Abstract
Repair of DNA inter-strand cross-links in mammalian cells involves several biochemically distinctive processes, including the release of one of the cross-linked strands and translesion DNA synthesis (TLS). In this report, we investigated in vitro TLS activity of psoralen DNA inter-strand cross-link by three DNA repair polymerases, DNA polymerase beta, kappa and iota. DNA polymerase beta is capable of bypassing a psoralen cross-link with a low efficiency. Cell extracts prepared from DNA polymerase beta knockout mouse embryonic fibroblast showed a reduced bypass activity of the psoralen cross-link and purified DNA polymerase beta restored the bypass activity. In addition, DNA polymerase iota mis-incorporated thymine across the psoralen cross-link and DNA polymerase kappa extended these mis-paired primer ends, suggesting that DNA polymerase iota may serve as an inserter and DNA polymerase kappa may play a role as an extender in the repair of psoralen DNA inter-strand cross-links. The results demonstrated here indicate that multiple DNA polymerases could participate in TLS steps in mammalian DNA inter-strand cross-link repair.
INTRODUCTION
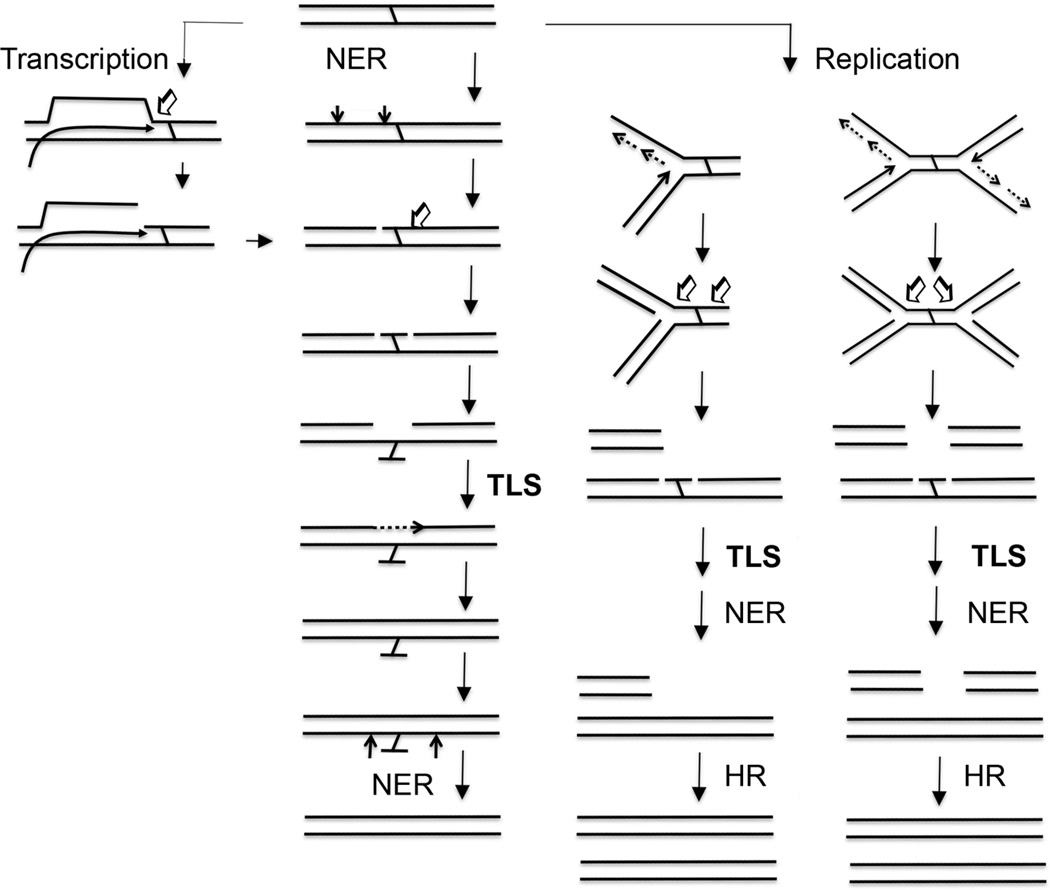
DNA inter-strand cross-links (ICLs) are repaired by multiple pathways in mammalian cells1 (Figure 1). Each pathway requires the release of one of the cross-linked strands (unhooking of a cross-link) followed by translesion DNA synthesis (TLS) of the unhooked (yet cross-linked) strand. Several DNA polymerases have been implicated to accomplish this TLS process, including DNA polymerase nu, kappa and zeta; however, recent studies indicate redundancy among TLS polymerases in ICL repair2, 3.
Figure 1.
Proposed ICL repair pathways in humans. DNA interstrand cross-links (ICLs) are repaired by multiple pathways. In each pathway, translesion DNA synthesis (TLS) plays a critical role. (1) Nucleotide excision repair (NER) makes dual incisions at the 5’ side of an ICL and leaves a nick. A subsequent incision by a nuclease/exonuclease releases one of the cross-linked strands (unhooking of an ICL). A gap generated is filled by a TLS polymerase to restore duplex DNA. A second round of NER removes a lesion with the unhooked ICL. DNA repair synthesis and DNA ligation completes a repair process. (2) When RNA polymerase encounters an ICL during transcription, RNA polymerase stalls near the site of the ICL and generates a DNA-RNA-protein complex. This complex might recruit NER and/or nucleases to generate a nick at 5’ to the ICL. Once the nick is introduced, the ICL will be repaired by the same process as the NER-mediated ICL repair. (3) During DNA replication, a replication fork is blocked near the site of the ICL and generates a “Y-shape” structure. A structure-specific endonuclease such as XPF-ERCC1 generates a nick at one side of the ICL and a second endonuclease makes a second incision at the other side of the ICL to unhook the ICL. These actions generate one broken end and a gap with an unhooked ICL. TLS fills the gap and restores duplex DNA. After NER removes a lesion with an unhooked ICL, homologous recombination (HR) repairs the broken end and restores a replication fork. A second scenario is initiated by a merging of two replication forks at an ICL. When two opposing replication forks meet at an ICL, an “X-shape” structure is generated. Endonucleases generate incisions at the 5’ and 3’-sides of an ICL to unhook the ICL. These actions result in the formation of two broken ends and a gap with an unhooked ICL. TLS fills the gap and restores duplex DNA. After NER removes a lesion with an unhooked ICL, HR repairs the broken ends. Endonucleases and other repair factors involved and exact repair intermediate structures in each pathway are the matters of investigation.
DNA polymerase beta participates in base excision repair (BER). Interestingly, DNA polymerase beta-overexpressing cells are resistant to several chemotherapeutics, including cisplatin. Suppression of DNA polymerase beta altered the cellular sensitivity to cisplatin, but not to oxaliplatin, indicating a role of DNA polymerase beta in the repair of cisplatin-ICLs 4. In addition, DNA polymerase beta bypasses 8-oxoguanine, UV photo products and cis-platin intra-strand cross-link (G^G) when the lesion is placed in a gap 5. These reports suggest that DNA polymerase beta is involved in a TLS step in ICL repair.
DNA polymerase kappa inserts nucleotides opposite of a variety of DNA bases and has a unique property to bypass minor groove adducts that form at the N2-position of guanine in vitro6, 7. DNA polymerase kappa can function during the gap-filling step of nucleotide excision repair (NER), in place of DNA polymerase delta, under certain conditions 8. Importantly, DNA polymerase kappa bypasses N2-N2 guanine ICLs, which mimic mitomycin C ICLs, in vitro9. Also, the suppression of DNA polymerase kappa results in sensitivity to mitomycin C, suggesting a role of DNA polymerase kappa for the bypass of minor groove ICLs in human cells9.
DNA polymerase iota is a highly error-prone DNA polymerase, especially opposite from pyrimidines10. DNA polymerase iota has also the ability to bypass various types of DNA adducts in vitro11. It has been demonstrated that the sequential action of DNA polymerase iota and DNA polymerase kappa promotes the efficient bypass of some lesions, in which DNA polymerase iota incorporates the nucleotide opposite the lesion site and DNA polymerase kappa performs the extension reaction 12.
We investigated in vitro TLS activity of DNA polymerase beta, kappa and iota using a psoralen ICL as a template. DNA polymerase beta incorporated a dNMP opposite a psoralen ICL and extended the primer to a much lesser extent. We also detected DNA polymerase beta-dependent bypass activity of a psoralen ICL in cell extracts from mouse embryonic fibroblasts. DNA polymerase kappa and iota did not demonstrate a bypass activity of a psoralen ICL; however, the sequential action of DNA polymerase iota and DNA polymerase kappa promotes the bypass of a psoralen ICL. Moreover, DNA polymerase iota mis-incorporated TMP opposite of a psoralen ICL with a similar efficiency of the incorporation of dAMP. Because psoralen modifies thymines to form an ICL, the mis-incorporation of TMP opposite a psoralen ICL results in a T-to-A transversion, which is the main point mutation induced by psoralen ICLs. These results suggest that DNA polymerase beta, kappa and iota might be involved in TLS steps in mammalian ICL repair pathways.
EXPERIMENTAL PROCEDURES
Substrate preparation
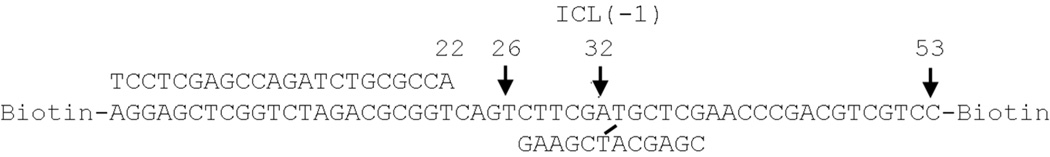
A 12-mer oligonucleotide (5'-GAAGCTACGAGC-3') with a psoralen furan-side mono-adduct at the T (100 pmol) was annealed to 100 pmol of a 53-mer oligonucleotide (5’- biotin-CCTGCTGCAGCCCAAGCTCGTAGCTTCTGACTGGCGCAGATCTGGCTCGAGGAbiotin-3') (underlined portion complementary to 12-mer). The partially duplexed DNA was exposed to UVA (366 nm) for 10 min to convert the mono-adduct to an ICL. The cross-linked substrate was purified from a 10% denaturing polyacrylamide gel and annealed to a 5’-32P-labeled 22-mer primer (5'-TCCTCGAGCCAGATCTGCGCCA-3'). The substrate was then purified from a 6% non-denaturing polyacrylamide gel. Under these conditions, the cross-linked substrate is well separated from the non-cross-linked substrate. Typically, about 0.3 % of the substrate was non-cross-linked13, 14. For the undamaged substrate, a 32P-labeled 22-mer primer was annealed to the 53-mer template (Figure 2).
Figure 2.
A defined substrate with a single psoralen ICL. A psoralen ICL is located in the middle of a 12-mer oligonucleotide annealed to a 53 nt-length template. Biotin moieties are linked both to 5’- and 3’- ends of the 53 nt length template.
The psoralen modified 12-mer was a generous gift from Dr. John Hearst. The 5’- and 3’-biotin-labeled 53-mer was purchased from OPERON and the 22-mer was synthesized at the Molecular Biology Core at the University of Nebraska Medical Center.
In vitro DNA polymerase assay
Reaction mixtures (10 mM HEPES-KOH pH 7.9, 7.5% glycerol, 2.5 mM KCl, 5 mM MgCl2, 0.1 mg/ml BSA, 250 μM of each dNTP, 2 nM of the substrate, and the indicated concentration of DNA polymerase in 20 µl) were incubated at 37 °C for the indicated time. The amount of DNA polymerase beta or DNA polymerase kappa used was determined to provide similar primer-extension efficiency on an undamaged template. The reactions were terminated by the addition of 25 mM EDTA, followed by phenol/chloroform extraction. The reaction products were isolated by ethanol precipitation and analyzed on a 10% denaturing polyacrylamide gel. The dried gel was exposed to a PhosphorImage screen, an image was obtained by scanning the screen with the Typhoon 9410 (GE Healthcare), and the products were quantified by ImageQuant. The amount of products longer than 34 nt was used to calculate the percent bypass activity.
For DNA polymerase iota, a reaction was performed in iota buffer (40 mM Tris-HCl pH 8.0, 0.15 mM MnCl2, 0.2 mg/ml BSA, 2.5 % glycerol, 250 µM of each dNTP, 60 pM of the substrate, and the indicated amount of DNA polymerase iota) at 37°C for the indicated time. For the steady-state kinetic study, DNA polymerase iota (2 nM) was incubated with 200 pM of the substrate at 37 °C for 3 min in the presence of a range of concentrations (1, 3, 10, 30, and 100 µM) of each dNTP.
In vitro TLS assay with cell extracts
Nuclear extracts were prepared from mouse embryonic fibroblast (MEF) cells as described15, 16. Nuclear extract (3 µg) from wild type MEF cells or DNA polymerase beta-knockout MEF cells was incubated with 0.3 nM of the substrate in 20 µl of the reaction buffer (12.5 mM HEPES-K-OH pH 7.5, 50 mM NaCl, 5 mM MgCl2, 250 µM dNTPs, 2 mM ATP, 0.1 mg/ml BSA, 5% glycerol, and the indicated amount of purified DNA polymerase) at 37°C for 30 min. After phenol/chloroform extraction, the reaction products were isolated by ethanol precipitation and analyzed on a 10% denaturing polyacrylamide gel.
Cell-lines and purified proteins
MEF cells with wild type DNA polymerase beta and DNA polymerase beta-knockout17 were generous gifts from Dr. Joan Sweasy at Yale University. MEF cells were grown in DMEM medium with 10% FBS at 37°C and 5% CO2.
Human GST-tagged DNA polymerase iota was expressed in yeast and purified as described in Makarova et al18. Purified recombinant DNA polymerase beta and kappa19 were generously provided by Dr. Leonid V. Gening at the Institute of Molecular Genetics of Russian Academy of Science, and purified human 9-1-1 complex20 was a generous gift from Dr. Laura Lindsey-Boltz and Dr. Aziz Sancar at the University of North Carolina at Chapel Hill.
RESULTS
Bypass of a psoralen ICL by DNA polymerase beta in vitro
One of cross-linked strands is proposed to be released by various mechanisms (Figure 1). To restore duplex DNA for a subsequent repair process, bypass of the released fragment that is attached to the other strand is pre-requisite (Figure 1). However, an exact DNA structure that is utilized by TLS polymerases (how many nucleotides are attached after unhooking and how far the unhooked strand is located from a primer end) is not known. We prepared a 53-nt length template DNA with a 12-mer oligonucleotide attached via psoralen ICL and placed a four-nucleotide gap between a primer end and a 5’-end of an unhooked strand (Figure 2). Similar types of substrate DNA have been used to study the TLS activity of an ICL9, 13, 14, 21. A primer-extension to the attached oligonucleotide by a DNA polymerase results in the production of a 26-nt length fragment. An inhibition of a polymerase-mediated chain elongation by a psoralen ICL will generate a 32-nt length fragment (designated as ICL(−1)) and an incorporation of a dNMP opposite a psoralen ICL generates a 33-nt length fragment (designated as ICL(0)). Fragments larger than 34-nt in length generated by a DNA polymerase are considered to be bypass products.
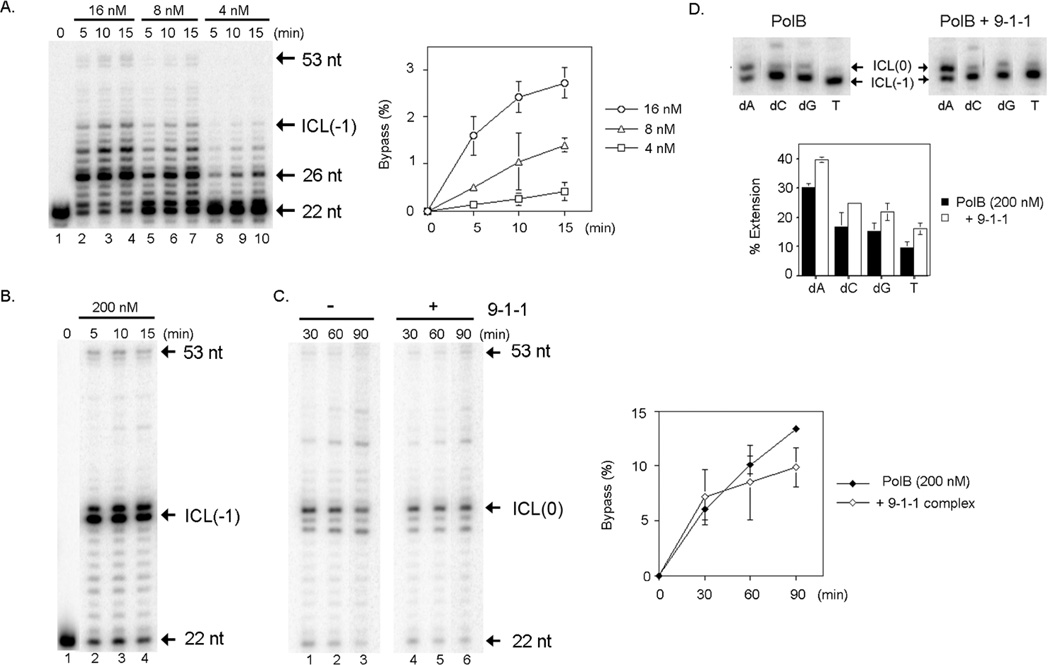
DNA polymerase beta is implicated to play a role in ICL repair in mammalian cells. We examined the in vitro bypass of a psoralen ICL by DNA polymerase beta (Figure 3). Purified human DNA polymerase beta was incubated with a defined substrate with an unhooked psoralen ICL at 37°C for the indicated time. We detected a low efficiency bypass of the psoralen by DNA polymerase beta, although a major product by DNA polymerase beta was a 32-nt length fragment showing that DNA polymerase beta was stalled at one nucleotide before the psoralen ICL (Figure 3A). An increased concentration of the DNA polymerase beta improved the bypass efficiency (Figure 3B). Interestingly, the DNA polymerase beta produced two major fragments with a 32-nt and a 33-nt length with a higher concentration (Figures 3B and C). These results demonstrated that DNA polymerase beta is capable of incorporating a dNMP opposite the psoralen ICL. We conclude that the DNA polymerase beta is capable of bypassing a psoralen ICL in vitro.
Figure 3.
Bypass of an unhooked psoralen ICL by DNA polymerase beta. (A) DNA polymerase beta bypasses a psoralen ICL in vitro. DNA polymerase beta was incubated with 2 nM of a defined substrate with a psoralen ICL in the presence of 250 µM dNTPs at 37°C. The reaction mixtures were incubated for 5 min (lanes 2, 5, and 8), 10 min (lanes 3, 6, and 9), and 15 min (lanes 4, 7, and 10). Lane 1, no polymerase; lanes 2–4, 16 nM DNA polymerase beta; lanes 5–7, 8 nM DNA polymerase beta; lanes 8–10, 4 nM DNA polymerase beta. 22 nt, primer; ICL(-1), 32 nt fragment (a termination product at one nucleotide before the ICL), and 53 nt, a fully extended product. Fragments larger than 34 nt were considered as bypass products. Bypass efficiency was determined and plotted as a graph next to the gel. The error bars represent standard deviations obtained from three independent experiments. (B) DNA polymerase beta incorporates nucleotides opposite a psoralen ICL. An increased concentration of DNA polymerase beta (200 nM) was incubated with 2 nM of the substrate in the presence of 250 µM of dNTPs at 37°C for 5 min (lane 1), 10 min (lane 2), and 15 min (lane 3). 22 nt, primer; ICL(-1), 32 nt fragment (a termination product at one nucleotide before the ICL). (C) A checkpoint clamp 9-1-1 complex has no effect on the ICL bypass activity of DNA polymerase beta in vitro. The 9-1-1 complex (25 nM) was pre-incubated with 2 nM of the substrate on ice for 5 min. After adding DNA polymerase beta (200 nM), the reaction mixture was incubated at 37°C for 30 min (lanes 1 and 4), 60 min (lanes 2 and 5), and 90 min (lanes 3 and 6). Lanes 1–3, without 9-1-1 complex; lanes 4–6, with 9-1-1 complex. 22 nt, primer; ICL(-1), 32 nt fragment (a termination product at one nucleotide before the ICL); ICL(0), 33 nt fragment (product of incorporation of a nucleotide opposite the ICL). Bypass efficiency was determined and plotted as a graph next to the gel image. The error bars represent standard deviations obtained from three independent experiments. (D) Preferential incorporation of dAMP opposite a psoralen ICL by DNA polymerase beta. The ICL(-1) substrate was prepared by extending the 22 nt-primer of the substrate to 32 nt by Klenow fragment. The ICL(-1) substrate (2 nM) was incubated with DNA polymerase beta in the presence of a single dNTP (1 mM) at 37 °C for 20 min. Left panel, without 9-1-1 complex, right panel, with 9-1-1 complex (25 nM). The percentage of primer extension was determined in each lane and plotted as a graph below the gel image. The error bars represent standard deviations from three independent experiments.
Since DNA polymerase beta incorporates a dNMP opposite a psoralen ICL, we examined whether one of the four nucleotides is preferentially incorporated opposite a psoralen ICL. The reaction products generated by E. coli Klenow fragment were used as a template for this purpose (referred as “ICL(−1) substrate”). After the reaction with the Klenow fragment that stops at one nucleotide before a psoralen ICL, the products were isolated and used to determine a specificity of nucleotide incorporation opposite the psoralen ICL. DNA polymerase beta was incubated with the ICL(−1) substrate in the presence of only one dNTP. DNA polymerase beta incorporated a nucleotide opposite from a psoralen ICL in the order of dAMP ≫ dCMP = dGMP ≫ TMP (Figure 3D). Because psoralen modifies thymines to form an ICL, DNA polymerase beta incorporates the proper nucleotide dAMP opposite a psoralen ICL.
It was reported that accessory factors of DNA polymerases such as PCNA influence efficacy in bypass of DNA damage and preference of incorporation of dNTP opposite DNA damage22, 23. A checkpoint clamp, RAD9-RAD1-HUS1 (9-1-1) complex, stimulates the polymerase activity of DNA polymerase beta in vitro 24. We investigated the impact of the 9-1-1 complex on the bypass activity of a psoralen ICL by DNA polymerase beta. The 9-1-1 complex stimulated the polymerase activity of DNA polymerase beta on a non-damaged template under the conditions used, as reported (Supplemental Information 1). The addition of the 9-1-1 complex in the DNA polymerase beta-mediated TLS reaction did not improve the bypass efficiency (Figure 3C, lanes 4–6, graph) nor alter the specificity of the nucleotide incorporation opposite a psoralen ICL, although the complex stimulated the overall efficiency of dNTP incorporations (Figure 3D, graph).
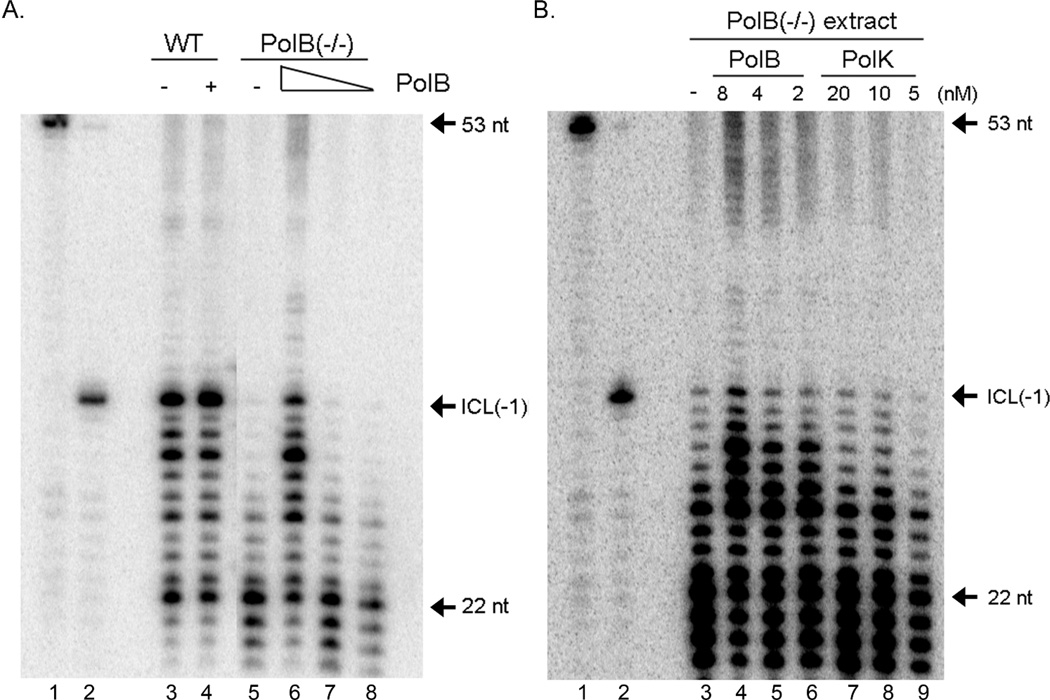
To study a contribution of DNA polymerase beta in a TLS step of ICL repair, we examined the in vitro bypass activity of a psoralen ICL using cell-free extracts (Figure 4). Nuclear extracts were prepared from wild type mouse embryonic fibroblasts (MEFs) or DNA polymerase beta −/− MEFs. The substrate DNA was incubated with nuclear extracts and then bypass activity was examined. Wild type nuclear extracts showed a bypass activity (Figure 4A, lane 3), while DNA polymerase beta−/− nuclear extracts displayed a significantly reduced bypass activity (Figure 4A, lane 5). Furthermore, the purified DNA polymerase beta was able to restore the bypass activity in the DNA polymerase beta−/− nuclear extracts (Figure 4A, lanes 6–8; 4B, lanes 4–6). Importantly, purified DNA polymerase kappa, which showed a similar polymerase activity to the DNA polymerase beta used (Supplemental Information 2 and 3), failed to recover the reduced bypass activity in the DNA polymerase beta−/− nuclear extracts (Figure 4B, lanes 7–9). In addition to the products longer than 34 nt in length, we also detected fragments longer than 53 nt-length products in the reaction with extracts (Supplemental Information 4). These products were generated after ligation of different sizes of extended primers that contained cross-links. Thus, they were not the products of TLS (Supplemental Information 4). These results demonstrate the existence of DNA polymerase beta-dependent TLS of a psoralen ICL in vitro.
Figure 4.
DNA polymerase beta-dependent bypass of a psoralen ICL in mouse embryonic fibroblast cell extract in vitro. Only the products shorter than 53 nt in length were shown (see Supplemental Information 4 for the entire gel image). (A) In vitro bypass of a psoralen ICL is compromised in nuclear extract from DNA polymerase beta deleted MEF cells. Nuclear extract (3 µg) from wild type (WT) or DNA polymerase beta deleted (PolB−/−) MEF cells was incubated with 2 nM of the substrate at 37 °C for 30 min. Bypass products (longer than ICL(-1)) were significantly less in the reaction with nuclear extract from DNA polymerase beta deleted MEF cells (lane 5) compared to the ones with extract from wild type MEF cells (lane 3). The addition of purified human DNA polymerase beta (lane 6–8) successfully restored the bypass activity of the ICL in the nuclear extract from DNA polymerase beta deleted MEF cells. The same amount of purified DNA polymerase beta did not influence the bypass activity in nuclear extract from wild type MEF cells (compare lanes 3 and 4). Lane 1, 53 nt marker; lane 2, 32 nt marker: lane 3, WT extract; lanes 4, WT extract with purified PolB; lane 5, PolB−/− extract; and Lanes 6–9, PolB−/− extract with purified PolB. (B) DNA polymerase kappa cannot substitute DNA polymerase beta for in vitro bypass of a psoralen ICL in nuclear extract. Purified human DNA polymerase kappa failed to restore bypass activity in nuclear extract from DNA polymerase beta deleted MEF cells (lanes 7–9). DNA polymerase beta that showed similar DNA polymerase activities (Supplemental Information 2 and 3) to DNA polymerase kappa complemented the defect in bypass activity in nuclear extract from DNA polymerase beta deleted MEF cells (lanes 4–6).
Bypass of a psoralen ICL by the sequential action of DNA polymerases iota and kappa
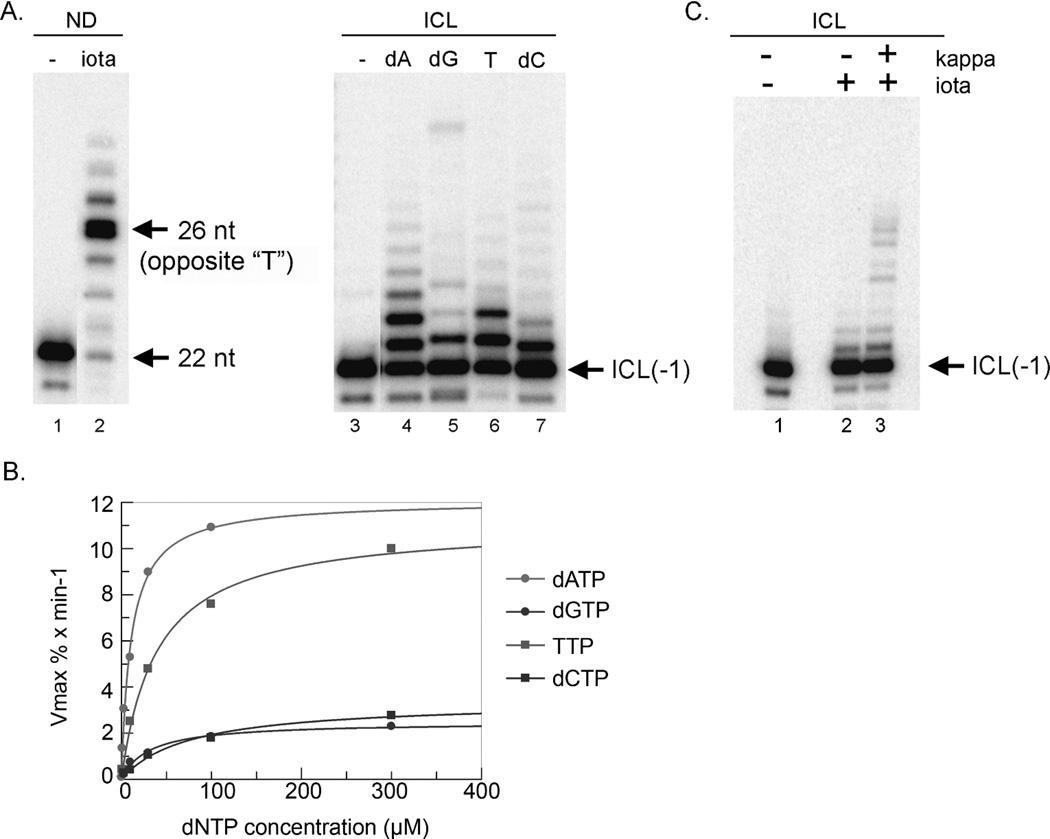
DNA polymerase iota is a highly error-prone DNA polymerase with a low processivity10, 25. Interestingly, DNA polymerase iota tends to stall after incorporating dGMP opposite thymine on a template (Figure 5A, lane 2)18, 25. Those biochemical properties prompted us to examine whether DNA polymerase iota can insert a nucleotide opposite a psoralen ICL. Indeed, DNA polymerase iota incorporated a nucleotide opposite a psoralen ICL on the ICL(−1) substrate (Figure 5A, lanes 4–7). We further investigated the specificity of the nucleotide incorporation opposite a psoralen ICL by steady-state kinetics experiments. In contrast to the preferential incorporation of dAMP opposite a psoralen ICL by DNA polymerase beta (Figure 3D), DNA polymerase iota efficiently incorporated dAMP and TMP opposite a psoralen ICL (Figure 5B, Table I). This result is very surprising because a signature property of DNA polymerase iota is the preferential misincorporation of dGMP opposite a thymine on a template. The data might indicate a structural change induced by a psoralen modification on thymine, which alters a specificity of incorporation opposite a psoralen-modified thymine.
Figure 5.
A potential role of DNA polymerase iota in bypass of a psoralen ICL. (A) DNA polymerase iota preferentially incorporates TMP opposite a psoralen ICL in vitro. Purified GST-tagged DNA polymerase iota tends to stall at a template thymine (non-damaged) (lane 2). The ICL(-1) substrate (60 pM) was incubated with purified GST-tagged DNA polymerase iota in the presence of a single dNTP (100 µM) at 37°C for 3 min. DNA polymerase iota incorporates TMP and dAMP opposite a psoralen ICL (lanes 6 and 4, respectively). (B) Steady-state kinetic experiments. TMP and dAMP were incorporated opposite a psoralen ICL by DNA polymerase iota. (C) Sequential action of DNA polymerase iota and kappa in bypass of a psoralen ICL. The ICL(-1) substrate (60 pM) was incubated with GST-tagged DNA polymerase iota at 37°C for 15 min and then DNA polymerase kappa was added and incubated for an additional 15 min. DNA polymerase kappa extended the iota-generated mismatched primers (compare lanes 2 and 3).
Table 1.
Kinetic measurement of nucleotide incorporation opposite psoralen ICL by DNA polymerase iota
| dNTP |
Vmax (fmol/min; mean ± SD) |
Km (µM; mean ± SD) |
Vmax/Km | *finc |
|---|---|---|---|---|
| dATP | 12.1 ± 0.5 | 10.9 ± 1.5 | 1.1 | |
| dGTP | 2.5 ± 0.1 | 30.1 ± 5.4 | 0.08 | 7.3 × 10−2 |
| dCTP | 3.4 ± 0.3 | 75.5 ± 16.4 | 0.04 | 3.6 × 10−2 |
| TTP | 11.1 ± 0.4 | 39.4 ± 4.8 | 0.28 | 0.25 |
finc = (Vmax/Km) incorrect / (Vmax/Km) correct
DNA polymerase kappa was shown to bypass an N2-N2 guanine minor groove ICL efficiently9. The N2-N2 guanine ICL was especially part of a two-nucleotide fragment attached on a template. DNA polymerase kappa did not bypass a psoralen ICL in our substrate (Supplemental Information 3B), which is part of the12-nt attached on a template, in agreement with the notion that longer nucleotides attached to a template are inhibitory for the bypass of the N2-N2 guanine minor groove ICL. Having the data that DNA polymerase iota incorporates a nucleotide opposite a psoralen ICL and the known extension activity from a mismatched primer-end by DNA polymerase kappa26, we examined the impact of a sequential action of DNA polymerases iota and kappa (Figure 5C). DNA polymerase iota was incubated with the ICL(−1) substrate in the presence of four dNTPs at 37°C for 15 min. Then, DNA polymerase kappa was added to the reaction and incubated for an additional 15 min at 37°C. DNA polymerase kappa extended a primer-end opposite a psoralen generated by DNA polymerase iota (Figure 5C, lanes 2 and 3). DNA polymerase kappa does not incorporate a nucleotide opposite a psoralen ICL at the concentration used in these reactions (Supplemental Information 3B). These results showed that a sequential action of DNA polymerases iota and kappa bypasses a psoralen ICL.
DISCUSSION
DNA interstrand cross-links (ICLs) are repaired by multiple pathways, DNA replication-coupled and DNA replication-independent pathways1. The repair factors required and the detailed mechanisms for these pathways are different among species27, 28. However, the following two principles of the repair are very similar. A release (or separation) of one of the cross-linked strands is the pre-requisite step for all of the pathways reported. A gap generated opposite the released cross-linked strand that is still attached to the complementary strand should be filled by a translesion DNA synthesis (TLS) DNA polymerase to be further processed to complete the repair. DNA replication-coupled ICL repair has been studied using Xenopus egg extracts29, 30. It has been demonstrated that REV7 mediates a TLS reaction to bypass a cisplatin-ICL in the replication-coupled ICL repair30. Genetic and biochemical studies indicate the involvement of various TLS polymerases, including DNA polymerases eta, kappa and zeta in DNA replication-independent ICL repair pathways in mammalian cells2, 3. In addition, DNA polymerase nu is implicated to play a role in human ICL repair14, 31. In this report, we provided biochemical evidence that DNA polymerases beta, kappa and iota may participate in ICL repair in mammalian cells. When a gap is generated at the 5’ side of an ICL during the processing of the ICL, DNA polymerase beta might have better chance to access the gap and bypass the ICL (Figures 3 and 4). Once DNA polymerases stalled at one nucleotide before an ICL, DNA polymerase iota might incorporate a nucleotide and DNA polymerase kappa may extend a primer end generated by DNA polymerase iota for TLS steps in ICL repair.
Our results showed that DNA polymerase beta incorporates a proper nucleotide, dAMP, opposite a psoralen ICL, which are modified thymines at a 5’-TA-3’ site (Figure 3D). It is interesting to note that DNA polymerase beta shows a similar preference of the incorporation opposite UV photo products, cyclobutane pyrimidine dimer (CPD) and the (6–4) photo product at a 5’-TT-3’ site. DNA polymerase beta incorporates the proper dAMP opposite these UV photoproducts and it also incorporates dCMP to a lesser extent32. DNA polymerase beta also incorporates the proper dAMP opposite another modified thymine, thymine glycol33, while it mis-incorporates TMP 34opposite the modified guanine, cisplatin GpG intra-strand cross-link35, 36. Because psoralen ICL, UV photo products and thymine glycol are modified at the 5, 6-position of thymines, the base pairing capacity of the modified thymine with adenine may not be influenced. Therefore, DNA polymerase beta that has a relatively high fidelity incorporates dAMP opposite these lesions. The bypass efficiency of a DNA lesion by a given DNA polymerase will be determined by the base pairing potential of the lesion and a proper fitting of the base pairing in the active pocket of the polymerase. DNA polymerase beta might be suitable for bypass of thymine-derived DNA damage.
In sharp contrast to non-damaged thymine in the template, DNA polymerase iota prefers to insert dAMP and TMP and seems to reject dGMP opposite from a psoralen ICL (Figure 5B). The mechanistic aspects of these incorporations opposite a psoralen ICL are not clear. It was proposed that the active site of DNA polymerase iota was narrow so an incorporation of purines using Watson-Crick base pairing is not efficient and DNA polymerase iota might use non-canonical base-pairings, Hoogsteen or wobble base-pairings34, 37, 38. Therefore, one possible mechanism of incorporation of a nucleotide opposite a psoralen ICL by DNA polymerase iota is the formation of a Hoogsteen base pair between an incoming nucleotide and a template psoralen ICL. We used 7-deaza-dATP to test this possibility. Thymine on a template forms a Hoogsteen base pair with an incoming dATP with the syn configuration through N6 and N7. The 7-Deaza-dATP cannot form a Hoogsteen base pair with a thymine. DNA polymerase iota incorporated 7-deaza-dAMP opposite a psoralen ICL as efficiently as dAMP (Supplemental Information 5). The results indicate that DNA polymerase iota does not use a Hoogsteen base pair, and probably uses wobble-base paring to incorporate dAMP opposite a psoralen ICL.
Because the 9-1-1 complex stimulates DNA polymerase beta24, we also examined whether the complex has any impact on DNA polymerase kappa and iota. The 9-1-1 complex slightly stimulated DNA polymerase iota (Supplemental Information 6A, lanes 1 and 2), but not DNA polymerase kappa (data not shown). The DNA polymerase iota did not alter the specificity of nucleotide incorporation opposite a psoralen ICL in the presence of the 9-1-1 complex (Supplemental Information 6B). Interestingly, the DNA polymerase iota incorporated dAMP and dCMP better opposite a non-modified thymine or cytosine on a template in the presence of the 9-1-1 complex (Supplemental Information 6C and 6D). We do not know the significance of these phenomena. However, the results might suggest that the 9-1-1 complex changes the efficiency of nucleotide incorporation by the DNA polymerase iota, depending on the structure of a base on a template.
Unlike DNA polymerase beta and iota, DNA polymerase kappa does not insert a nucleotide opposite a psoralen ICL, which is a part of a 12-mer oligonucleotide under the conditions we used. In addition to the bypass activities of minor groove adducts, DNA polymerase kappa is capable of extending a primer from mis-paired ends, suggesting its role as an extender in a two-step TLS of DNA damage26. It was shown that the sequential action of DNA polymerases iota and kappa enables efficient and error-free bypass of a minor groove adduct, γ-hydroxy-1, N2-propano-2’-deoxyguanosine12. DNA polymerase iota incorporates a correct nucleotide dCMP opposite the lesion and DNA polymerase kappa carries out an efficient extension of the primer generated. Analogous to this mechanism, we demonstrated that DNA polymerase kappa extends mis-paired primers generated by the incorporation of nucleotides opposite a psoralen ICL. Each dNTP was incorporated opposite from a psoralen ICL by an exonuclease-deficient Klenow (exo-) and the mis-paired primers were used as a template. DNA polymerase kappa was able to extend all four primers (Supplemental Information 7). Our results might indicate that DNA polymerase kappa plays two separate roles in TLS of ICLs, bypassing ICLs and extending a primer generated by other DNA polymerase.
Psoralen ICLs are known to induce T to A transversions at 5’-TA-3’ sites in mammalian cells39–42. These data demonstrate that a mis-incorporation of TMP opposite a psoralen ICL causes a T to A transversion. Our results showed that DNA polymerase iota preferentially incorporates dAMP and TMP opposite a psoralen ICL, while DNA polymerase beta and kappa incorporate dAMP opposite a psoralen ICL (Figures 5A and B, Table I). DNA polymerase iota might be a corresponding polymerase to induce psoralen-induced T to A transversions. It is important to investigate whether inactivation of DNA polymerase iota in mammals alters the mutational specificity induced by psoralen ICLs. “Psoralen plus UVA treatment” (PUVA treatment) has been a major therapy for psoriasis. Patients, in many cases, require repeated PUVA treatment for many years. The PUVA treated patients have a risk of developing squamous cell carcinoma. While the exact mechanism of the formation of squamous cell carcinoma is not known, psoralen-induced mutagenesis should contribute to the development significantly. The development of a specific inhibitor of the DNA polymerase iota might help reduce the risk of developing squamous cell carcinoma in PUVA treatment.
DNA interstrand cross-links are repaired by multiple pathways in mammalian cells. An intermediate structure, in which a TLS polymerase is involved, is unclear. Our model substrate DNA contains a four-nucleotide gap between a primer end and the 5’-end of cross-linked 12-mer through a psoralen. The primer is degraded significantly with the DNA polymerase beta−/− extracts and the addition of purified DNA polymerase beta averts these degradations (Figure 4). These results indicate that our substrate with a four-nucleotide gap was not recognized and utilized by other DNA polymerases under the conditions used. In contrast, DNA polymerase beta prefers a substrate with a gap,43 therefore, our substrate is suitable to investigate the role of DNA polymerase beta in TLS of ICLs. It has been reported that DNA polymerase kappa increases the efficiency of the bypass of N2-N2 guanine ICLs by shortening the size of the attached oligonucelotide9. When the ICL is in a two-nucleotide fragment, DNA polymerase kappa polymerizes this template with a similar efficiency to polymerize the non-damaged template. These results emphasize a significant contribution of the DNA structure of a template to bypass the ICLs. The identification of an intermediate structure for a TLS step in each ICL repair pathway should help understand which DNA polymerase is involved in the TLS step.
Supplementary Material
ACKNOWLEDGEMENTS
Funding Source Statement: This work was supported by the National Institutes of Health Grant GM080458 (to T.B.).
We thank Dr. Joan Sweasy (Yale University) for the MEF cells (DNA polymerase beta deletion and wild type), Dr. Leonid V. Gening (Institute of Molecular Genetics of Russian Academy of Science) for the purified recombinant human DNA polymerase beta and kappa, and Dr. Laura Lindsey-Boltz and Dr. Aziz Sancar (University of North Carolina at Chapel Hill) for the purified human 9-1-1 complex.
ABBREVIATIONS
- ICLs
DNA interstrand cross-links
- TLS
translesion DNA synthesis
- PolB
DNA polymerase beta
- PolK
DNA polymerase kappa
Footnotes
SUPPORTING INFORMATION
These materials are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45(1):23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30(5):1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho TV, Scharer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51(6):552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 4.Iwatsuki M, Mimori K, Yokobori T, Tanaka F, Tahara K, Inoue H, Baba H, Mori M. A platinum agent resistance gene, POLB, is a prognostic indicator in colorectal cancer. J Surg Oncol. 2009;100(3):261–266. doi: 10.1002/jso.21275. [DOI] [PubMed] [Google Scholar]
- 5.Yamtich J, Sweasy JB. DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta. 2010;1804(5):1136–1150. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804(5):1113–1123. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73(1):134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8(6):640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 9.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283(25):17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol Cell Biol. 2000;20(19):7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 12.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24(13):5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zietlow L, Bessho T. DNA polymerase I-mediated translesion synthesis in RecAindependent DNA interstrand cross-link repair in E. coli. Biochemistry. 2008;47(19):5460–5464. doi: 10.1021/bi702343y. [DOI] [PubMed] [Google Scholar]
- 14.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48(49):11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem. 2003;278(7):5250–5254. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- 16.Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat Struct Mol Biol. 2006;13(8):729–733. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- 17.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379(6561):183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 18.Makarova AV, Grabow C, Gening LV, Tarantul VZ, Tahirov TH, Bessho T, Pavlov YI. Inaccurate DNA synthesis in cell extracts of yeast producing active human DNA polymerase iota. PLoS One. 2011;6(1):e16612. doi: 10.1371/journal.pone.0016612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gening LV, Klincheva SA, Reshetnjak A, Grollman AP, Miller H. RNA aptamers selected against DNA polymerase beta inhibit the polymerase activities of DNA polymerases beta and kappa. Nucleic Acids Res. 2006;34(9):2579–2586. doi: 10.1093/nar/gkl326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A. 2001;98(20):11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klug AR, Harbut MB, Lloyd RS, Minko IG. Replication bypass of N2- deoxyguanosine interstrand cross-links by human DNA polymerases eta and iota. Chem Res Toxicol. 2012;25(3):755–762. doi: 10.1021/tx300011w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crespan E, Hubscher U, Maga G. Error-free bypass of 2-hydroxyadenine by human DNA polymerase lambda with Proliferating Cell Nuclear Antigen and Replication Protein A in different sequence contexts. Nucleic Acids Res. 2007;35(15):5173–5181. doi: 10.1093/nar/gkm568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447(7144):606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 24.Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich- Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32(11):3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 2004;3(11):1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci U S A. 2002;99(4):1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25(6):1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McVey M. Strategies for DNA interstrand crosslink repair: insights from worms, flies, frogs, and slime molds. Environ Mol Mutagen. 2010;51(6):646–658. doi: 10.1002/em.20551. [DOI] [PubMed] [Google Scholar]
- 29.Le Breton C, Hennion M, Arimondo PB, Hyrien O. Replication-fork stalling and processing at a single psoralen interstrand crosslink in Xenopus egg extracts. PLoS One. 2011;6(4):e18554. doi: 10.1371/journal.pone.0018554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134(6):969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30(4):1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Servant L, Cazaux C, Bieth A, Iwai S, Hanaoka F, Hoffmann JS. A role for DNA polymerase beta in mutagenic UV lesion bypass. J Biol Chem. 2002;277(51):50046–50053. doi: 10.1074/jbc.M207101200. [DOI] [PubMed] [Google Scholar]
- 33.Belousova EA, Maga G, Fan Y, Kubareva EA, Romanova EA, Lebedeva NA, Oretskaya TS, Lavrik OI. DNA polymerases beta and lambda bypass thymine glycol in gapped DNA structures. Biochemistry. 2010;49(22):4695–4704. doi: 10.1021/bi901792c. [DOI] [PubMed] [Google Scholar]
- 34.Choi JY, Lim S, Eoff RL, Guengerich FP. Kinetic analysis of base-pairing preference for nucleotide incorporation opposite template pyrimidines by human DNA polymerase iota. J Mol Biol. 2009;389(2):264–274. doi: 10.1016/j.jmb.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaisman A, Warren MW, Chaney SG. The effect of DNA structure on the catalytic efficiency and fidelity of human DNA polymerase beta on templates with platinum-DNA adducts. J Biol Chem. 2001;276(22):18999–19005. doi: 10.1074/jbc.M007805200. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann JS, Pillaire MJ, Maga G, Podust V, Hubscher U, Villani G. DNA polymerase beta bypasses in vitro a single d(GpG)-cisplatin adduct placed on codon 13 of the HRAS gene. Proc Natl Acad Sci U S A. 1995;92(12):5356–5360. doi: 10.1073/pnas.92.12.5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430(6997):377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 38.Kirouac KN, Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota. Embo J. 2009;28(11):1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage E, Drobetsky EA, Moustacchi E. 8-Methoxypsoralen induced mutations are highly targeted at crosslinkable sites of photoaddition on the non-transcribed strand of a mammalian chromosomal gene. Embo J. 1993;12(2):397–402. doi: 10.1002/j.1460-2075.1993.tb05671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang SC, Lin JG, Chiou CC, Chen LY, Yang JL. Mutation specificity of 8- methoxypsoralen plus two doses of UVA irradiation in the hprt gene in diploid human fibroblasts. Carcinogenesis. 1994;15(2):201–207. doi: 10.1093/carcin/15.2.201. [DOI] [PubMed] [Google Scholar]
- 41.Gunther EJ, Yeasky TM, Gasparro FP, Glazer PM. Mutagenesis by 8-methoxypsoralen and 5-methylangelicin photoadducts in mouse fibroblasts: mutations at crosslinkable sites induced by offoadducts as well as cross-links. Cancer Res. 1995;55(6):1283–1288. [PubMed] [Google Scholar]
- 42.Nataraj AJ, Black HS, Ananthaswamy HN. Signature p53 mutation at DNA cross-linking sites in 8-methoxypsoralen and ultraviolet A (PUVA)-induced murine skin cancers. Proc Natl Acad Sci U S A. 1996;93(15):7961–7965. doi: 10.1073/pnas.93.15.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem Rev. 2006;106(2):361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.