Abstract
Neurodegenerative disorders, such as Alzheimer’s disease, are associated with changes in multiple neuroimaging and biological measures. These may provide complementary information for diagnosis and prognosis. We present a multi-modality classification framework in which manifolds are constructed based on pairwise similarity measures derived from random forest classifiers. Similarities from multiple modalities are combined to generate an embedding that simultaneously encodes information about all the available features. Multimodality classification is then performed using coordinates from this joint embedding. We evaluate the proposed framework by application to neuroimaging and biological data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Features include regional MRI volumes, voxel-based FDG-PET signal intensities, CSF biomarker measures, and categorical genetic information. Classification based on the joint embedding constructed using information from all four modalities out-performs classification based on any individual modality for comparisons between Alzheimer’s disease patients and healthy controls, as well as between mild cognitive impairment patients and healthy controls. Based on the joint embedding, we achieve classification accuracies of 89% between Alzheimer’s disease patients and healthy controls, and 75% between mild cognitive impairment patients and healthy controls. These results are comparable with those reported in other recent studies using multi-kernel learning. Random forests provide consistent pairwise similarity measures for multiple modalities, thus facilitating the combination of different types of feature data. We demonstrate this by application to data in which the number of features differ by several orders of magnitude between modalities. Random forest classifiers extend naturally to multi-class problems, and the framework described here could be applied to distinguish between multiple patient groups in the future.
Keywords: Alzheimer’s disease, mild cognitive impairment, multi-modality classification, magnetic resonance imaging, positron emission tomography, cerebrospinal fluid biomarkers, genetics
1. Introduction
Changes in multiple biomarkers may provide complementary information for the diagnosis and prognosis of neurodegenerative disorders such as Alzheimer’s disease (AD). At present, the clinical diagnosis of AD is based on assessments of cognition and behaviour, which start to decline in the later disease stages. Recently published revisions to the diagnostic criteria (Albert et al., 2011; McKhann et al., 2011; Sperling et al., 2011) incorporate suggestions that biological and neuroimaging measures of structural and molecular changes in the brain may be better suited for the early detection of AD, as well as for monitoring its progression. Automated classification of individual patients based on multiple biomarkers could provide valuable support for clinicians, when considered alongside cognitive assessment scores and traditional visual image analysis. This could be particularly useful for monitoring the progression of patients with mild cognitive impairment (MCI), which is often a transitional stage between the cognitive decline associated with normal ageing, and that of established AD.
The neuropathological changes associated with the development of AD begin many years before cognitive symptoms become apparent. According to the amyloid cascade hypothesis, the disease process begins with the formation of insoluble β-amyloid (Aβ) plaques, whose presence triggers the hyperphosphorylation of tau protein, ultimately leading to cell death (Selkoe, 1991). Surrogate measures of the levels of Aβ and tau in the brain may be obtained from the cerebrospinal fluid (CSF). Various studies have shown reduced CSF Aβ and elevated CSF tau in AD patients compared with cognitively normal individuals (for example, Motter et al. (1995); Vandermeeren et al. (1993)). MCI patients tend to have CSF Aβ and tau levels between those expected of AD patients and healthy controls, with AD-like biomarker levels associated with an increased likelihood of progression to AD (Hansson et al., 2006). Positron emission tomography (PET) imaging with radiotracers for amyloid provides an alternative method for assessing intracranial Aβ deposition (Klunk et al., 2004).
PET imaging with the radiotracer [18F]-fluorodeoxyglucose (FDG) can be used to assess brain function in terms of the rate of cerebral glucose metabolism. Both AD and MCI are associated with significantly reduced glucose metabolism in affected regions, including temporal and parietal lobes, and the posterior cingulate cortex (Herholz et al., 2002; Langbaum et al., 2009). Reduced metabolism in AD patients is predictive of their cognitive decline and histopathological diagnosis (Hoffman et al., 2000; Silverman et al., 2001), and in MCI patients can predict their progression to AD (Anchisi et al., 2005). Changes in metabolism can be detected on FDG-PET before corresponding structural changes are visible (Aisen et al., 2010). FDG-PET-based classification techniques can discriminate both AD and MCI patients from healthy controls (Hinrichs et al., 2009).
The progressive structural damage caused by AD can be non-invasively assessed by using magnetic resonance imaging (MRI) to measure cerebral atrophy or ventricular expansion. Temporal lobe atrophy is closely associated with AD, and histological studies show that the hippocampus, amygdala and entorhinal cortex are particularly vulnerable to AD pathology (Braak and Braak, 1998). Accelerated hippocampal atrophy compared with healthy controls has been measured using MRI in both AD and MCI patients (Schuff et al., 2009; van de Pol et al., 2007). A recent study (Cuingnet et al., 2011) comparing ten MRI-based high-dimensional classification methods reported good results in discriminating between AD patients and healthy controls (up to 81% sensitivity and 95% specificity). Two methods relied on only hippocampal shape or volume, while the remainder were whole-brain approaches, using either cortical thickness measures, or voxel-wise tissue class probabilities. Four of the ten methods achieved slightly better accuracy than a random classifier in distinguishing MCI patients who later progressed to AD from MCI patients remaining stable over 18 months.
Although age is the most significant risk factor for AD (Rocca et al., 1991), genetic and environmental factors also play a role. The ApoE gene is the only one so far shown to be associated with sporadic late-onset AD (Dawbarn and Allen, 2007). There are three major alleles of this gene: ε2, ε3 and ε4. The most common is ε3, while ε4 is associated with an increased risk of developing AD, and ε2 with a reduced risk (Corder et al., 1993). More extensive AD pathology is generally observed in carriers of the ApoE ε4 allele than in non-carriers (Roses and Saunders, 1997). Genetics can therefore impact the biological and neuroimaging biomarkers. For example, AD carriers of the ApoE ε4 allele typically have reduced CSF Aβ, elevated CSF tau, and accelerated hippocampal atrophy on MRI compared with non-carriers (Tapiola et al., 2000; Schuff et al., 2009). Cognitively normal carriers of the ApoE ε4 allele display reduced glucose metabolism on FDG-PET in AD-typical regions (Langbaum et al., 2009).
The above biomarker patterns, however, are not specific to AD. For example, reduced CSF Aβ and elevated CSF tau are also associated with Lewy body dementia and vascular dementia (Andreasen et al., 2001; Blennow and Hampel, 2003). Temporal lobe atrophy is also observed in hippocampal sclerosis and temporal lobe epilepsy (Keihaninejad et al., 2012), and an AD-like metabolic pattern can also indicate Creutzfeldt-Jacob disease (Hoffman et al., 1990). It has been suggested that there may be some complementary information between modalities which can be exploited to produce more powerful combined classifiers (Walhovd et al., 2010; Landau et al., 2010). The potential utility of biomarker combinations is also supported by the recently revised diagnostic criteria (Albert et al., 2011; McKhann et al., 2011; Sperling et al., 2011). For research applications, these incorporate a biomarker-based probability of AD etiology which is highest when evidence of both amyloid deposition and neuronal degeneration or injury are observed. This support has resulted in an increasing interest in using multi-modality imaging and biological data for classification. Studies reporting multi-modality AD classification are far less common than those based on data from a single modality (Weiner et al., 2012). However, two independent studies (Zhang et al., 2011; Hinrichs et al., 2011) using multi-kernel learning report that classification based on multi-modality data is superior to that based on any individual modality. Classification accuracies of 93% between AD patients and healthy controls, and 76% between MCI patients and healthy controls were reported in Zhang et al. (2011), based on a combination of FDG-PET, MRI and CSF measures.
We present a framework for multi-modality classification based on pairwise similarity measures derived from random forests (Breiman, 2001). The similarities are used to construct a manifold representation from labelled training data and then to infer the clinical labels of test data mapped into this space. Classical multidimensional scaling (MDS) (Torgerson, 1952) is applied to learn a manifold on which to perform classification, resulting in the construction of a coordinate embedding in which the distances between points preserve their pairwise similarities. MDS is commonly used to provide low-dimensional visualisations of similarity relationships, including those derived from random forests (Hastie et al., 2011). Random forest-derived similarities have been successfully applied in unsupervised clustering tasks, for example those involving high-dimensional genetic or tissue microarray data (Shi and Horvath, 2006; Shi et al., 2005). Here, random forests are used to derive supervised similarity measures, with the aim of generating manifolds that are optimal for the task of clinical group discrimination. The proposed method facilitates the incorporation of multi-modality data, since similarities derived from several datasets may be combined to generate an embedding that simultaneously encodes information from all features.
Manifold learning techniques based on pairwise similarities between images have been applied in a variety of neuroimaging studies. For example, Laplacian eigenmaps (Belkin and Niyogi, 2003) have been used to generate an embedding of brain MR images based on similarities derived from overlaps of their structural segmentations (Aljabar et al., 2008). Isomap (Tenenbaum et al., 2000) has also been used to estimate the manifold structure of brain MR images, using distance measures based on nonrigid transformations between image pairs (Gerber et al., 2010). A framework for fusing manifold learning steps based on different pairwise similarity measures was presented in Aljabar et al. (2010). The method proposed here uses random forests to derive consistent pairwise similarity measures for multiple modalities, thus facilitating the combination of different types of feature data. We evaluate the proposed multi-modality classification framework by application to neuroimaging and biological data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). These data include FDG-PET and MR imaging data, CSF biomarker measures, and categorical genetic information. This work is an extension of preliminary studies involving neuroimaging data which have been previously presented at a workshop (Gray et al., 2011).
2. Methodology
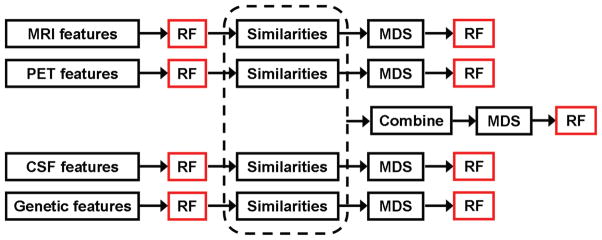
A schematic overview of the proposed approach is illustrated in Figure 1. A random forest classifier was applied to the feature data from each modality independently, firstly to obtain single-modality classification results for comparison, but also to derive the similarities required for manifold learning. The resulting similarity matrices were combined, and classical MDS was applied to generate a joint embedding for multi-modality classification. Full details of the data collection and feature extraction are presented in Section 3.1.
Figure 1.
Schematic overview of the proposed methodology. Each random forest (RF) step provides a classification result whose performance will be reported. Random forests are used both to derive the pairwise similarity measures for each feature set, and also to perform the single- and multi-modality classification experiments.
2.1. Random forests for classification
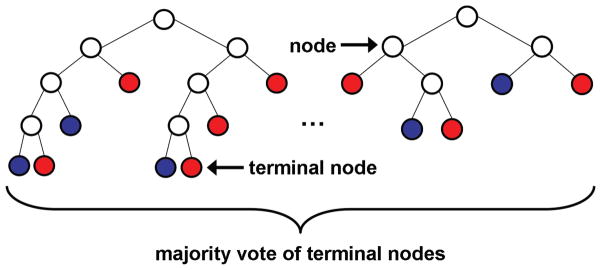
A random forest (Breiman, 2001) is an ensemble classifier consisting of many decision trees, where the final predicted class for a test example is obtained by combining the predictions of all individual trees, as illustrated in Figure 2. Random forests combine bootstrap aggregation (bagging) (Breiman, 1996) and random feature selection (Amit and Geman, 1997; Ho, 1998) to construct a collection of decision trees exhibiting controlled variation. We used the R implementation of random forests, a port of Leo Breiman and Adele Cutler’s original Fortran code by Andy Liaw and Matthew Wiener, version 4.6–6 (http://cran.r-project.org/web/packages/randomForest).
Figure 2.
Illustration of a random forest, showing two trees in detail. Each node is partitioned based on a single feature, and each branch ends in a terminal node. Terminal nodes provide a prediction for the class of a test example based on the path taken through the tree. The colour of a terminal node indicates its class prediction. The final predicted class for a test example is obtained by combining the predictions of all individual trees.
The training set for each individual tree in a random forest is constructed by sampling N examples at random with replacement from the N available examples in the dataset. This is known as bootstrap sampling, and bagging describes the aggregation of predictions from the resulting collection of trees. As a result of the bootstrap sampling procedure, approximately one third of the available N examples are not present in the training set of each tree. These are referred to as the “out-of-bag” data of the tree, for which internal test predictions can be made. By aggregating the predictions of the out-of-bag data across all trees, an internal estimate of the generalisation error of the random forest can be determined.
At each node in a tree, d ≪ D features are randomly selected from the D available features in the dataset, and the node is partitioned using the best possible binary split. A parent node np is partitioned into child nodes nl and nr according to the Gini index (Breiman et al., 1984), which measures the likelihood that an example would be incorrectly labelled if it were randomly classified according to the distrbution of labels within the node. For a binary split, the Gini index of a node n may be expressed as , where pc is the relative proportion of examples belonging to class c present in node n. The best possible binary split is the one which maximises the improvement in the Gini index ΔIG(np) = IG(np) − plIG(nl) − prIG(nr), where pl and pr are the proportions of examples in node np that are assigned to child nodes nl and nr, respectively. The Gini index can also be used to assess the relative importances of features for classification. A measure of the importance of an individual feature may be computed by summing the decreases in the Gini index occuring at all nodes in the forest which are partitioned based on that feature.
2.2. Manifold learning based on random forest similarities
Random forests can provide measures of the similarity between pairs of examples in the dataset. Each of the N examples is represented by a feature vector, all of which are passed down each tree in the forest. The similarities are initialised to zero, and if examples i and j finish in the same terminal node of a tree, their similarity sij is increased by one. The final pairwise similarity measures are normalised by the total number of trees in the forest.
The similarities thus form a N × N matrix with elements sij, and corresponding distance matrix elements dij = 1 − sij (Cox and Cox, 2001). Manifold learning techniques may be applied to find an appropriate coordinate embedding for the feature vectors, such that the distance relationships between them are preserved. A review of the most popular manifold learning techniques, as applied to medical imaging, is provided in Aljabar et al. (2012). MDS (Torgerson, 1952) is commonly used in conjunction with random-forest derived similarity measures for low-dimensional data visualisation, as well as unsupervised clustering tasks. In this work, MDS is applied to similarity measures derived in a supervised manner from random forests, with the aim of generating manifolds that are optimal for the task of clinical group discrimination.
Using MDS, the matrix of coordinates X is derived by performing an eigen-value decomposition on the matrix of scalar products
Retaining only the eigenvectors corresponding to the k largest-valued eigenvalues leads to a k-dimensional embedding for the data. A goodness-of-fit parameter G, describing the extent to which the selected k eigenvectors represent the full matrix, can be useful in selecting an appropriate dimensionality for the embedding. The measure of goodness-of-fit used in this work is given by
where the eigenvalues λj are sorted in decreasing order (Mardia et al., 1979).
To generate an embedding that simultaneously incorporated information from multiple modalities, a joint similarity matrix S was defined as a linear combination of the similarity matrices from each of the four modalities Si. Each modality was assigned a weighting factor αi, such that , where . To ensure the best combination of the four modalities for classification, the ai parameters were optimised as part of the training process. This was achieved by performing a grid-search within the training data to select the optimum modality weightings. The classifier was then trained using the selected set of parameters, before having its performance assessed on the test data.
3. Experiments and Results
We applied the proposed methodology to neuroimaging and biological data from 147 ADNI participants for whom baseline 1.5 T MRI, FDG-PET and CSF biomarker measures were available, as well as ApoE genotype information.
3.1. Neuroimaging and biological feature data
Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.ucla.edu). The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. A more detailed description of the ADNI study is provided in Appendix A.
We use imaging and biological data from 147 ADNI participants, including 37 AD patients, 75 MCI patients, and 35 healthy controls (HC). These represent all participants for whom ApoE genotype information and baseline 1.5 T MRI, FDG-PET and CSF measures of Aβ, tau and phosphorylated tau (ptau) were available, subject to the exclusions described in Appendix B. The MCI patients were divided into those who have progressed to AD (pMCI) and those whose diagnoses have so far remained stable (sMCI) based on changes in clinical status occurring over 20 ± 11 months (range 6 – 36). Clinical and demographic information for the study population is provided in Table 1.
Table 1.
Clinical and demographic information for the study population. For each group, the total number of subjects (N) and number of females (F) are shown, along with the average age, average mini mental state examination (MMSE) score, and clinical dementia rating (CDR).
| N(F) | Age (mean ± std. dev.) | MMSE Score (mean ± std. dev.) | CDR (%) | |||
|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | ||||
| AD | 37(14) | 76.8 ± 6.6 | 23.5 ± 2.0 | 0 | 49 | 51 |
| pMCI | 34(12) | 76.1 ± 7.3 | 26.5 ± 1.7 | 0 | 100 | 0 |
| sMCI | 41(12) | 75.7 ± 6.5 | 27.3 ± 1.8 | 0 | 100 | 0 |
| HC | 35(12) | 74.5 ± 5.2 | 28.9 ± 1.2 | 100 | 0 | 0 |
3.1.1. MRI feature extraction
Automatic whole-brain segmentations into 83 anatomical regions were prepared in native MRI space using multi-atlas propagation with enhanced registration (MAPER), an approach that has been previously described and validated for use in AD (Heckemann et al., 2010). The segmentations are available to download through the ADNI website, and full details of the procedure and morphometric analysis are presented in Heckemann et al. (2011). The required atlas data consisted of manually segmented T1-weighted MR volumes from 30 young, healthy adults, as described in Hammers et al. (2003). Protocols for the manual delineation are described in Hammers et al. (2003) and Gousias et al. (2008).
To match the requirements of MAPER, additional pre-processing was applied for brain extraction and tissue classification. For brain extraction, intracranial masks were generated using binary masks covering intracranial white and grey matter as the starting point, as described in Heckemann et al. (2011). The binary masks had been generated with a semi-automatic procedure as part of a separate project using MIDAS (Freeborough et al., 1997; Leung et al., 2011). Individual tissue probability maps for grey matter, white matter and CSF were obtained using FSL FAST (http://www.fmrib.ox.ac.uk/fsl). Binary maximum-probability tissue class labels were employed to mask the anatomical segmentations. All regions except ventricles, central structures, cerebellum and brainstem were masked with a grey matter label. Lateral ventricles were masked with a CSF label. Regional volumes were normalised by the intracranial volume, resulting in 83 volumetric region-based features per image.
3.1.2. FDG-PET feature extraction
Each FDG-PET image was motion-corrected as necessary, converted to a 30-minute static image, examined for major artefacts, and affinely aligned with the corresponding MRI using the Image Registration Toolkit (IRTK; http://www.doc.ic.ac.uk/~dr/software). An affine transformation was preferred over a rigid one because it can account for scaling or voxel size errors remaining after phantom correction of the MRI (Clarkson et al., 2009).
Using the “Segment” module of SPM5 (http://www.fil.ion.ucl.ac.uk/spm), each MRI was linearly and non-linearly deformed to the Montreal Neurological Institute (MNI) template (Ashburner and Friston, 2005). The resulting transformation parameters were applied to the MRI-space PET images using trilinear interpolation. The MNI-space PET images (voxel sizes 2×2×2mm) were then smoothed to a common isotropic spatial resolution of 8 mm full-width-at-half-maximum (FWHM) using scanner-specific kernels (Joshi et al., 2009), and then by an additional 8 mm FWHM isotropic Gaussian kernel. The smoothed images were intensity normalised to account for inter-subject variability in overall radioactivity using an independently-derived cluster of relatively preserved regions (Yakushev et al., 2009). The apriori brainmask available in SPM5 was thresholded at 50% probability, and applied to each normalised FDG-PET image to exclude voxels outside the brain. Signal intensities were extracted from all remaining voxels, resulting in 239,304 voxel-based features per image.
3.1.3. Biological features
The ADNI Biomarker Core provides biological data for the study participants. These data include CSF measures of Aβ, tau and ptau, as well as ApoE genotype information determined from a blood sample. Details of the biofluid collection and processing are provided in Trojanowski et al. (2010). The genetic feature data for each participant consisted of a single categorical variable describing their ApoE genotype. These data are summarised in Table 2.
Table 2.
Summarised biological and genetic information for the study population. CSF measures are expressed as mean ± std. dev., and genetic information is expressed as the proportion of each diagnostic group possessing each of the five ApoE allele pairs.
| CSF measures (pg/mL) | ApoE genotype (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Aβ | tau | ptau | ε3, ε3 | ε3, ε4 | ε4, ε4 | ε2, ε3 | ε2, ε4 | |
| AD | 141 ± 43 | 127 ± 64 | 46 ± 24 | 24 | 41 | 30 | 0 | 5 |
| pMCI | 152 ± 47 | 96 ± 41 | 37 ± 12 | 41 | 41 | 18 | 0 | 0 |
| sMCI | 167 ± 55 | 105 ± 81 | 35 ± 22 | 39 | 39 | 12 | 10 | 0 |
| HC | 208 ± 56 | 69 ± 28 | 26 ± 16 | 48 | 23 | 0 | 26 | 3 |
3.2. Classification experiments
Classification performance was assessed between three clinically relevant pairs of diagnostic groups (AD/HC, MCI/HC, pMCI/sMCI). Robust estimates of classifier performance were obtained using a stratified repeated random sampling approach. The mean accuracy, balanced accuracy, sensitivity and specificity were evaluated over 100 runs in which 75% of the data were randomly selected for training, with the remaining 25% used as test data. Balanced accuracy is the arithmetic mean of the sensitivity and specificity. This provides a more meaningful performance metric for groups of unequal sizes.
3.2.1. Single-modality classification results
A random forest classifier was applied to the feature data from each of the four modalities independently, and the single-modality classification results obtained are presented in Table 3.
Table 3.
Single-modality classification accuracy (acc.), balanced accuracy (bacc.), sensitivity (sens.) and specificity (spec.) based on the application of a random forest classifier to the original imaging and biological feature data. Results are expressed as mean (standard error).
| CSF | MRI | FDG-PET | Genetic | ||
|---|---|---|---|---|---|
| AD/HC | Acc. (%) | 76.5 (0.8) | 81.6 (0.8) | 86.0 (0.7) | 72.6 (0.9) |
| Bacc. (%) | 76.8 (1.3) | 81.8 (1.3) | 86.0 (1.2) | 72.7 (1.3) | |
| Sens. (%) | 73.0 (1.3) | 79.8 (1.3) | 86.8 (1.1) | 71.3 (1.3) | |
| Spec. (%) | 80.5 (1.3) | 83.8 (1.3) | 85.1 (1.3) | 74.1 (1.4) | |
| MCI/HC | Acc. (%) | 63.1 (0.8) | 66.9 (0.9) | 66.5 (0.8) | 73.8 (0.5) |
| Bacc. (%) | 63.8 (1.4) | 68.9 (1.3) | 66.9 (1.3) | 60.7 (0.9) | |
| Sens. (%) | 62.0 (1.1) | 63.7 (1.2) | 65.7 (1.1) | 94.7 (0.5) | |
| Spec. (%) | 65.5 (1.7) | 74.0 (1.4) | 68.1 (1.5) | 26.6 (1.2) | |
| pMCI/sMCI | Acc. (%) | 52.9 (1.0) | 55.1 (1.0) | 52.6 (1.0) | 47.3 (0.9) |
| Bacc. (%) | 53.5 (1.6) | 55.5 (1.7) | 53.1 (1.7) | 42.4 (2.4) | |
| Sens. (%) | 58.1 (1.6) | 59.1 (1.8) | 57.4 (2.0) | 32.1 (2.1) | |
| Spec. (%) | 48.8 (1.5) | 51.9 (1.6) | 48.8 (1.5) | 52.6 (2.7) |
Before performing classification experiments, the number of trees grown in each forest, t, and the number of features randomly selected at each tree node, d, had to be chosen. We used t = 5, 000 for all the experiments that follow, since stable estimates of the out-of-bag classification error were consistently observed for t ≳ 1, 000. We used for all experiments, following the recommendation of Liaw and Wiener (2002). The value of d was also consistently observed to have little effect on the out-of-bag classification error estimate.
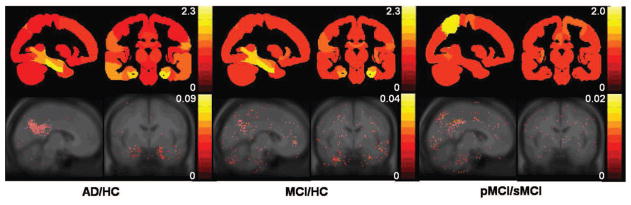
As described in Section 2.1, estimates of the relative importances of the features for classification may be extracted from the random forest. Feature importances for the two imaging modalities are shown in Figure 3. The most important features for MRI include volumes of the hippocampus, amygdala, and other medial temporal lobe structures. The most important features for FDG-PET include signal intensities of voxels located in the posterior cingulate gyrus, parietal lobe, posterior temporal lobe, and around the hippocampus.
Figure 3.
Feature importances for discriminating between the three clinical group pairs using region-based MRI (top), and voxel-based FDG-PET (bottom). For MRI, regional feature importances are superimposed onto slices of a maximum probability brain atlas which has been masked in the same way as the anatomical segmentations. For FDG-PET, important voxels are overlaid onto a MNI-space average MR image.
3.2.2. Single-modality similarity-based classification results
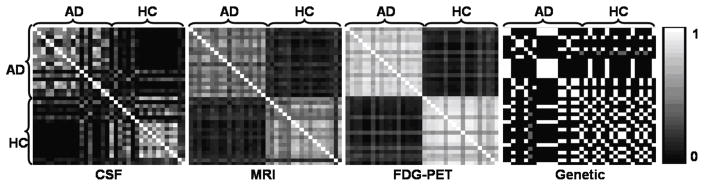
The random forest classifiers described in Section 3.2.1 were used to derive pairwise similarity measures for each of the four modalities, as described in Section 2.2. Examples of the similarity matrices are shown in Figure 4.
Figure 4.
Similarity matrices for each of the four modalities for the AD/HC experiment. Matrices are symmetric, and each entry represents the similarity between a pair of subjects based on the input feature data.
MDS was applied to each similarity matrix, and a goodness-of-fit value of 90% was used to determine an appropriate dimensionality for the resulting embeddings. A random forest classifier was then applied to the embedded feature data from each of the four modalities independently. The single-modality classification results obtained are presented in Table 4, along with the dimensionality of each embedding.
Table 4.
Single-modality classification accuracy (acc.), balanced accuracy (bacc.) sensitivity (sens.) and specificity (spec.) based on the application of a random forest classifier to the embedded imaging and biological feature data. Results are expressed as mean (standard error). The mean dimensionality of each embedding (k) is also shown.
| CSF | MRI | FDG-PET | Genetic | ||
|---|---|---|---|---|---|
| AD/HC | Acc. (%) | 76.1 (0.8) | 82.5 (0.7) | 86.4 (0.7) | 72.6 (0.9) |
| Bacc. (%) | 76.3 (1.3) | 82.1 (1.4) | 86.5 (1.2) | 72.7 (1.3) | |
| Sens. (%) | 72.8 (1.3) | 88.6 (1.2) | 85.8 (1.2) | 71.3 (1.3) | |
| Spec. (%) | 79.8 (1.4) | 75.6 (1.5) | 87.1 (1.3) | 74.1 (1.4) | |
| k | 13 | 22 | 9 | 2 | |
| MCI/HC | Acc. (%) | 61.7 (0.8) | 67.3 (1.0) | 53.5 (0.7) | 73.8 (0.5) |
| Bacc. (%) | 61.7 (1.3) | 69.1 (1.4) | 60.2 (1.2) | 60.7 (0.9) | |
| Sens. (%) | 61.6 (1.1) | 64.3 (1.3) | 42.3 (1.1) | 94.7 (0.5) | |
| Spec. (%) | 61.8 (1.5) | 73.9 (1.4) | 78.0 (1.3) | 26.6 (1.2) | |
| k | 25 | 47 | 35 | 2 | |
| pMCI/sMCI | Acc. (%) | 52.1 (1.0) | 58.4 (1.0) | 53.0 (1.0) | 43.5 (0.9) |
| Bacc. (%) | 52.7 (1.7) | 58.3 (1.7) | 52.8 (1.7) | 41.2 (2.4) | |
| Sens. (%) | 57.9 (1.6) | 56.9 (1.6) | 50.6 (1.8) | 27.4 (2.0) | |
| Spec. (%) | 47.5 (1.7) | 59.7 (1.8) | 54.9 (1.6) | 55.0 (2.7) | |
| k | 21 | 38 | 35 | 1 |
No consistent differences are observed between the balanced accuracies based on the embedded feature data shown in Table 4, and those based on the original feature data shown in Table 3.
3.2.3. Multi-modality similarity-based classification results
MDS was applied to the joint similarity matrix constructed using information from all four modalities, and a goodness-of-fit value of 90% was again used to determine an appropriate dimensionality for the resulting embedding. A random forest classifier was applied to the embedded feature data, and the multimodality classification results obtained are presented in Table 5. The balanced accuracies based on multi-modality classification are significantly (p < 0.01) higher than those based on any individual modality for both the AD/HC and MCI/HC experiments. For the pMCI/sMCI experiment, however, the balanced accuracy based on multi-modality classification is not significantly different from that based on MRI information alone. Table 5 additionally shows results based on the application of a random forest classifier to the combined feature set generated by a simple concatenation of features from all four modalities. These show no significant differences from the results based on FDG-PET alone.
Table 5.
Multi-modality classification accuracy (acc.), balanced accuracy (bacc.) sensitivity (sens.) and specificity (spec.) based on the application of a random forest classifier to the jointly embedded imaging and biological feature data, as well as the concatenated set of all features. Results are expressed as mean (standard error). The mean dimensionality of each embedding (k) is also shown.
| Combined embedding | Concatenated features | ||
|---|---|---|---|
| AD/HC | Acc. (%) | 89.0 (0.7) | 86.2 (0.7) |
| Bacc. (%) | 89.0 (1.2) | 87.1 (1.1) | |
| Sens. (%) | 87.9 (1.2) | 85.1 (1.4) | |
| Spec. (%) | 90.0 (1.1) | 86.1 (1.3) | |
| k | 18 | - | |
| MCI/HC | Acc. (%) | 74.6 (0.8) | 66.3 (0.8) |
| Bacc. (%) | 72.7 (0.8) | 65.3 (1.1) | |
| Sens. (%) | 77.5 (1.0) | 68.5 (1.5) | |
| Spec. (%) | 67.9 (1.7) | 66.9 (1.3) | |
| k | 20 | - | |
| pMCI/sMCI | Acc. (%) | 58.0 (0.9) | 53.0 (1.1) |
| Bacc. (%) | 57.9 (1.7) | 57.3 (1.9) | |
| Sens. (%) | 57.1 (1.8) | 49.6 (1.4) | |
| Spec. (%) | 58.7 (1.5) | 53.5 (1.7) | |
| k | 29 | - |
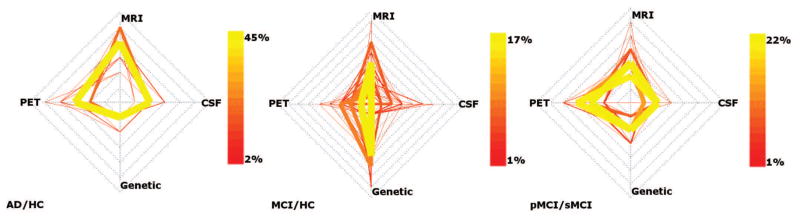
For each of the classification experiments, the distribution of modality weighting parameters selected over the 100 runs is illustrated in Figure 5.
Figure 5.
Cobweb plots showing the distribution of parameters selected over the 100 leave-25%-out runs for all three classification experiments. The four spokes of each plot represent the four modalities, and each coloured line connecting the four spokes represents a set of parameter values. The colour and weight of each line represents the percentage of runs in which the associated parameter set was selected.
4. Discussion and Conclusion
We have presented a framework for multi-modality classification based on pairwise similarity measures derived from random forests. The similarities are used to construct a manifold representation from labelled training data and then to infer the clinical labels of test data mapped into this space. Random forests are used to derive similarity measures in a supervised manner, with the aim of generating manifolds that are optimal for the task of clinical group discrimination. The proposed method facilitates the incorporation of multi-modality data, since consistent similarities may be derived from several datasets, and combined to generate an embedding that simultaneously encodes information from all features. Multi-modality classification may then be performed using coordinates from this joint embedding. The applicability of the method to a large and diverse dataset is demonstrated using imaging and biological information from the ADNI study. This includes FDG-PET and MR imaging data, CSF biomarker measures, and categorical genetic information.
Classification based on the joint embedding constructed using information from all four modalities is superior to classification based on any individual modality for comparisons between AD patients and HC, as well as between MCI patients and HC. This finding is in agreement with other recent studies (Zhang et al., 2011; Hinrichs et al., 2011), and supports previous suggestions that there is some complementary information between modalities which can be exploited to produce more powerful combined classifiers (Walhovd et al., 2010; Landau et al., 2010). Classification performance is commonly reported in terms of accuracy, but here we perform statistical comparisons between experiments based on the balanced accuracy. This provides a more meaningful performance metric for groups of unequal sizes. In terms of accuracy, we achieve 89% classification between AD patients and HC, and 75% between MCI patients and HC.
Direct comparisons with existing work are complicated by factors such as the inclusion of different subjects and modalities, as well as the use of different methods for feature extraction and cross-validation. In assessing the proposed framework, we consider comparisons between the single- and multimodality classification performance more important, since these are based on the same method, data, and processing pipeline. Our multi-modality classification results are, however, comparable with those reported in other recent studies based on ADNI data. Zhang et al. (2011) combine MRI, FDG-PET and CSF data to achieve classification accuracies of 93% between AD patients and HC, and 76% between MCI patients and HC. Hinrichs et al. (2011) combine MRI, FDG-PET, CSF, genetic and cognitive data to achieve an accuracy of 92% between AD patients and HC. Both studies employ multi-kernel classifiers based on support vector machines, which are relatively well-established in the field (Weiner et al., 2012). Random forests have not been extensively applied in neuroimaging research, although they are becoming more widely used for applications including classification (Hope et al., 2008; Chincarini et al., 2011) and segmentation (Geremia et al., 2011; Iglesias et al., 2011). Classification of neurodegenerative disease based on the combination of imaging and non-imaging information is therefore a new application for random forests, and we are encouraged by the fact that our results are comparable with those of more established techniques.
The ability of random forests to extract consistent pairwise similarity measures for multiple modalities facilitates the combination of different types of feature data. We demonstrate this using datasets in which the number of features differ between modalities by several orders of magnitude. A simple concatenation of all features was shown not to optimally combine these data (Table 5), since the high-dimensional FDG-PET features dominate the results.
In previous work (Gray et al., 2011), we obtained comparable multi-modality classification results using only information extracted from the two neuroimaging modalities (accuracies of 90% for AD/HC, and 76% for MCI/HC). The lack of improvement over these previously reported results is likely to be attributable to the considerable reduction in size of the subject group as a result of our requirement for CSF biomarker information. The subject group for this study is approximately half the size of that used in Gray et al. (2011). The present work additionally employs a more robust form of cross-validation, using a stratified repeated random sampling approach, as opposed to the single round of ten-fold cross-validation employed in Gray et al. (2011).
In the context of a neuroimaging application, estimates of the importances of the features for classification are valuable because this allows assessment of whether the features that contribute most to the classifier correspond to regions or structures with a biologically plausible connection to pathology. In this work, the most important features for discriminating between clinical groups correspond with those known to be visibly affected in AD on both FDG-PET and structural MR imaging (Hampel et al., 2008; Patwardhan et al., 2004). Important features for distinguishing between AD patients and HC are more localised to affected areas, with the more challenging distinctions between MCI patients and HC, or pMCI and sMCI patients, requiring features spread across a larger part of the brain. The motivation for extracting region-based features from the MR images and voxel-based features from the FDG-PET images was to demonstrate that these two different types of imaging features could be readily combined using the proposed method.
The lack of significant difference between classification performance based on the original feature data and that based on the embedding coordinates for each individual modality is expected, since a random forest is already a nonlinear classifier. However, a difference is observed for the comparison between MCI patients and HC based on the voxel-wise FDG-PET features. This may be due to the inhomogeneity of the MCI group, which comprises both pMCI and sMCI patients. It is possible that the high-dimensional voxel-based FDG-PET features are sensitive to differences in the pattern of glucose metabolism between these two groups, resulting in a reduced classification performance based on the associated embedding coordinates.
Random forests are ensemble-based classifiers that are often applied to high-dimensional datasets. In this work, random forests are also applied to low-dimensional biological data so that consistent pairwise similarity measures may be obtained for all modalities. In the case of a single feature, such as the categorical genetic information, a random forest reduces to bootstrap aggregation.
Visualisation of the parameters selected to combine similarities for multimodality classification (Figure 5) provides some interesting insights into the relationships among the modalities. The figure indicates the optimum way in which to combine MRI, FDG-PET, CSF and genetic information within the framework described. For distinguishing between AD patients and HC, for example, it appears that FDG-PET and MR imaging features provide the most complementary information. For distinguishing MCI patients from HC, genetic information appears to have a relatively high importance. However, these indications may not necessarily have a direct biological interpretation, and are likely not sufficient to draw any firm conclusions about the relative importances of the four modalities. The optimum modality weightings for distinguishing between AD patients and HC are more stable than for the other two experiments. The weighting parameters selected for distinguishing MCI patients from HC may be more variable because the heterogeneity of the MCI group makes their selection dependent on the proportions of pMCI and sMCI patients in the training set. The selected modality weightings are not equal for any experiment, supporting the need to optimise the relative contributions of the four modalities for classification. The figure suggests an interesting avenue for further research, in that estimates of inter-modality correlations could help to determine the amount of complementary information between them. This could facilitate decisions on how to acquire the maximum amount of diagnostically relevant information for a patient using a minimum number of assessments.
The classification performance between pMCI and sMCI patients is not significantly improved by combining multi-modality information in this study. Other studies which apply multi-modality classification to this challenging problem have incorporated longitudinal information to improve performance (Hinrichs et al., 2011; Zhang et al., 2012). Our previous work based on FDG-PET (Gray et al., 2012) has also shown that incorporating longitudinal information can be beneficial to improve the ability to distinguish between these two groups. This will be an important issue to address in the future. It is important to consider, however, that progression from MCI to AD occurs at a rate of 10–15% per year (Petersen et al., 1999), with up to 80% of MCI patients developing AD over a six year period (Petersen, 2004). Longer clinical follow-up is therefore required to properly assess the utility of any classification method in separating pMCI from sMCI patients. This may be made possible by the continuation of the ADNI study in the form of ADNI-GO and ADNI-2. Further information about these studies is available via the ADNI website (http://adni.loni.ucla.edu/about/about-the-study/).
We have identified several areas for further research. Methodologically, the approach is generalisable, in that the manifold learning step could be performed using an alternative technique, and similarities could be combined using more sophisticated formulae. We applied MDS for manifold learning in this work because its application to random forest-derived similarities is straightforward, and relatively common in the literature (Hastie et al., 2011). However, MDS may not be the optimal embedding approach, and there is potential to improve the performance of this step by pursuing a thorough exploration of alternative techniques. Similarly, we chose to additively combine similarity matrices for simplicity, and there is potential for improvement by incorporating a more sophisticated formula. Random forest classifiers extend naturally to multi-class problems, and the framework described here could be applied to distinguish between multiple patient groups in the future. Exploration of the multi-class setting would better represent a real-world clinical scenario, including patients with other forms of dementia. In addition, the implementation of random forests used in this work could be modified to produce uncertainty information about the predicted diagnostic labels. This may be more useful to clinicians than a simple binary prediction, and could be achieved by having each leaf node store a probabilistic distribution of labels, rather than a point estimate. Criminisi et al. (2012), for example, describes this and other extensions to the original random forests algorithm, and presents a unified model of random decision forests for classification, regression, density estimation, manifold learning, and semi-supervised learning. Further investigations into different data fusion strategies may also be interesting. Although the feature level fusion approach proposed in this work can capture interdependencies between modalities, the application of a data level fusion technique to the neuroimaging data may be advantageous in terms of capturing local inter-modality correlations.
Supplementary Material
Research Highlights.
multi-modal classification based on pairwise similarities derived from random forests
framework evaluated by application to neuroimaging and biological data from ADNI
random forests provide consistent pairwise similarity measures for various modalities
classification based on MR volumes, FDG-PET intensities, CSF biomarkers, genetic data
multi-modality classification out-performs that based on any individual modality
Acknowledgments
The authors would like to thank Igor Yakushev (University of Mainz, Germany) for provision of the reference cluster image used for FDG-PET intensity normalisation, as well as Casper Nielsen and Kelvin Leung (Dementia Research Centre, University College London) for provision of the MIDAS brain masks used for MRI anatomical segmentation. Katherine R. Gray received a studentship from the Engineering and Physical Sciences Research Council. Rolf A. Heckemann was supported by a research grant from the Dunhill Medical Trust. The Image Registration Toolkit was used under licence from Ixico Ltd.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, John-son and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Appendix A. The Alzheimer’s Disease Neuroimaging Initiative
The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organisations, as a $60 million, five-year public-private partnership. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California - San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations. Subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit approximately 200 cognitively normal older individuals to be followed for three years, 400 MCI patients to be followed for three years, and 200 early AD patients to be followed for two years. Further up-to-date information is available on the ADNI information website (http://www.adni-info.org).
Appendix B. Exclusions
In this work, we use imaging and biological data from 147 ADNI participants. These represent all participants for whom ApoE genotype information and baseline 1.5 T MRI, FDG-PET and CSF measures of Aβ, tau and phosphorylated tau were available, subject to the following exclusions. FDG-PET images acquired using either the Siemens HRRT or BioGraph HiRez scanners were excluded due to differences in the pattern of FDG metabolism that were discovered during the ADNI quality control process. Further information is available on the ADNI PET Core website (http://www.loni.ucla.edu/twiki/bin/view/ADNI/ADNIPETCore). A small number of subjects (n = 14) whose imaging data could not be processed as required, or whose diagnosis did not clearly fall into one of the four diagnostic groups (AD, pMCI, sMCI, HC), were also excluded. ADNI subject identifiers for these participants are provided as supplementary material, along with more detailed reasons for their exclusion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, Walter S, Trojanowski JQ, Shaw LM, Beckett LA, Jack CR, Jr, Jagust W, Toga AW, Saykin AJ, Morris JC, Green RC, Weiner MW the Alzheimer’s Disease Neuroimaging Initiative. Clinical core of the Alzheimer’s disease neuroimaging initiative: progress and plans. Alzheimer’s and Dementia. 2010;6:239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC, Snyder PJ, Carrillo MC, Thies B, Phelps CH. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s and Dementia. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljabar P, Rueckert D, Crum WR. Automated morphological analysis of magnetic resonance brain imaging using spectral analysis. Neuroimage. 2008;43:225–235. doi: 10.1016/j.neuroimage.2008.07.055. [DOI] [PubMed] [Google Scholar]
- Aljabar P, Wolz R, Rueckert D. Manifold learning for medical image registration, segmentation, and classification. In: Suzuki K, editor. Machine learning in computer-aided diagnosis: medical imaging intelligence and analysis. IGI Global; 2012. [Google Scholar]
- Aljabar P, Wolz R, Srinivasan L, Counsell S, Boardman JP, Murgasova M, Doria V, Rutherford M, Edwards AD, Hajnal JV, Rueckert D. Combining morphological information in a manifold learning framework: application to neonatal MRI. 13th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI’10); 2010. pp. 1–8. [DOI] [PubMed] [Google Scholar]
- Amit Y, Geman D. Shape quantization and recognition with randomized trees. Neural Computation. 1997;9:1545–1588. [Google Scholar]
- Anchisi D, Borroni B, Franceschi M, Kerrouche N, Kalbe E, Beuthien-Beumann B, Cappa S, Lenz O, Ludecke S, Marcone A, Mielke R, Ortelli P, Padovani A, Pelati O, Pupi A, Scarpini E, Weisenbach S, Herholz K, Salmon E, Holthoff V, Sorbi S, Fazio F, Perani D. Heterogeneity of brain glucose metabolism in mild cognitive impairment and clinical progression to Alzheimer disease. Archives of Neurology. 2005;62:1728–1733. doi: 10.1001/archneur.62.11.1728. [DOI] [PubMed] [Google Scholar]
- Andreasen N, Minthon L, Davidsson P, Vanmechelen E, Vanderstichele H, Winblad B, Blennow K. Evaluation for CSF-tau and CSF-Abeta42 as diagnostic markers for Alzheimer disease in clinical practice. Archives of Neurology. 2001;58:373–379. doi: 10.1001/archneur.58.3.373. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Computation. 2003;15:1373–1396. [Google Scholar]
- Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease. Lancet Neurology. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Evolution of neuronal changes in the course of Alzheimer’s disease. Journal of Neural Transmission Supplement. 1998;53:127–140. doi: 10.1007/978-3-7091-6467-9_11. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Machine Learning. 1996;24:123–140. [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Wadsworth; Belmont: 1984. [Google Scholar]
- Chincarini A, Bosco P, Calvini P, Gemme G, Esposito M, Olivieri C, Rei L, Squarcia S, Rodriguez G, Bellotti R, Cerello P, De Mitri I, Retico A, Nobili F. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage. 2011;58:469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- Clarkson MJ, Ourselin S, Nielsen C, Leung KK, Barnes J, Whitwell JL, Gunter JL, Hill DLG, Weiner MW, Jack CR, Jr, Fox NC the Alzheimer’s Disease Neuroimaging Initiative. Comparison of phantom and registration scaling corrections using the ADNI cohort. Neuroimage. 2009;47:1506–1513. doi: 10.1016/j.neuroimage.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cox TF, Cox MAA. Multidimensional scaling. Chapman and Hall; 2001. [Google Scholar]
- Criminisi A, Shotton J, Konukoglu E. Decision forests: a unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Foundations and Trends in Computer Graphics and Vision. 2012;7:81–227. [Google Scholar]
- Cuingnet R, Gérardin E, Tessieras J, Auzias G, Lehéricy S, Habert M, Chupin M, Benali H, Colliot O the Alzheimer’s Disease Neuroimaging Initiative. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Dawbarn D, Allen SJ, editors. Neurobiology of Alzheimer’s disease. 3rd edition Oxford University Press; New York: 2007. [Google Scholar]
- Freeborough PA, Fox NC, Kitney RI. Interactive algorithms for the segmentation and quantitation of 3-D MRI brain scans. Computer Methods and Programs in Biomedicine. 1997;53:15–25. doi: 10.1016/s0169-2607(97)01803-8. [DOI] [PubMed] [Google Scholar]
- Gerber S, Tasdizen T, Fletcher TP, Whitaker R the Alzheimer’s Disease Neuroimaging Initiative. Manifold modeling for brain population analysis. Medical Image Analysis. 2010;14:643–653. doi: 10.1016/j.media.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia E, Clatz O, Menze BH, Konukoglu E, Criminisi A, Ayache N. Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. Neuroimage. 2011;57:378–390. doi: 10.1016/j.neuroimage.2011.03.080. [DOI] [PubMed] [Google Scholar]
- Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, Hammers A. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage. 2008;40:672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D. Random forest-based manifold learning for classification of imaging data in dementia. Machine Learning in Medical Imaging Workshop at 14th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI’11); 2011. pp. 159–166. [Google Scholar]
- Gray KR, Wolz R, Heckemann RA, Aljabar P, Hammers A, Rueckert D for the Alzheimer’s Disease Neuroimaging Initiative. Multi-region analysis of longitudinal FDG-PET for the classification of Alzheimer’s disease. Neuroimage. 2012;60:221–229. doi: 10.1016/j.neuroimage.2011.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Human Brain Mapping. 2003;19:224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H, Burger K, Teipel SJ, Bokde AL, Zetterberg H, Blennow K. Core candidate neurochemical and imaging biomarkers of Alzheimer’s disease. Alzheimer’s and Dementia. 2008;4:38–48. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Hansson O, Zetterberg H, Buchlave P, Londos E, Blennow K, Minthon L. Association between CSF biomarkers and incipient Alzheimer’s disease in patients with mild cognitive impairment: a follow-up study. Lancet Neurology. 2006;5:228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. Springer; New York: 2011. corrected 5th printing of 2nd edition. [Google Scholar]
- Heckemann RA, Keihaninejad S, Aljabar P, Gray KR, Nielsen C, Rueckert D, Hajnal JV, Hammers A the Alzheimer’s Disease Neuroimaging Initiative. Automatic morphometry in Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;56:2024–2037. doi: 10.1016/j.neuroimage.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A the Alzheimer’s Disease Neuroimaging Initative. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage. 2010;51:221–227. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- Herholz K, Salmon E, Perani D, Baron JC, Holthoff V, Frolich L, Schonknecht P, Ito K, Mielke R, Kalbe E, Zundorf G, Delbeuck X, Pelati O, Anchisi D, Fazio F, Kerrouche N, Desgranges B, Eustache F, Beuthien-Baumann B, Menzel C, Schroder J, Kato T, Arahata Y, Henze M, Heiss WD. Discrimination between Alzheimer dementia and controls by automated analysis of multicenter FDG PET. Neuroimage. 2002;17:302–316. doi: 10.1006/nimg.2002.1208. [DOI] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Mukherjee L, Xu GF, Chung MK, Johnson SC the Alzheimer’s Disease Neuroimaging Initiative. Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. Neuroimage. 2009;48:138–149. doi: 10.1016/j.neuroimage.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC the Alzheimer’s Disease Neuroimaging Initiative. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. The random subspace method for constructing decision forests. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1998;20:832–844. [Google Scholar]
- Hoffman EJ, Cutler PD, Digby WM, Mazziotta JC. 3-D phantom to simulate cerebral blood-flow and metabolic images for PET. IEEE Transactions on Nuclear Science. 1990;37:616–620. [Google Scholar]
- Hoffman JM, Welsh-Bohmer KA, Hanson M, Crain B, Hulette C, Earl N, Coleman RE. FDG PET imaging in patients with pathologically verified dementia. Journal Of Nuclear Medicine. 2000;41:1920–1928. [PubMed] [Google Scholar]
- Hope TA, Gregson PH, Linney NC, Schmidt MH, Abdolell M. Selecting and assessing quantitative early ultrasound texture measures for their association with cerebral palsy. IEEE Transactions on Medical Imaging. 2008;27:228–236. doi: 10.1109/TMI.2007.906089. [DOI] [PubMed] [Google Scholar]
- Iglesias JE, Liu CY, Thompson PM, Tu Z. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Transactions on Medical Imaging. 2011;30:1617–1634. doi: 10.1109/TMI.2011.2138152. [DOI] [PubMed] [Google Scholar]
- Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage. 2009;46:154–159. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keihaninejad S, Heckemann RA, Gousias IS, Hajnal JV, Duncan JS, Aljabar P, Rueckert D, Hammers A. Classification and lateralization of temporal lobe epilepsies with and without hippocampal atrophy based on whole-brain automatic MRI segmentation. PLoS ONE. 2012;7:e33096. doi: 10.1371/journal.pone.0033096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang YM, Blomqvist G, Holt DP, Bergstrom M, Savitcheva I, Huang GF, Estrada S, Ausén B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, Långström B. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Annals of Neurology. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- Landau SM, Harvey D, Madison CM, Reiman EM, Foster NL, Aisen PS, Petersen RC, Shaw LM, Trojanowski JQ, Jack CR, Jr, Weiner MW, Jagust WJ. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75:230–238. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbaum JBS, Chen K, Lee W, Reschke C, Bandy D, Fleisher AS, Alexander GE, Foster NL, Weiner MW, Koeppe RA, Jagust WJ, Reiman EM the Alzheimer’s Disease Neuroimaging Initiative. Categorical and correlational analyses of baseline fluorodeoxyglucose positron emission tomography images from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Neuroimage. 2009;45:1107–1116. doi: 10.1016/j.neuroimage.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Barnes J, Modat M, Ridgway GR, Bartlett JW, Fox NC, Ourselin S the Alzheimer’s Disease Neuroimaging Initiative. Brain MAPS: an automated, accurate and robust brain extraction technique using a template library. Neuroimage. 2011;55:1091–1108. doi: 10.1016/j.neuroimage.2010.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- Mardia KV, Kent JT, Bibby JM. Multivariate analysis. 5th edition Academic Press; London: 1979. [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack, CR, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carrillo MC, Thies B, Weintraub S, Phelps CH. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s and Dementia. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motter R, Vigopelfrey C, Kholodenko D, Barbour R, Johnsonwood K, Galasko D, Chang L, Miller B, Clark C, Green R, Olson D, Southwick P, Wolfert R, Munroe B, Lieberburg I, Seubert P, Schenk D. Reduction of beta-amyloid peptide42 in the cerebrospinal fluid of patients with Alzheimer’s disease. Annals of Neurology. 1995;38:643–648. doi: 10.1002/ana.410380413. [DOI] [PubMed] [Google Scholar]
- Patwardhan MB, McCrory DC, Matchar DB, Samsa GP, Rutschmann OT. Alzheimer disease: operating characteristics of PET – a meta-analysis. Radiology. 2004;231:73–80. doi: 10.1148/radiol.2311021620. [DOI] [PubMed] [Google Scholar]
- Petersen RC. Mild cognitive impairment as a diagnostic entity. Journal of Internal Medicine. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Archives of Neurology. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- van de Pol LA, van der Flier WM, Korf ESC, Fox NC, Barkhof F, Scheltens P. Baseline predictors of rates of hippocampal atrophy in mild cognitive impairment. Neurology. 2007;69:1491–1497. doi: 10.1212/01.wnl.0000277458.26846.96. [DOI] [PubMed] [Google Scholar]
- Rocca WA, Hofman A, Brayne C, Breteler MMB, Clarke M, Copeland JRM, Dartigues JF, Engedal K, Hagnell O, Heeren TJ, Jonker C, Lindesay J, Lobo A, Mann AH, Mls PK, Morgan K, O’Connor DW, Droux AdS, Sulkava R, Kay DWK, Amaducci L. Frequency and distribution of Alzheimer’s disease in Europe: a collaborative study of 1980–1990 prevalence findings. The EURODEM-Prevalence Research Group. Annals of Neurology. 1991;30:381–390. doi: 10.1002/ana.410300310. [DOI] [PubMed] [Google Scholar]
- Roses AD, Saunders AM. ApoE, Alzheimer’s disease, and recovery from brain stress. Cerebrovascular Pathology in Alzheimer’s Disease. 1997;826:200–212. doi: 10.1111/j.1749-6632.1997.tb48471.x. [DOI] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW the Alzheimer’s Disease Neuroimaging Initiative. MRI of hippocampal volume loss in early Alzheimer?s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. The molecular pathology of Alzheimer’s disease. Neuron. 1991;6:487–498. doi: 10.1016/0896-6273(91)90052-2. [DOI] [PubMed] [Google Scholar]
- Shi T, Horvath S. Unsupervised learning with random forest predictors. Journal of Computational and Graphical Statistics. 2006;15:118–138. [Google Scholar]
- Shi T, Seligson D, Belldegrun AS, Palotie A, Horvath S. Tumor classification by tissue microarray profiling: random forest clustering applied to renal cell carcinoma. Modern Pathology. 2005;18:547–557. doi: 10.1038/modpathol.3800322. [DOI] [PubMed] [Google Scholar]
- Silverman DHS, Small GW, Chang CY, Lu CS, de Aburto MAK, Chen W, Czernin J, Rapoport SI, Pietrini P, Alexander GE, Schapiro MB, Jagust WJ, Hoffman JM, Welsh-Bohmer KA, Alavi A, Clark CM, Salmon E, de Leon MJ, Mielke R, Cummings JL, Kowell AP, Gambhir SS, Hoh CK, Phelps ME. Positron emission tomography in evaluation of dementia: Regional brain metabolism and long-term outcome. JAMA - Journal Of The American Medical Association. 2001;286:2120–2127. doi: 10.1001/jama.286.17.2120. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s and Dementia. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapiola T, Pirttila T, Mehta PD, Alafuzoff I, Lehtovirta M, Soininen H. Relationship between apoE genotype and CSF beta-amyloid (1–42) and tau in patients with probable and definite Alzheimer’s disease. Neurobiology of Aging. 2000;21:735–740. doi: 10.1016/s0197-4580(00)00164-0. [DOI] [PubMed] [Google Scholar]
- Tenenbaum J, de Silva V, Langford J. A global geometric framework for non-linear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- Torgerson WS. Multidimensional scaling: I. Theory and method. Psychometrika. 1952;17:401–419. doi: 10.1007/BF02289530. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter WZ, Weiner MW, Jack CR, Jr, Jagust W, Toga AW, Lee VMY, Shaw LM. Update on the biomarker core of the Alzheimer’s Disease Neuroimaging Initiative subjects. Alzheimer’s and Dementia. 2010;6:230–238. doi: 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeeren M, Mercken M, Vanmechelen E, Six J, Vandevoorde A, Martin JJ, Cras P. Detection of tau proteins in normal and Alzheimer’s disease cerebrospinal fluid with a sensitive sandwich enzyme-linked immunosorbent assay. Journal of Neurochemistry. 1993;61:1828–1834. doi: 10.1111/j.1471-4159.1993.tb09823.x. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Dale AM, McEvoy LK, Brewer J, Karow DS, Salmon DP, Fennema-Notestine C. Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiology of Aging. 2010;31:1107–1121. doi: 10.1016/j.neurobiolaging.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Saykin AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ the Alzheimer’s Disease Neuroimaging Initiative. The Alzheimers Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer’s and Dementia. 2012;8:S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushev I, Hammers A, Fellgiebel A, Schmidtmann I, Scheurich A, Buchholz HG, Peters J, Bartenstein P, Lieb K, Schreckenberger M. SPM-based count normalization provides excellent discrimination of mild Alzheimer’s disease and amnestic mild cognitive impairment from healthy aging. Neuroimage. 2009;44:43–50. doi: 10.1016/j.neuroimage.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Zhang D, Shen D the Alzheimer’s Disease Neuroimaging Initiative. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS ONE. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D the Alzheimer’s Disease Neuroimaging Initiative. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.