag, Zrenner, Went & Nathans, 1992a)]; replacement of serine by proline at codon 214 [S214P (Weitz et al., 1992a)]; replacement of proline by serine at codon 264 [P264S (Weitz, Went & Nathans, 1992b)]; replacement of arginine by glutamine at codon 283 [R283Q (Baraas, Carroll, Gunther, Chung, Williams, Foster & Neitz, 2007)]. The deficiency affects males and females equally and shows an autosomal dominant inheritance pattern. There is variability in phenotype among individuals with the same genotype, as well as between individuals with different genotypes. There are, for example, reports of individuals who behave as congenital tritanopes (Miyake, Yagasaki & Ichikawa, 1985, Weitz et al., 1992a). On the other hand there are reports of larger error scores on color-vision tests in older versus younger tritan subjects (Baraas et al., 2007, Went & Pronk, 1985). It has also been demonstrated that one of the mutations, R283Q, is progressive in nature, causing an S-cone dystrophy in the eldest person with the mutation (Baraas et al., 2007).
Here, we present the results from color-vision testing and genetic analysis of a family with a hitherto unknown mutation that gives rise to a single amino-acid substitutions in the S-opsin gene [replacement of threonine by isoleucine at codon 190 (T190I)]. The mutation is in extracellular loop II (EII), a region of the S opsin that corresponds to the intradiscal domain of rhodopsin. Measurements of color-discrimination threshold at different levels of luminance reveal that subjects heterozygous for the T190I OPN1SW mutation behaved as mild tritans under light levels in the range of 12.3 to 92.3 Trolands (Td), but they behaved as if their S-cones did not function at all when light levels were in the range of 1.2 to 9.2 Td.
Subjects and Methods
Subjects
Thirteen members of the family with the proband for the T190I mutation, aged 21–85 yrs, and 64 normal trichromatic females, aged 18–38 yrs, participated in all or parts of this study.
The experiments were conducted in accordance with principles embodied in the Declaration of Helsinki (Code of Ethics of the World Medical Association) and was approved by the Regional Committee for Medical and Health Research Ethics (South-Eastern Norway), and by IRBs at the Medical College of Wisconsin and the University of Washington. All subjects, except the first author, were unaware of the purpose of experiment 2.
All subjects had normal ophthalmic examinations and normal or corrected-to-normal visual acuity (logMAR 0.0 or better). The three eldest in the proband’s family (K1603, K1604 and K1613) were tested in their homes and ophthalmic examinations were not carried out.
Experiment 1a: Genetic analyses
Twelve family members and fifteen unrelated controls gave whole blood for genetic analysis. Whole blood was obtained from subjects and genomic DNA was extracted using the PureGene kit (Gentra Systems, Minneapolis). The five exons that comprise the amino acid coding region of the S-opsin gene were amplified in the polymerase chain reaction (PCR) and directly sequenced. Exons 1, 3 and 4 were amplified and sequenced using primers and conditions described previously (Gunther et al., 2006). Exons 2 and 5 were amplified and sequenced using the same thermal cycling parameters as for the other three exons. The primers for exon 2 were 5' GCCCATTATTCTCACATTTCACC and 5'CACCACTGCCCTGCACTCT, and for exon 5 were 5'TCTGCCAAGGTTATCTCCAATTG and 5' AAAATTTAATTCTAGCTGTTGCAAAC. Sequencing reactions were analyzed on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, California). Fifteen normal trichromatic controls that participated in experiment 1a and 1b of the study who did not make tritan errors served as a control population on the genetic analysis. Exon 3 of their OPN1SW genes were sequenced to specifically look for the T190I mutation, in addition, exons 1, 2 and 4 were also sequenced.
The 1000 Genome Project database (Consortium, 2010) and the MutationTaster (Schwarz, Rodelsperger, Schuelke & Seelow, 2010) was used to test whether the mutation is predicted to be a common human polymorphism or disease causing. The human OPN1SW was aligned with the paralogs OPN1LW, OPN1MW, RHO, OPN3, OPN4 and the following orthologs: human, chimp, rhesus, cat, mouse, xenopus, and zebrafish. Multiple sequence alignment was obtained using the Ensembl database (http://www.ensembl.org/index.html).
Subjects K1540 and K1544 were obligate carriers of a protan color vision defect as their father has a confirmed protan color-vision deficiency. Their carrier status was confirmed genetically using a previously described method that takes advantage of the fact that normal color vision in males in typified by X chromosome opsin gene arrays that have an OPN1LW (L gene) in the first position whereas protan color vision defects are characterized by arrays with an M gene in the first position. Female carriers of a protan defect can be identified genetically on the basis of having an L gene in the first position on one X-chromosome and an OPN1MW (M gene) in the first position on the other X (Kainz, Neitz & Neitz, 1998). PCR was used to specifically amplify the first gene in the X-chromosome opsin gene arrays on the two X chromosomes of subjects K1540 and K1544, and the PCR product was used to sequence exon 5.
Experiment 1b: Color-vision testing (E1)
Nine subjects from the family with the proband for the T190I mutation accepted to be tested with the Farnsworth-Munsell 100-Hue (FM100-Hue) test and the HRR pseudo-isochromatic plates [4th edition, 2002 (Bailey, Neitz, Tait & Neitz, 2004)]; both tests were performed under standard photopic conditions (1000 lux) using a lamp with daylight spectra. Six of these were tested with the Cambridge Colour Test (CCT) under standard conditions (Regan, Reffin & Mollon, 1994) and four of these were also tested with Rayleigh and Moreland anomaloscopy and luminance matching (HMC Oculus Anomaloscope MR, Typ 47700, Oculus Optikgeräte GmbH, Germany).
Sixty-four female normal trichromats, which includes the fifteen that gave whole blood for genetic analysis, who made no errors on the Neitz Color Vision Test (Neitz & Neitz, 2001), the HRR pseudo-isochromatic plates and the Ishihara pseudo-isochromatic plates, and with no known family history of color-vision deficiencies, were included to define normal values for color-discrimination thresholds measured with the CCT under standard conditions (Regan et al., 1994) and Moreland anomaloscopy. CCT standard condition was defined as measured binocularly with natural pupil size in an otherwise darkened room. Subjects adapted to the darkness in the room for 1–2 minutes prior to testing. See Experiment 2 for further details about the Cambridge Colour Test.
The dominant wavelengths of the blue and green colors of the upper test field of the Moreland match on the anomaloscope were measured to be 436 nm and 492 nm respectively with the PR650 Spectra Colorimeter (Photo Research Inc., Chatsworth, MA). These values were used to determine the appropriate Kraats & van Norren age function (van de Kraats & van Norren, 2007) to fit the data collected for the 64 normal trichromatic females (Fig. 3). The expected normal mean for each age was calculated from this function (Table 3). The Moreland match was determined with the procedure suggested by Linksz (Linksz, 1964).
Figure 3.
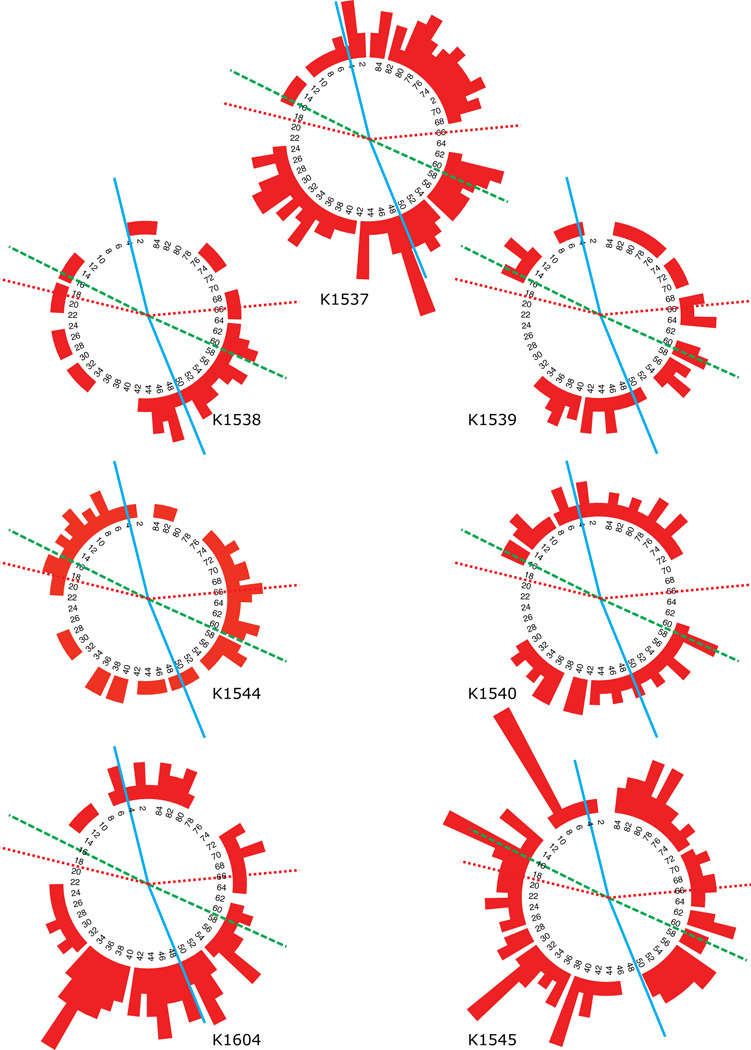
Results of the Farnsworth-Munsell 100-Hue test for subject (a) K1537 (proband’s mother), and the six subjects from generation III (b) K1538 (proband), (c) K1539 (proband’s sister), (d) K1544 (affected cousin), (e) K1540 (affected cousin), (f) K1604 (affected cousin) and (g) K1545 (unaffected cousin). Numbers in inner circle is cap numbers and the lines show color confusion axes centered on the middle of the cap number range of the protan (dotted), deutan (dashed) and tritan (solid) confusion axes. Deviations from the inner circle are the pattern of sorting errors.
Table 3.
Moreland anomaloscopy match-midpoints [MP (with mean MP values for each age)], matching-ranges (MR) and binocular tritan color-discrimination thresholds (standard condition: TCD) for (a) five individual tritans and (b) ten individual normal trichromats. K1545 is unaffected member of the family with the mutation. Subjects K1545 and K1604 declined to do Moreland anomaloscopy. The entries in bold are significantly different from the mean values based on 64 female normal trichromats (Z-score analysis).
| (a) | |||||
|---|---|---|---|---|---|
| Pedigree generation |
Subject ID | Age [yr] | MP (mean for age) |
MR | TCD |
| III | K1540 | 24 | 51 (52.8) | 22 | 280 |
| III | K1604 | 34 | not done | 167 | |
| III | K1539 | 34 | 48 (47.2) | 20.8 | 79 |
| III | K1538 | 36 | 44.9 (45.9) | 9.8 | 217 |
| II | K1537 | 54 | 46.9 (33.4) | 19.2 | 487 |
| (b) | |||||
|---|---|---|---|---|---|
| Pedigree generation |
Subject ID | Age [yr] | MP (mean for age) |
MR | TCD |
| E068(i) | 19 | 56.5 (55.1) | 3.9 | 100 | |
| K1563 | 22 | 51.5 (53.8) | 4.1 | 91 | |
| K1551 | 24 | 53.4 (52.8) | 7.9 | 111 | |
| E039(i) | 26 | 55.7 (51.8) | 2.4 | 101 | |
| III | K1545 | 30 | not done | 73 | |
| K1573 | 33 | 49.6 (47.8) | 10.6 | 74 | |
| K1561 | 35 | 49 (46.6) | 15.7 | 125 | |
| K1565 | 38 | 50 (44.6) | 9 | 108 | |
| K1555 | 39 | 48.3 (43.9) | 3.1 | 75 | |
| K1571 | 39 | 46.7 (43.9) | 6.3 | 55 | |
genetics not performed
Experiment 2: Color-discrimination thresholds (E2)
In the second experiment the subjects’ color-discrimination thresholds were measured with the CCT (Regan et al., 1994), but now monocularly with a pupil aperture and at two different levels of retinal illuminance in an otherwise darkened room after light-adaptation and after dark-adaptation. Fourteen subjects, aged 20–40 yrs, agreed to participate: five females from generation III of the proband’s family (four affected and one unaffected) and nine unrelated normal female trichromatic controls. The unaffected member (K1545) of the proband’s family was included with the group of normal trichromatic female controls. The nine unrelated normal trichromatic female controls were identified in the first experiments; that is they behaved as normal trichromats on all standard color-vision tests and did not have a mutation in the S-opsin.
The stimulus employed in the CCT is an array of spatially discrete round patches of varying size and luminance, and the luminance of each patch varies from trial to trial. In the following experiment it was set to vary between 2.0 and 15.0 cd m-2. The patches that make up the background have a single chromaticity. The patches that make up the target have a chromaticity that is varied systematically along three different axes in the CIE 1976 (u′, v′) chromaticity diagram. The average luminance of the target and the background is the same creating luminance noise so that the stimulus effectively is pseudo-isochromatic. The target has the form of a Landolt-C of outer size 4.3-deg with a gap that subtends 1-deg visual angle 3.1 m from the eye. Test stimuli were generated via a VSG ViSaGe graphics system (Cambridge Research Systems Ltd., Rochester, UK) and presented on a 22-inch CRT monitor (LaCie Electron 22blueIV, LaCie Group, France). The frame rate of the monitor was 100 Hz. The luminance and chromaticity of the monitor was checked with the OptiCal luminance meter (Cambridge Research System Ltd., Rochester, UK) and PR650 Spectra Colorimeter (Photo Research Inc., Chatsworth, MA). Errors in the displayed CIE 1931 (x, y, Y) coordinates of a few test patches of different chromaticities were < 0.005 in (x, y) and < 10 % in Y. A head-and-chin rest was used to maintain head position and the viewing distance.
Color-discrimination thresholds were measured in three directions along the protan, deutan and tritan confusion axes. The CCT standard color-vector length is 0.1100 units in the CIE 1976 (u′, v′) diagram. In this experiment the vector length was extended to 0.1650 units to allow for greater sensitivity. A staircase procedure is implemented in the CCT, and all three directions were tested in one experiment with a separate staircase running for each direction (trivector test). It would take the subject less than 3 min to complete a trivector test. Any one staircase is terminated after 11 reversals, and the mean of the last six reversals is taken as the threshold (For further details please see Ref. Regan et al., 1994). Each interval lasted for 3 sec, and a distinct auditory tone could be heard after each interval. Thresholds were measured three times at each of four conditions: light-adapted and dark-adapted under two different retinal illuminance ranges (12.3–92.3 Td and 1.2–9.2 Td), but always in the same sequence.
The procedure was as follows: Condition 1: subjects were first bleached for 1 min with a tungsten-halogen lamp covered by a large diffuser (28 000 Td) followed by a 4-min waiting period before testing on the cone plateau. Threshold measurements were made with the preferred eye wearing optimal correction in a well-fitted trial frame and a pupil aperture of 2.8 mm. The other eye was occluded. Condition 2: they were bleached again followed by a 4-min waiting period and tested, but now with a 1.0 ND filter added in front of the pupil aperture. Condition 3: they were dark adapted for 30 min, and tested again still with the 1.0 ND filter. Condition 4: dark adaptation was topped-up for 5 min and they were tested, but now without the 1.0 ND filter. Pupil size was measured during bleaching, and all subjects had pupil diameters larger than 3 mm. Threshold for the initial binocular trivector tests with natural pupils are included in the results for comparison (retinal illuminance of 25.1–188.4 Td assuming an average pupil diameter of 4 mm).
The orientation of the gap in the Landolt-C varies randomly from trial to trial (up, down, left or right). The subject’s task was to maintain steady fixation and to indicate the position of the gap. Subjects gave their responses by pushing the correct button on a response box.
Results
Experiment 1a: Genetic analyses
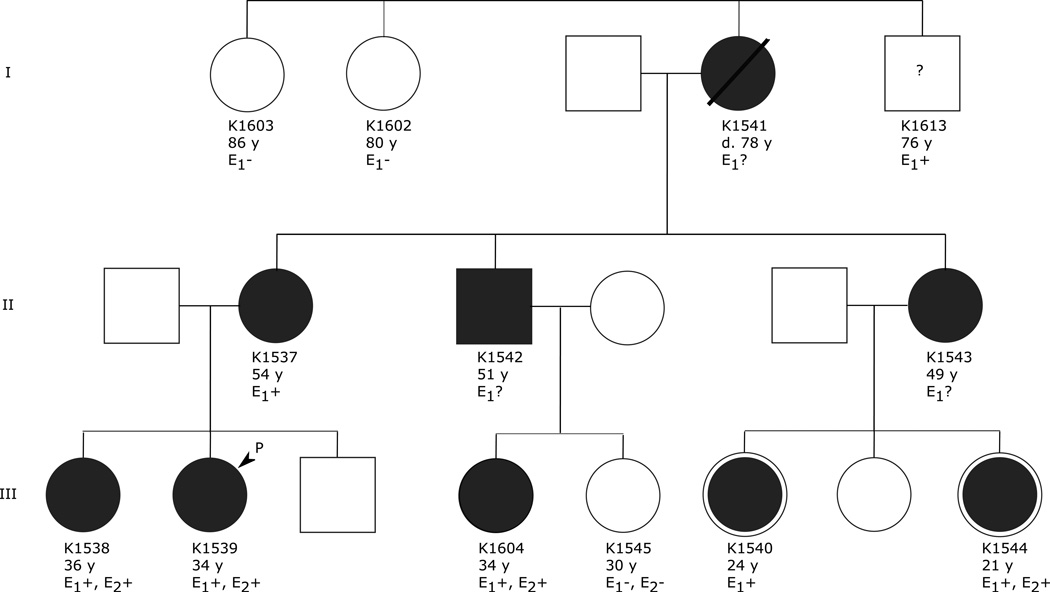
Figure 1 shows the pedigree of the family. Tritan subjects K1540 and K1544 are obligate carriers of a protan color-vision deficiency. Genetic confirmation of this was obtained by sequencing exon 5 of the first gene in the X-chromosome opsin gene arrays for each subject. Female carriers of protan defects have a mixture of OPN1LW and OPN1MW opsin gene exon 5 in this assay (Kainz et al., 1998), as did both subjects, thereby confirming that they are indeed protan carriers. With regard to the OPN1SW gene, nine (K1604, K1541, K1537, K1543, K1542, K1538, K1539, K1540, K1544) of twelve family members were heterozygous for a nucleotide change that substituted the nonpolar, neutral amino acid isoleucine (I) in place of the polar amino acid threonine (T) that is normally present at amino acid position 190 of the S-opsin (Fig. 2). Three of the family members (K1602, K1603, and K1545) and all 15 control subjects were homozygous for threonine. One of the normal trichromatic subjects (K1551) was heterozygous for the intron 3 G-deletion, which is thought to be unrelated to a tritan deficiency (Weitz et al., 1992a).
Figure 1.
Pedigree of the T190I family. Roman numerals (I, II, III) indicate the generation. Numbers refer to subject ID of family members that accepted the invitation to participate in the study. The letter E indicates that the subject has been evaluated with the Farnsworth-Munsell 100-Hue test and the Hardy-Rand-Rittler pseudo-isochromatic plates (E1); the Cambridge Colour Test to measure color-discrimination thresholds along the protan, deutan and tritan confusion axes for light-adapted and dark-adapted conditions (E2); positive results (+), negative results (−) or the results are unavailable because the subject declined to participate in this part of the study (?). Diagonal line indicates deceased individual with age at death (d.) below the symbol. Affected members; black filled symbols, unaffected; empty symbols. The arrow indicates proband III: 1539. Individuals III: 1540 and III: 1544 are both obligate carriers of protan color-vision deficiency.
Figure 2.
Model of the S-opsin. Circles represent amino acids. The light grey circle indicates the location of the T190I substitution in extracellular loop II (E-II) found in the family in this study. The other five reported mutations causing tritan color-vision deficiency are indicated by black filled circles: L56P, G79R, S214P, P264S, R283Q. The figure is modified from (Stenkamp, Filipek, Driessen, Teller & Palczewski, 2002) with permission.
All sequences in the 1000 Genome Project Database show that the T190I mutation is not a common polymorphism as there was no variation at codon 190. The MutationTaster (Schwarz et al., 2010) predicts that the mutation is disease causing with an approximate score of 2.43 and that it perturbs a functional domain of the protein. In addition, it is evident from Table 1 that the orthologous sequences near T190 are identical across all species (human, chimp, rhesus, cat, mouse, xenopus, zebrafish).
Table 1.
OPN1SW and orthologs compared near T190. T190 is the boxed residue. Dots indicate where orthologous sequences are identical to human. The paralogs are OPN1LW, OPN1MW, RHO, OPN3, OPN4. The corresponding positions in OPN1LW, OPN1MW, RHO (all human sequences) are conserved as hydroxyl bearing amino acids (Serine for RHO, OPN1LW, and OPN1MW). The multiple sequence alignment was obtained using the Ensembl database (http://www.ensembl.org/index.html).
| human | P | D | W | Y | T | V | G | T | K | Y | R | S | E | S | Y | T |
| chimp | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| rhesus | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| cat | • | • | • | • | • | • | • | • | • | • | • | • | • | Y | • | • |
| mouse | • | • | • | • | • | • | • | • | • | • | • | • | • | Y | • | • |
| xenopus | • | • | • | • | • | • | • | • | • | • | • | • | • | Y | • | • |
| zebrafish | • | • | • | • | • | K | S | E | E | • | N | • | • | • | • | • |
Experiment 1b: Color-vision testing (E1)
Table 2 shows the error scores of the FM100-Hue test and plate 6 of the HRR pseudo-isochromatic plates for affected and unaffected members of the proband’s family. FM100-Hue error scores increase with increasing age and it is only subject K1537, the proband’s mother, who’s performance results in a significant tritan axis on the Farnsworth-Munsell 100-Hue test (Fig. 3) and a selectivity index larger than 2.0 (Vingrys & King-Smith, 1988). She also behaves as if she has a mild to moderate tritan deficiency on Moreland anomaloscopy and the CCT (standard condition) (Table 3). Plate 6 is the second tritan screening plate of the HRR test. Five out of six tritan subjects with a confirmed mutation made errors on one of the symbols on plate 6 on the HRR test. They could identify symbols on all the other screening plates, but none of them could detect the triangle symbol on plate 6. Subject K1539 can identify the triangle symbol after a while, but she refers to it as “weak” compared with the circle symbol on the same plate. Only the youngest tritan subject (K1540) can identify all symbols including the triangle on plate 6.
Table 2.
Farnsworth-Munsell 100-Hue error scores [(ES) with expected values for each age (Kinnear & Sahraie, 2002)], midpoint cap (MPC) and selectivity index (SI)], and whether errors were made on the triangle on plate 6 of the Hardy-Rand-Rittler pseudo-isochromatic plates (4th edition, 2002) for six affected and three unaffected (in bold) family members. None made errors on plates 5, 7–10, or the circle symbol on plate 6.
| Pedigree generation |
Subject ID | Age [yr] when tested |
FM 100- Hue ES |
FM 100-Hue MPC/SI(iii) |
HRR plate 6 |
|---|---|---|---|---|---|
| III | K1544 | 21 | 84 (76) | 64/1.73 | seen |
| III | K1540 | 24 | 90 (78) | 43/1.97 | not seen |
| III | K1545 | 30 | 161 (80) | 16/1.17 | seen |
| III | K1604 | 34 | 140 (80) | 42/1.61 | not seen |
| III | K1539 | 34 | 65 (80) | 53/1.57 | weak |
| III | K1538 | 36 | 107 (80) | 67/1.21 | not seen |
| II | K1537 | 54 | 164 (130) | 51/2.56 | not seen |
| I | K1613(i) | 76 | 198 (195) | 55/1.37 | not seen |
| I | K1602 | 80 | 181 (195(ii)) | 67/1.62 | seen |
| I | K1603 | 86 | 286 (195(ii)) | 55/1.50 | seen |
genetics not performed;
oldest age published is 79 yrs [see Ref. (Kinnear & Sahraie, 2002)];
selectivity index <2.0 is probably normal (Vingrys & King-Smith, 1988).
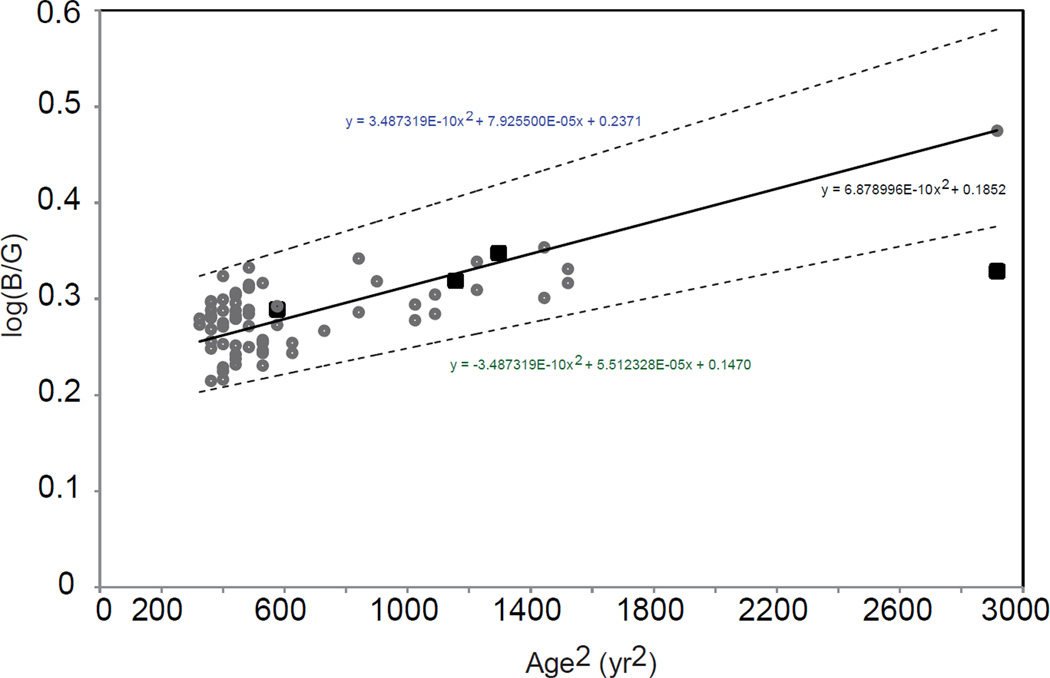
Figure 4 show the Moreland match midpoints (log B/G) plotted as a function of age squared (years2) for 64 normal trichromats (filled circles) and four with the T190I mutation (filled squares). The Moreland match midpoints were distributed normally in the groups of normal trichromats (Kolmogrov-Smirnov and Shapiro-Wilk W). The solid line is the Kraats & van Norren age function (van de Kraats & van Norren, 2007) and the dashed lines are 95% confidence limits (1.96 × SD). The three youngest tritan subjects had match midpoints that were within the normal range, whereas the oldest tritan subject’s match midpoint was outside the normal range. The two youngest, however, had matching ranges that were significantly different from the mean values based on 64 female normal trichromats (Z-score analysis).
Figure 4.
Moreland match midpoints (log B/G) plotted as a function of age squared (years2) for 64 normal trichromats (filled circles) and four with the T190I mutation (filled squares). The solid line is the Kraats & van Norren age function (van de Kraats & van Norren, 2007) calculated to fit the normal data and the dashed lines are 95% confidence limits (1.96 × SD). The confidence limits may be too narrow for the older age groups as they are based on subjects aged 18–38 yrs.
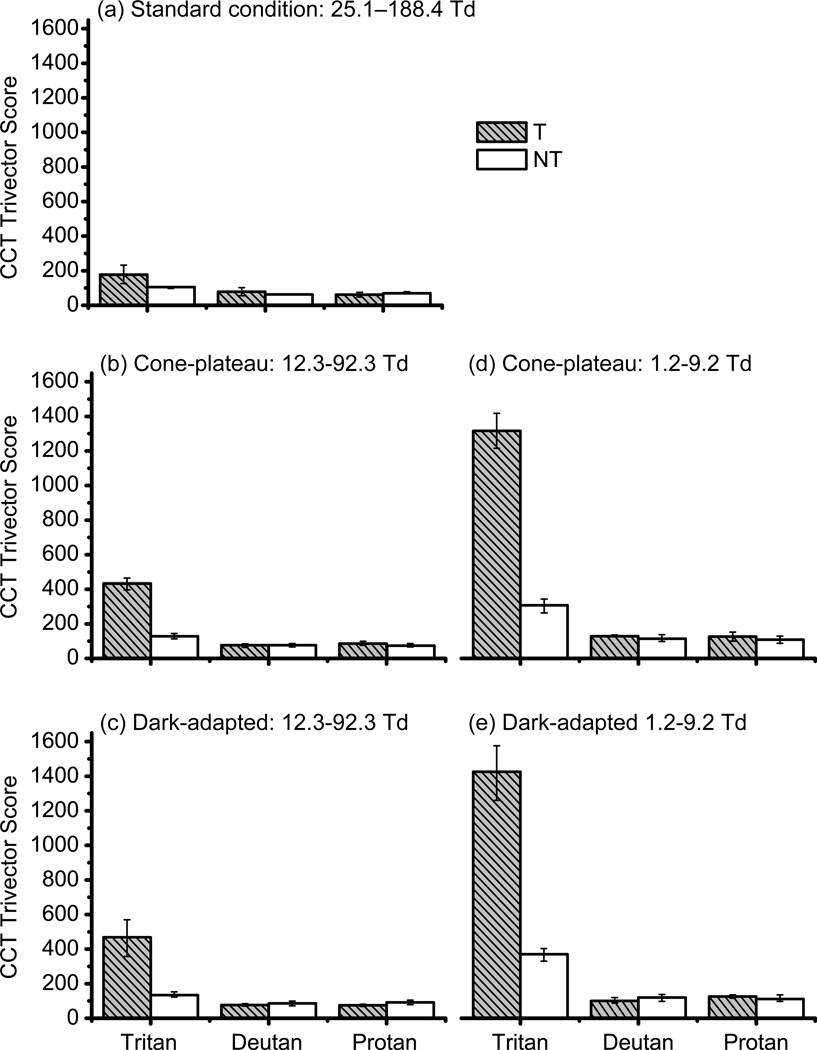
Figure 5(a) show color-discrimination thresholds along the protan, deutan and tritan confusion axes for the CCT standard condition (binocular with natural pupils). The normal mean (±1 SD/SE) color-discrimination thresholds for the 64 normal trichromats were 63.0(±19.2/2.4), 62.7(±20.7/2.6), 87.4 (23.9/3.0) respectively. There was no significant difference in mean thresholds between tritans (filled bars) and normal trichromats (open bars) for the protan and deutan confusion axes. Four out of five tritan females with the T190I mutation had significantly higher thresholds along the tritan confusion axes (Table 3, column labeled TCD: entries are in bold). Individual results for both the Moreland match and the color discrimination thresholds for the normal trichromatic and tritan subjects are shown in Table 3. Z-score analysis (>2.5 standard deviations from the normal mean) revealed that the subjects with tritan mutations had either Moreland matching range or both Moreland matching range and tritan color discrimination threshold (standard condition) that were significantly different from normal trichromats (Table 3, column labeled MP and MR: entries are in bold). Thus, within the family, there is a very strong association between the presence of the T190I mutation and making tritan errors on one or more standard color-vision tests (p = 0.0045, Fisher's exact test). When the 15 normal control subjects were included in the analysis, the association between making tritan errors and having the T190I mutant S opsin was even stronger (p > 0.0001, Fisher's exact test).
Figure 5.
Mean color-discrimination thresholds along the protan, deutan and tritan confusion axes for binocular condition with natural pupils [(a) standard condition, retinal illuminance range 25.1–188.4 Td – assuming an average pupil diameter of 4 mm] and for each of the four experimental conditions. The two rows of panels are for light-adapted and dark-adapted conditions and the two columns of panels are for the different illuminant ranges, (b) and (c) 12.3–92.3 Td, (d) and (e) 1.2– 9.2 Td. The two bars are for the four tritans (filled) and ten normal trichromats (open) respectively. Ordinate (y-axis) show mean thresholds for given confusion axis expressed as distance in CIE 1976 (u’, v’) space multiplied by 10^3. Maximum threshold was 1650. Error bars are standard errors.
Experiment 2: Color-discrimination thresholds (E2)
Figure 5(b–e) shows color-discrimination thresholds along the protan, deutan and tritan confusion axes for each of the four experimental conditions: light-adapted (on the cone-plateau) and dark-adapted conditions and two different illuminant ranges. The results of subject K1544, who is also an obligate carrier of a protan color-vision deficiency, is included in the tritan group as her behavior was no different from the three other tritan subjects. Similarly, the result of subject K1545, who is unaffected, is included in the control group as her behavior was no different from the other nine normal trichromatic control subjects. There were no differences in thresholds within subjects across the three times they were tested on each condition (Friedman test), and there were no difference in mean thresholds along the protan and deutan confusion axes between the those with the tritan mutation and the normal controls. There were, however, differences in mean thresholds along the tritan confusion axis for each of the four conditions with the tritan subjects having significantly higher thresholds than the normal controls (Mann-Whitney U: p < 0.05).
The effect of rod influence on color-discrimination thresholds can be calculated by subtracting mean light-adapted (LA) thresholds from mean dark-adaptated (DA) thresholds (c.f. Knight, Buck, Fowler & Nguyen, 1998). Table 4 shows the calculated mean effect of rod influence on color-discrimination thresholds along the (a) tritan, (b) deutan and (c) protan confusion axes (and standard errors) for the four tritan subjects with the T190I mutation and the ten normal trichromatic subjects who performed these experiments. Discrimination thresholds are elevated by rods along the tritan confusion axes for the tritan subjects at both 9.2 Td and 92.3 Td, but only at 9.2 Td for normal trichromats (entries are in bold). Rods have little influence on thresholds along the protan confusion axes for both tritans and normal trichromats, but discrimination thresholds are reduced by rods along the deutan confusion axis for tritan subjects at 9.2 Td.
Table 4.
Effect of rod influence [dark-adaptation (DA) minus light-adapted (LA)] on color-discrimination thresholds along the (a) tritan, (b) deutan, and (c) protan confusion axes (and standard errors) for four tritan and ten normal trichromatic subjects (one unaffected member of the proband’s family and nine unrelated).
| Tritan axis | Tritans | Normal trichromats | ||||
| Light level | DA (SE) | LA (SE) | DA-LA | DA (SE) | LA (SE) | DA-LA |
| 9.2 Td | 1427 (134) | 1309 (84) | 119 | 400 (39) | 304 (36) | 97 |
| 92.3 Td | 487 (84) | 426 (34) | 61 | 146 (15) | 136 (15) | 10 |
| Deutan axis | Tritans | Normal trichromats | ||||
| Light level | DA (SE) | LA (SE) | DA-LA | DA (SE) | LA (SE) | DA-LA |
| 9.2 Td | 105 (42) | 147 (41) | −43 | 132 (17) | 129 (18) | 3 |
| 92.3 Td | 81 (6) | 85 (12) | −4 | 93 (14) | 85 (12) | 8 |
| Protan axis | Tritans | Normal trichromats | ||||
| Light level | DA (SE) | LA (SE) | DA-LA | DA (SE) | LA (SE) | DA-LA |
| 9.2 Td | 132 (33) | 146 (42) | −14 | 124 (19) | 126 (21) | −2 |
| 92.3 Td | 82 (9) | 93 (21) | −11 | 103 (16) | 85 (12) | 18 |
Discussion
This is the sixth OPN1SW mutation to be associated with S-cone dysfunction. All mutations in the OPN1SW gene that result in an amino acid substitution that have been observed to date are disease causing (Baraas et al., 2007, Gunther et al., 2006, Weitz et al., 1992a, Weitz et al., 1992b). Members of the family with the T190I mutation all fail the tritan plates on HRR pseudo-isochromatic plates, and members of the family who lack the mutation pass the tritan plates (Table 2). This is evidence that the mutation that arose in this family is not benign. The MutationTaster algorithm evaluates disease-causing potential of sequence alterations and it predicts that the I190T substitution is disease causing and that it perturbs a functional domain of the protein. The T190I mutation is not a common polymorphism on the OPN1SW gene, as all sequences in the 1000 genomes database show the T190 do not vary at this position. In addition, T190 is highly conserved in the OPN1SW opsin gene across all species (Table 1).
The results from color-vision testing (E1) reveal that the individuals with the T190I S-opsin mutation show a pattern of behavior consistent with a mild to moderate S-cone dysfunction in daylight. The number of errors on color-vision tests increases with increasing age, as observed with the R283Q S-opsin mutation—the 57 year old with the R283Q mutation was a tritanope in daylight (Baraas et al., 2007), but the 54 yr old in this study exhibit a more moderate degree of tritan color-vision deficiency under similar conditions (Table 2: subject ID K1537). Results from the tritan color discrimination threshold (CCT standard condition) reveal that all but one had significantly higher thresholds than normal controls; the young subjects showed a mild tritan deficiency and the older a moderate tritan deficiency. The CCT color discrimination thresholds for the normal controls were slightly higher than those reported earlier for the age group 20–59 yrs (Paramei, 2012). The reason may be the shorter response time employed here (3 sec versus 8 sec, however, the increase in thresholds between age 20+ and 50+ is as small as 12 units, which is smaller than the SD for the age group 20–29 yrs. Thus, normal aging cannot explain the mild to moderate tritan deficiencies observed in the family members with the T190I mutation. On the contrary, the tritan color-vision deficiency associated with the T190I mutation may indeed go unnoticed if younger subjects only are tested with the FM100-Hue test (Table 2 and Fig. 3) under standard photopic conditions or even with Moreland anomaloscopy (Fig. 4).
The performance of subjects with the T190I mutation were significantly different from that of normal trichromats along the tritan vector for both light-adapted and dark-adapted conditions and for both levels of retinal illumination (E2: Fig. 5). Normal trichromats behaved as if they had a mild tritan color-vision deficiency at low luminance levels, whereas those with the mutation became dichromatic at the same luminance levels. Subjects with the T190I mutation behaved as if they were tritanopes; that is S-cones with loss of sensitivity at low light levels. Their thresholds were on average four times higher compared with the normal controls (Fig. 5) under similar conditions. Some of the affected subjects may have even greater loss in sensitivity, but there is a ceiling effect with regards to maximum color contrast available within the gamut of a calibrated CRT monitor. Thus, more elaborate tests of spectral sensitivity would be required to quantify this more precisely. Nevertheless, the sensitivity loss in the affected individuals bears resemblance to congenital stationary night blindness caused by mutations in the rod pigment rhodopsin that are thought to constitutively activate transducin, thereby reducing the sensitivity of the rod photoreceptors (Jin, Cornwall & Oprian, 2003). Similarly, the reduced S-cone sensitivity at low light levels in individuals with the T190I mutation might be explained by constitutive activation of cone transducin by the mutant S opsin.
This is the first mutation in extra-cellular loop II of the S-cone opsin gene that is associated with tritan color-vision deficiency; the other five known mutations are in the transmembrane domains. A mutation at the analogous position of rhodopsin (T153M) was previously found in a 49 year old human who showed a mild phenotype in experiments using a standard electroretinogram (ERG) protocol to quantify the dark-adapted waveform (Cideciyan, Hood, Huang, Banin, Li, Stone, Milam & Jacobson, 1998). Two parameters, maximum amplitude and sensitivity, of the rod and cone components of the ERG were estimated, and these values were used to estimate rod outer segment area. At least in some regions of the retina, both rod and cone components fell within the normal range, although the rod component was near the lower limits of the normal range, and rod outer segment area was estimated to be about 80% of normal. The authors argued that the relatively mild phenotype is consistent with a simple loss of function mutation present in a single dose. The findings in individuals with the T190I mutation in S opsin are also consistent with a loss of function in a single dose.
It has been hypothesized that if the function of S-cones is reduced then rods may play a more active part in color discrimination for larger test fields (8 deg) (Pokorny, Smith & Went, 1981). That rods impair discrimination along the tritan color axis for normal trichromats (Table 4) has been reported previously (Brown, 1951, Knight et al., 1998, Knight, Buck & Pereverzeva, 2001, Stabell & Stabell, 1977). In our experiments, the effect of the rod impairment is more or less the same for normal trichromats as for tritans, except that the effect of rod impairment starts at the highest level of luminance (92.3 Td) for tritans (c.f.Hough, 1968). Thus, the results presented here do not support the idea that when S-cone function is reduced, rods play a larger role in color discrimination. The implication is that normally functioning S cones are needed for blue-green color discrimination at mesopic light levels.
Highlights.
Novel OPN1SW mutation (T190I) and chromatic contrast tested at different levels of retinal illuminance.
Strong association between making tritan errors and having the T190I OPN1SW mutation.
The T190I OPN1SW mutation cause abnormal S-cone function.
Acknowledgements
The authors would like to thank Jack D. Moreland for advice on calculating the age function for the Moreland match. Supported by the Research Council of Norway Grants 176541/V10 and 182768/V10 (RCB), NIH Core Grant for Vision Research EY01730, and Research to Prevent Blindness (University of Washington).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey JE, Neitz M, Tait DM, Neitz J. Evaluation of an updated HRR color vision test. Visual Neuroscience. 2004;21(3):431–436. doi: 10.1017/s0952523804213463. [DOI] [PubMed] [Google Scholar]
- Baraas RC, Carroll J, Gunther KL, Chung M, Williams DR, Foster DH, Neitz M. Adaptive optics retinal imaging reveals S-cone dystrophy in tritan color-vision deficiency. Journal of the Optical Society of America A-Optics Image Science and Vision. 2007;24(5):1438–1447. doi: 10.1364/josaa.24.001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WRJ. The influence of luminance level on visual sensitivity to color differences. Journal of the Optical Society of America. 1951;41(10):684–688. doi: 10.1364/josa.41.000684. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Hood DC, Huang Y, Banin E, Li ZY, Stone EM, Milam AH, Jacobson SG. Disease sequence from mutant rhodopsin allele to rod and cone photoreceptor degeneration in man. Proceedings of the National Academy of Sciences of the U S A. 1998;95(12):7103–7108. doi: 10.1073/pnas.95.12.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther KL, Neitz J, Neitz M. A novel mutation in the short-wavelength-sensitive cone pigment gene associated with a tritan color vision defect. Visual Neuroscience. 2006;23:403–409. doi: 10.1017/S0952523806233169. [DOI] [PubMed] [Google Scholar]
- Hough EA. The spectral sensitivity functions for parafoveal vision. Vision Research. 1968;8(11):1423–1430. doi: 10.1016/0042-6989(68)90088-6. [DOI] [PubMed] [Google Scholar]
- Jin S, Cornwall MC, Oprian DD. Opsin activation as a cause of congenital night blindness. Nature Neuroscience. 2003;6(7):731–735. doi: 10.1038/nn1070. [DOI] [PubMed] [Google Scholar]
- Kainz PM, Neitz M, Neitz J. Molecular genetic detection of female carriers of protan defects. Vision Research. 1998;38(21):3365–3369. doi: 10.1016/s0042-6989(97)00366-0. [DOI] [PubMed] [Google Scholar]
- Kinnear PR, Sahraie A. New Farnsworth-Munsell 100 hue test norms of normal observers for each year of age 5–22 and for age decades 30–70. British Journal of Ophthalmology. 2002;86(12):1408–1411. doi: 10.1136/bjo.86.12.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Buck SL, Fowler GA, Nguyen A. Rods affect S-cone discrimination on the Farnsworth-Munsell 100-hue test. Vision Research. 1998;38(21):3477–3481. doi: 10.1016/s0042-6989(97)00414-8. [DOI] [PubMed] [Google Scholar]
- Knight R, Buck SL, Pereverzeva M. Stimulus size affects rod influence on tritan chromatic discrimination. Color Research & Application. 2001;26:S65–S68. [Google Scholar]
- Linksz A. An essay on color vision. New York: Grune & Stratton; 1964. [Google Scholar]
- Miyake Y, Yagasaki K, Ichikawa H. Differential diagnosis of congenital tritanopia and dominantly inherited juvenile optic atrophy. Archives of Ophthalmology. 1985;103(10):1496–1501. doi: 10.1001/archopht.1985.01050100072022. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Neitz M, Neitz J. A new mass screening test for color-vision deficiencies in children. Color Research and Application. 2001;26:S239–S249. [Google Scholar]
- Paramei GV. Color discrimination across four life decades assessed by the Cambridge Colour Test. Journal of the Optical Society of America A-Optics Image Science and Vision. 2012;29(2):A290–A297. doi: 10.1364/JOSAA.29.00A290. [DOI] [PubMed] [Google Scholar]
- Pokorny J, Smith VC, Went LN. Color matching in autosomal dominant tritan defect. Journal of Optical Society of America. 1981;71(11):1327–1334. doi: 10.1364/josa.71.001327. [DOI] [PubMed] [Google Scholar]
- Regan BC, Reffin JP, Mollon JD. Luminance noise and the rapid determination of discrimination ellipses in colour deficiency. Vision Research. 1994;34(10):1279–1299. doi: 10.1016/0042-6989(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Stabell U, Stabell B. Wavelength discrimination of peripheral cones and its change with rod intrusion. Vision Research. 1977;17(3):423–426. doi: 10.1016/0042-6989(77)90034-7. [DOI] [PubMed] [Google Scholar]
- Stenkamp RE, Filipek S, Driessen CAGG, Teller DC, Palczewski K. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochimica et Biophysica Acta. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- van de Kraats J, van Norren D. Optical density of the aging human ocular media in the visible and the UV. Journal of the Optical Society of America A-Optics Image Science and Vision. 2007;24(7):1842–1857. doi: 10.1364/josaa.24.001842. [DOI] [PubMed] [Google Scholar]
- Vingrys AJ, King-Smith PE. A quantitative scoring technique for panel tests of color vision. Investigative Ophthalmology and Visual Science. 1988;29(1):50–63. [PubMed] [Google Scholar]
- Weitz CJ, Miyake Y, Shinzato K, Montag E, Zrenner E, Went LN, Nathans J. Human tritanopia associated with two amino acid substitutions in the blue-sensitive opsin. American Journal of Human Genetics. 1992a;50(3):498–507. [PMC free article] [PubMed] [Google Scholar]
- Weitz CJ, Went LN, Nathans J. Human tritanopia associated with a third amino acid substitution in the blue-sensitive visual pigment. American Journal of Human Genetics. 1992b;51(2):444–446. [PMC free article] [PubMed] [Google Scholar]
- Went LN, Pronk N. The genetics of tritan disturbances. Human Genetics. 1985;69(3):255–262. doi: 10.1007/BF00293036. [DOI] [PubMed] [Google Scholar]