Summary
Living cells use cell surface proteins, such as mechanosensors, to constantly sense and respond to their environment. However, the way in which these proteins respond to mechanical stimuli and assemble into large complexes remains poorly understood at the molecular level. In the past years, atomic force microscopy (AFM) has revolutionized the way in which biologists analyze cell surface proteins to molecular resolution. In this Commentary, we discuss how the powerful set of advanced AFM techniques (e.g. live-cell imaging and single-molecule manipulation) can be integrated with the modern tools of molecular genetics (i.e. protein design) to study the localization and molecular elasticity of individual mechanosensors on the surface of living cells. Although we emphasize recent studies on cell surface proteins from yeasts, the techniques described are applicable to surface proteins from virtually all organisms, from bacteria to human cells.
Key words: Atomic force microscopy, AFM, Cell adhesion, Cell surface, Cell imaging, Mechanosensing, Microscopy, Single molecule
Introduction
Proteins located on the cell surface have pivotal roles in adhesion, sensing of the environment, signaling, communication, transport, energy transformation, embryonic and tissue development, tumour metastasis and microbial infection (Sheetz, 2001; Discher et al., 2005; Vogel and Sheetz, 2006; Lecuit and Lenne, 2007). These complex functions rely on the assembly and interactions of specific proteins, including mechanosensors and cell adhesion molecules. Whereas much is known about the molecular biology of such surface proteins, their biophysical properties, such as their mechanics and clustering in the cell membrane, remain poorly understood at the molecular level.
Cells can react to external cues by engaging signal transduction pathways, which detect environmental changes at the cell surface and trigger the appropriate intracellular responses. Frequently, these signaling pathways are initiated by sensor proteins that span the plasma membrane and ultimately trigger a change in gene expression and, consequently, in the cell’s proteome (McIntosh et al., 2009; Rodicio and Heinisch, 2010). Such sensor proteins either bind specific ligands (e.g. hormones, adhesion molecules, etc.) or detect mechanical forces and related physical stimuli. Hence, mechanosensing and mechanotransduction, which convert these mechanical forces into biochemical signals, have key roles in regulating processes such as cell growth, differentiation, cell shape and cell death (Schwartz, 2009; Vogel and Sheetz, 2006; Brown and Discher, 2009).
Cell adhesion molecules also have key roles in mediating cellular processes such as cell–cell communication (Dalva et al., 2007), tissue development (Morgan et al., 2007), inflammation (Weber et al., 2007), cancer (Gray-Owen and Blumberg, 2006) and microbial infection (Verstrepen et al., 2004; Kolter and Greenberg, 2006; Telford et al., 2006; Sokurenko et al., 2008). There is growing evidence that the force-induced deformation of such proteins has an important role in modulating their cellular function. Prominent examples of such processes are ‘catch bonds’, i.e. bonds between receptors and ligands that are strengthened by tensile mechanical force (Sokurenko et al., 2008), as well as the force-induced exposure of cryptic peptide sequences, as observed in, for example, fibronectin and integrin (Vogel and Sheetz, 2006; Brown and Discher, 2009).
Atomic force microscopy (AFM) makes it possible to observe, manipulate and explore the cell surface at a molecular resolution, and therefore has produced a wealth of new opportunities in cell biology, including understanding the nanoscale organization and dynamics of cell membranes and cell walls, measuring cell mechanics and cell adhesion, unraveling the molecular elasticity of cellular proteins and the mechanisms by which they assemble into nanodomains in the membrane (Müller and Dufrêne, 2011; Müller et al., 2009). In this Commentary, we explain the basic principles of AFM, discuss current strategies for imaging live cells and for probing single functional proteins on their surfaces, provide a critical evaluation of the potential and limitations of the technique, and survey recent discoveries in cell surface biology that were driven by AFM. Although animal cell studies will be covered, we will especially focus on breakthroughs that have been made in yeast cell research, because this is where substantial progress in single-cell surface protein analysis has recently been made.
Atomic force microscopy
General principles
Instead of using an incident beam to visualize a sample – as would be the case in classical microscopy – AFM senses the small forces (in the piconewton range, ∼10−12 N) that act on the surface of a sample (Binnig et al., 1986; Müller and Dufrêne, 2011; Müller et al., 2009). Images of molecules or cells in buffer or growth medium are made by scanning a sharp tip over the sample surface and measuring the interaction force between the tip and the surface (Fig. 1A). The sample is mounted on a piezoelectric scanner, which ensures three-dimensional positioning with high accuracy. While the tip is being scanned across the sample in the x and y directions, the force between the tip and specimen can be recorded. The sharp tip is attached to a soft cantilever, and, as the cantilever bends, its deflection is detected by movement of a laser beam reflected from the tip. AFM topographic imaging is widely used in the life sciences, and has provided high-resolution images of biomolecules, membranes and cells in buffer at unprecedented resolution (Dufrêne, 2008a, Dufrêne, 2008b; Engel and Gaub, 2008; Müller and Dufrêne, 2011; Müller et al., 2009).
Fig. 1.
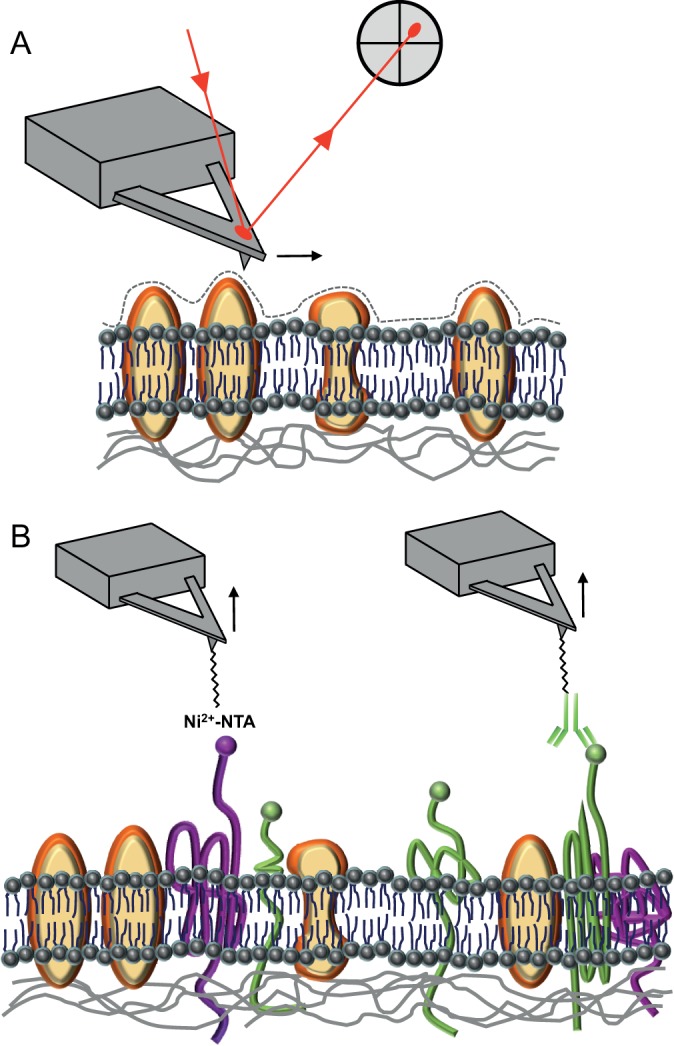
Atomic force microscopy. (A) In the imaging mode, a very sharp tip follows the contours of the cell surface with nanometer resolution. The lipid bilayer of the plasma membrane is shown, with inserted proteins as yellow objects. (B) In SMFS, the small interaction force between the AFM tip and cell surface molecules is measured, while the distance between the tip and cell is altered. The two examples show a tip labeled with a chemical group (Ni2+-NTA, left) or a ligand (e.g. an antibody, right) to detect, determine the location of and manipulate individual cell surface proteins, such as mechanosensors (shown in magenta and green, respectively).
AFM is also widely used to manipulate and analyze single biomolecules with a method called single-molecule force spectroscopy (SMFS) (Hinterdorfer and Dufrêne, 2006; Engel and Gaub, 2008; Puchner and Gaub, 2009; Müller et al., 2009; Dufrêne et al., 2011). Here, the tip is brought into proximity of and retracted from the biological sample, and the cantilever deflection measures the interaction force (Fig. 1B). The force–distance curves that are obtained with this procedure provide key insights into the localization, elasticity and binding strength of single molecules. As we will discuss below, the manipulation of single molecules on the surface of living cells often requires labeling the tip with chemical groups or bioligands using specific protocols (Fig. 1B).
Imaging living cells with AFM
Soon after its invention, AFM became a valuable tool for imaging cells (Butt et al., 1990; Radmacher et al., 1992). However, AFM imaging of single cells requires their firm attachment to a surface, which is not always a simple task. A straightforward approach is to exploit the ability of animal cells to spread and adhere to solid supports (Radmacher et al., 1992). Coating the substrate with adhesion proteins might be used to enhance immobilization, and this method has made it possible to observe, for example, actin filament dynamics beneath the plasma membrane of glial cells (Henderson et al., 1992). In some cases, chemical fixation using cross-linking agents such as glutaraldehyde might be required either to prevent cell damage or detachment from the support caused by the scanning tip, or as a means to obtain high-resolution images (Fig. 2A). Using these different protocols, various cell types have been investigated, including macrophages, CV-1 kidney cells, fibroblasts, Madin–Darby canine kidney (MDCK) cells, platelets and cardiomyocytes (Fig. 2A; Dufrêne, 2011; Jena and Horber, 2002).
Fig. 2.
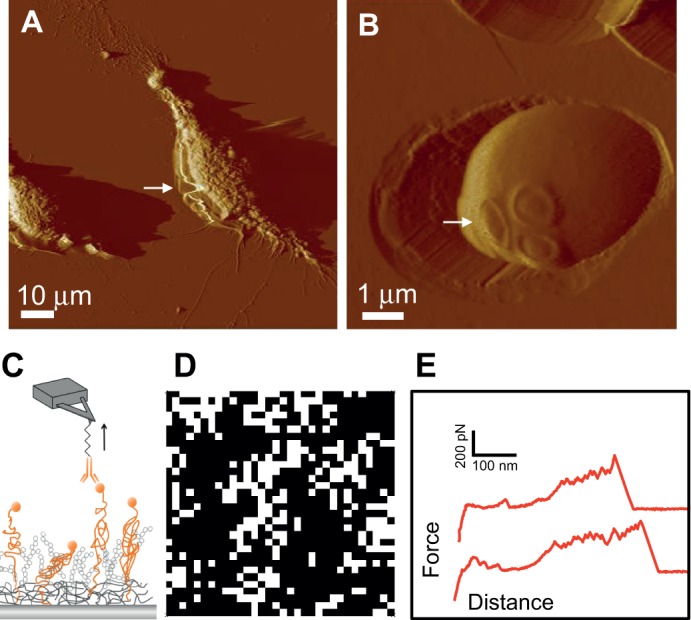
Atomic force microscopy in cell biology. (A,B) Imaging cells. AFM images of gently fixed macrophages spread on glass (A) and of a single yeast cell of Candida albicans trapped in a porous polymer membrane (B). Arrows highlight a common artifact, namely the alteration of the cell surface by the scanning tip. (C-E) Imaging and manipulating cell surface proteins: single-molecule force spectroscopy with AFM tips attached to specific antibodies makes it possible to detect single proteins on the cell surface (C), to map their distribution (D) and to measure their mechanical response in relation to function (E). The recognition map in (D) documents the detection of single cell-adhesion protein (white pixels), and the notion that they are concentrated into localized nanodomains. The force–distance curves in (E) document multiple force peaks that reflect the force-induced unfolding of individual proteins. Shown here are the results for the Candida albicans Als adhesion proteins, whose unfolding and clustering are believed to have a role in cell adhesion.
In recent years, much progress has been made with regards to live-cell imaging of various microbes (Fig. 2B; Dufrêne, 2008b; Dufrêne, 2011). In order to gain reliable high-resolution images of microbial cells, sample preparation is of crucial importance. Unlike animal cells, microbes have a well-defined shape and usually do not spread on surfaces under experimental conditions. As a result, the contact area between a cell and the support is very small, which often leads to cell detachment caused by the scanning tip. Therefore, several approaches have been developed to ensure more stable cell attachment (Dufrêne, 2008a). For instance, it is possible to attach the cells onto supports that have been functionalized with either positively charged macromolecules, like poly-L-lysine or polyethylenimine, or with molecules containing hydrophobic groups. This method has been successfully applied to lactic acid and Gram-negative bacteria, diatoms and fungi, and has yielded new insights into surface structure and elasticity of these organisms. Cells can also be immobilized mechanically within gelatin-coated supports or captured in the holes of porous polymer membranes. In the latter approach, a concentrated cell suspension is driven through a porous membrane with a pore size similar to that of the cells to be investigated (Kasas and Ikai, 1995) (Fig. 2B). This procedure is fairly simple and does not involve a macromolecular support, thus preventing the risk of contamination of the cell surface or the AFM tip.
Although AFM imaging offers key benefits over more conventional microscopy techniques, newcomers should be aware that the current technology is still limited by a number of problems that hamper its widespread use in cell biology (Box 1). Nonetheless, AFM can be employed to address a number of interesting cell biological questions, as we will discuss below.
Single-protein localization and manipulation
Unlike fluorescence microscopy, AFM topographic imaging lacks biochemical specificity (Table 1). Thus, specific molecules, such as a given receptor, cannot be unambiguously identified and located on cell surfaces. This is an important limitation, because the organization and interactions of the cell surface machinery is an important parameter for controlling its functions. However, the use of spatially resolved SMFS with AFM tips that are labeled with specific chemical groups or ligands now enables the detection and localization of single functional proteins on cell surfaces (Hinterdorfer and Dufrêne, 2006). The rationale behind this approach is to use SMFS to record arrays of force curves across the cell surface with a specifically labeled tip (Fig. 2C), estimate the specific molecular recognition force value for each force curve and then display it in terms of pixels, where the pixel brightness reflects the magnitude of the recorded force (Fig. 2D). The cell surface recognition map that can be obtained using this approach enables researchers to understand the distribution of a given receptor and to determine whether it is clustered or randomly distributed across the cell surface (Fig. 2D). However, it should be noted that, as is the case for cell imaging, single-protein analysis by SMFS is still a rather new method that suffers from a number of drawbacks (Box 1).
Table 1.
Comparison of high-resolution techniques for imaging single proteins in cells
| Technique | AFM | Super-resolution fluorescence microscopy (TIRF, PALM, STORM) | Electron microscopy (TEM) | |
| Topographic imaging | SMFS | |||
| Resolution | ≈50 nma | ≈20 nm | ≈20–50 nm | ≈1-10 nm |
| Live cell | Yes | Yes | Yes | No |
| Time for sample preparation | 10 minutes | 1 dayb | 1-3 hours | 2-5 days |
| Time for image acquisition | ≈5 minutes | 25-50 minutes | ≈5 minutes | 5-10 minutes |
| Image processing | 5 minutes | A few hours | Up to 24 hours | 30 minutes |
| Simultaneous detection of different proteins | No | No | Yes | Yesc |
| Protein tracking | No | No | Yes | No |
| Cost of equipment | €150,000-250,000 | €250,000-500,000 | €≈500,000 | |
| Operational costs | Low | Moderate | Moderate | High |
| Advantages | Localization and force response of single proteins; protein unfolding; dynamic processes; various environments (temperature, buffer, etc.) | Spatial-temporal resolution; only basic genetic manipulations required; high sample throughput; study of live protein interactions and their quantification | Imaging of the cell ultrastructures at very high resolution | |
| Disadvantages | Only the cell surface is analyzed; only a single cell at a time; slow temporal resolution; various sources of artifacts like cell or tip alteration | Frequently a bad signal-to-noise ratio; photobleaching; no information on physical properties of proteins; tendency of fluorescent protein tags to multimerize (i.e. interaction artifacts) | Fixation artifacts; no access to dynamics; no information on physical properties of proteins | |
Only whole cell analyses are considered, i.e. purified proteins or membranes are not included. TIRF, total internal reflection fluorescence; PALM, photoactivated localization microscopy; STORM, stochastic optical reconstruction microscopy; TEM, transmission electron microscopy.
In mammalian cells (a 10-nm resolution can be routinely achieved on microbial cells).
Including tip preparation.
Indirect; e.g. gold beads of different sizes.
SMFS cannot only be used for mapping single protein molecules on a given cell surface, but can also be employed to subject cell surface proteins to force in order to study their mechanical properties (Fig. 2E). Early studies have demonstrated that proteins can be subjected to controlled forces using this approach, and have yielded details of protein unfolding pathways (Rief et al., 1997). Since then, single-molecule AFM, combined with molecular dynamics simulations and protein engineering, has greatly enhanced our understanding of the mechanical behavior of a great variety of proteins (Engel and Gaub, 2008; Puchner and Gaub, 2009; Li and Cao, 2010; Marszalek and Dufrêne, 2012). Most of these single protein manipulation experiments have been conducted in vitro, where the system under study can be tightly controlled. However, this also means that molecules are removed from the native cellular context that controls their structural assembly and functional state (Dufrêne et al., 2011). In the following section, we will highlight some recent breakthrough studies in which SMFS has been successfully used to unravel the nanomechanics and clustering of single proteins on the surface of living cells.
Measuring mechanics of single proteins
Nanomechanics of membrane sensors
The biophysical mechanisms underlying cellular mechanosensing are poorly understood at the molecular level. In cell wall integrity (CWI) sensing and signaling in yeast, five plasma-membrane-spanning proteins (Wsc1, Wsc2, Wsc3, Mid2 and Mtl1) presumably detect mechanical forces acting on the cell wall and membrane, and subsequently trigger an intracellular signaling cascade, which ultimately results in the expression of genes encoding either cell wall biosynthetic enzymes or cell wall proteins (Levin, 2005; Rodicio and Heinisch, 2010). However, the lack of an appropriate probing technique meant that, until recently, it remained unclear how these membrane sensors responded to mechanical force.
SMFS studies with genetically manipulated Wsc1 in live Saccharomyces cerevisiae have recently unraveled the mechanical behavior of single sensor molecules in relation to their function (Dupres et al., 2009; Heinisch et al., 2010a). Central to this discovery was the use of genetic manipulations to address a conceptual problem: AFM is a surface-based technique, so how can it probe protein sensors (such as Wsc1) that are embedded within the cell wall? The yeast cell wall has a thickness of ∼100 nm (Backhaus et al., 2010), whereas the maximal length of the extracellular part of the Wsc1 sensor is 86 nm, meaning it does not reach the outermost cell surface. Therefore, the mechanosensors were artificially elongated to a length of 153 nm by inserting a stiff serine/threonine-rich region from the Mid2 sensor. Furthermore, the addition of a histidine tag to the N-terminal end of the protein allowed the proteins to be detected by a AFM tip modified with Ni2+-nitrilotriacetate (Ni2+-NTA). Force–extension curves obtained by pulling on the modified Wsc1 sensors revealed a fascinating behavior: the sensors display the characteristics of a Hookean spring (i.e. their force curve contains a linear region where force is directly proportional to extension) (Dupres et al., 2009). These nanospring properties, which contrast with the properties of most proteins (Rief et al., 1997; Marszalek and Dufrêne, 2012), have subsequently been shown to be mediated by the extracellular serine/threonine-rich region of the sensor (Dupres et al., 2009). Notably, this technology has also provided a tool for determining cell wall thickness in vivo in yeast, because sensor molecules of different lengths can only be detected once they reach the surface (Dupres et al., 2010).
Accordingly, SMFS opens new avenues for investigating how proteins respond to forces in living cells and how mechanosensing events proceed in vivo, thereby providing new insights into what has been suggested to be one of the most ancient sensory mechanisms in evolution (Kee and Robinson, 2008). An exciting new development in this direction will be the combination of SMFS with fluorescence microscopy. Pulling on a sensor from the outside with SMFS should lead to the recruitment of the corresponding intracellular signaling components to the cytoplasmic tail of the sensor, which can subsequently be observed by fluorescence microscopy. We also anticipate that the single protein manipulations described here should allow researchers to address the mechanics of a variety of other membrane sensors that would usually be hidden and not accessible by AFM per se. For instance, mammalian cell sensors, such as integrins, are usually covered by a polysaccharide and protein matrix, the glycocalyx, which prevents reliable SMFS experiments. Elongation and tagging of such proteins should provide a means to solve this issue.
Nanomechanics of cell adhesion proteins
During the past 15 years, SMFS has been widely used to measure the molecular interactions of cell adhesion molecules, including selectins, cadherins, integrins, oligosaccharides, proteoglycans and microbial adhesins (see Hinterdorfer and Dufrêne, 2006; Müller and Dufrêne, 2011, and references therein). Because these experiments were conducted on purified molecules, it is of great interest to probe single cell-adhesion proteins directly in living cells to further understand the interactions and functions of adhesion molecules in vivo.
Adhesion of the pathogen Candida albicans is mediated by cell-surface glycoproteins referred to as the Als family. Microscopic assays have revealed that Als-mediated adhesion involves an initial adhesion step, followed by force-induced protein conformational changes that result in the formation of strong amyloid-like bonds that can ‘cement’ the adhering cells together (Dranginis et al., 2007; Lipke et al., 2012). Again, the molecular details of this mechanism remained essentially unknown. SMFS was therefore used to study the adhesive and mechanical properties of Als5 in live cells (Alsteens et al., 2009). Pulling on single Als5 proteins using a ligand-modified AFM tip has revealed characteristic force signatures with multiple well-defined peaks, with each peak corresponding to the force-induced unfolding of an individual tandem-repeat domain. AFM measurements also revealed that the unfolding probability increases with the number of repeats and correlates with the level of cell–cell adhesion, which confirms that the modular domains have a role in many fungal adhesion molecules. Presumably, the force-induced unfolding of Als proteins leads to extended conformations in which previously masked hydrophobic groups become exposed, thus favoring hydrophobic interactions between opposing cells. These single-molecule measurements open up new avenues for understanding the mechanical properties of adhesion molecules in a variety of cell types. In particular, they should contribute to increasing our understanding of how force-induced conformational changes in proteins such as integrins modulate their binding strength (Vogel and Sheetz, 2006; Brown and Discher, 2009).
Imaging protein clustering
Understanding how membrane proteins assemble to form membrane micro- and nano-domains is another important question in cell biology. A key example of such clustering is the formation of adhesion domains (i.e. focal adhesion complexes) composed of aggregated proteins that mediate the early stage of adhesion (Vogel and Sheetz, 2006). The small size (∼10-500 nm) and dynamic nature of membrane domains make their direct visualization in live cells very challenging. However, AFM is emerging as a valuable tool to map the distribution of single-proteins on cell surfaces, thereby unraveling the formation and composition of such protein domains. Prominent examples include the mapping of the distribution of prostaglandin receptors on live CHO cells (Kim et al., 2006), the localization of vascular endothelial cadherin-binding sites (Chtcheglova et al., 2007), and the measurement of the organization and stiffness of neuron membrane domains containing glycosylphosphatidylinositol (GPI)-anchored proteins (Roduit et al., 2008). As discussed below, functional protein clusters were recently found to form and increase in size under stress, thereby activating cell signaling and cell adhesion in yeasts.
Stress-induced clustering of membrane sensors
Fluorescence microscopy has shown that GFP–Wsc1 sensors form membrane patches, but the structure–function relationships of such clusters has long remained a mystery (Wilk et al., 2010). To address this problem, spatially resolved SMFS was used to map the distribution of single Wsc1 sensors on the surface of live S. cerevisiae (Heinisch et al., 2010b). Molecular recognition images were recorded on cells expressing His-tagged elongated sensors using Ni2+-NTA tips. Many sensor molecules appeared to form clusters of ∼200 nm, which is in the range of the large patches observed by fluorescence microscopy for the microdomains containing the marker protein Can1 (MCC) in S. cerevisiae (Malinsky et al., 2010). Both the surface density of Wsc1 sensors and their tendency to cluster increases when cells are stressed by heat or low osmolarity, which suggests that clustering is a stress response that is intimately connected to signaling.
An analysis of three different Wsc1 mutants has also indicated that the Wsc1 cysteine-rich domain has a crucial function in sensor clustering and signaling. Although mutant proteins display surface density and spring behaviors that are similar to those of wild-type Wsc1, they are more evenly distributed over the yeast cell surface, instead of being clustered. In addition, Wsc1 sensor clustering mediated by the cysteine-rich domain is dependent on the formation of disulfide bridges (Dupres et al., 2011). As these mutant proteins produce non-functional sensors that are unable to activate the downstream signaling pathway, these findings demonstrate the importance of the cysteine-rich domain in sensor clustering and turnover (Heinisch et al., 2010b). The studies also suggest that in yeast, like in higher eukaryotes, the function of sensors is coupled to their localized enrichment in membrane patches that are referred to as ‘sensosomes’. We are confident that single-protein imaging by SMFS will be useful to study similar clustering of sensor proteins in mammalian cells, especially following the further development of high-speed SMFS mapping.
Force-induced clustering of cell adhesion proteins
A fascinating phenomenon that is portrayed by cell adhesion domains in mammalian cells is their ability to grow and strengthen under force (Bershadsky et al., 2006). In Candida albicans, macroscopic assays have suggested that adhesion-triggered changes in the conformation of Als5 propagate around the cell surface, thereby forming ordered adhesion domains (Lipke et al., 2012). This mechanism was recently experimentally confirmed by SMFS: mechanical force was shown to trigger the formation of cell adhesion nanodomains in living yeast cells (Alsteens et al., 2010). Pulling on single adhesins was shown to induce the formation of adhesion domains of 100-500 nm in size, and these force-induced nano-domains propagate over the entire cell surface. Als5-mediated remodeling is independent of cellular metabolic activity, because the protein shows the same behavior in heat-killed cells, which are no longer metabolically active. Remarkably, a single-site mutation in the conserved amyloid-forming sequence of the protein has revealed that amyloid interactions represent the driving force that underlies Als5 clustering (Garcia et al., 2011). Hence, the strength of yeast cell–cell adhesion results from the force-induced amyloid-like clustering of hundreds of proteins on the cell surface to form arrays of ordered multimeric binding sites (Lipke et al., 2012). These results highlight the role that functional amyloids can have in cell adhesion, both in lateral clustering (in cis) of many adhesion proteins to increase binding avidity, and in the potential to form stable amyloid-type bonds between cells (in trans), a mechanism that we expect to be detected in many more organisms as more cell adhesion systems are examined (Romero et al., 2010).
Interestingly, the above mechanical restructuring is reminiscent of shear-induced unfolding and refolding of proteins during blood clotting, where force induces the exposure of interaction sites in the blood glycoprotein von Willebrand factor (VWF), which has recently been demonstrated by single-molecule laser tweezer experiments (Kim et al., 2010). Shear-induced bond strengthening, called ‘catch bonding’, is also crucial in arresting leukocyte rolling and extravasation (Alon and Dustin, 2007). Clearly, as the SMFS technique further evolves and matures, it will contribute to the discovery of similar remarkable phenomena in many other cell surface proteins.
Conclusions
Understanding protein mechanics and protein clustering in vivo is a crucial challenge in cell biology, which, until now, has been difficult to tackle owing to the lack of high-resolution probing techniques. This Commentary has highlighted how the combination of single-molecule AFM with genetic manipulations is a powerful tool for probing functional receptors and sensors on cell surfaces, in a way that was not possible previously, and how this approach nicely complements other imaging techniques in cell biology (Table 1). This integrated platform can be applied to measuring the mechanical properties of single proteins, and can be used to image their localization, thus revealing whether they are isolated or form clusters.
Despite the key benefits of AFM, researchers should realize that the technology is still under development and that there are still a number of limitations that must be solved in order for the technique to become more widespread in the cell biology community (Box 1). Clearly, a detailed understanding of the principles and limitations of the different AFM modalities is essential before users start their first experiments. In SMFS, protocols have been developed for attaching biomolecules to AFM tips but they still require specific expertise that is usually not found in cell biology laboratories. In the future, defining simple standardized protocols for tip functionalization and data interpretation, and automation of force spectroscopy analyses, making them readily available to the biology community, will contribute to make SMFS accessible for cell biologists.
Undoubtedly, the main limitation of both AFM imaging and spatially resolved SMFS in cell biology is their rather slow temporal resolution (Box 1). Hopefully, current efforts in developing high-speed AFM techniques should soon provide access to millisecond resolutions using live cells. Another important direction for future research is to expand the range of potential applications for single molecule analysis, by combining AFM with advanced light microscopy techniques, such as stimulated emission depletion microscopy (Hell, 2009). Such combined imaging platforms would allow researchers to simultaneously identify, track, observe and force probe the individual constituents of the cell surface, thereby helping to answer many unresolved problems in cell biology.
Box 1. Advantages and limitations of AFM in cell biology
AFM shows distinct advantages over optical and electron microscopy techniques for cell surface biology (Table 1), namely (1) the ability to observe purified membranes and live cells at nanometer resolution and under physiological conditions (e.g. in buffer solution at room temperature and atmospheric pressure), (2) the capability to track structural dynamics and remodeling of the cell surface in response to environmental stimulants (e.g. drugs), and (3) the possibility to determine the location of and force-probe single constituents of the cell surface. Despite these key benefits, there are still a number of limitations and technological issues that must be solved before the full potential of the technique to address cell biological questions can be exploited (Table 1).
When observing cells using AFM topographic imaging, an important source of problems is the resolution and interpretation of the images, which directly depend on the imaging force, tip geometry and physical properties of the sample (Dufrêne, 2008a). Large forces acting between tip and sample during imaging can dramatically reduce the image resolution and cause molecular damage or displacement (Fig. 2A,B). This is particularly true for soft corrugated mammalian cells, where the tip generally deforms the cell surface, pushes cellular material along the scanning direction and easily becomes blind owing to contamination by loosely bound macromolecules. Therefore, it is essential to control the force that is applied during scanning (0.1-0.5 nN) by selecting appropriate buffer conditions. In addition, unlike microbial cells, which can be readily imaged in their native state (Fig. 2B), animal cells generally require chemical fixation in order to be imaged by AFM (Fig. 2A). The image resolution achieved with AFM on mammalian cells is generally not better than 50-100 nm. One approach to solve this problem is to use a nanopipette that does not contact the surface directly, which gives resolutions from live cells of ∼20 nm (Novak et al., 2009).
Another limitation of AFM imaging compared with fluorescence microscopy is its rather poor temporal resolution (typically ∼1 minute per image), which is much slower than the time scale at which dynamic processes usually occur in cell biology. However, remarkable advances are being made in developing high-speed AFM set-ups that can operate in the millisecond timescale and thus offer new possibilities to explore cellular dynamics (Shibata, 2010).
A crucial feature of SMFS is the functionalization of the AFM tips with biomolecules. This can be achieved using essentially two types of surface chemistries, which are based either on the strong chemisorption of thiols onto gold surfaces or on the covalent attachment of silanes or alcohols onto silicon oxide surfaces (reviewed by Hinterdorfer and Dufrêne, 2006). In addition, well-defined procedures are available to accurately measure the localization and forces of single proteins on live cells (Hinterdorfer and Dufrêne, 2006), but accurate data collection and interpretation often remain technically challenging. In this context, the main issues are those associated with the quality of the tip and its possible alteration during the course of an experiment. As with statistical validation, spatially resolved SMFS should be performed on different locations of the cell surface using different tips and the measurements should be repeated using many cells from independent cultures. Finally, an important drawback of AFM-based SMFS is that it probes cell surface biomolecules, but is unable to access the interior of living cells.
Supplementary Material
Footnotes
Funding
Work in the Y.F.D. group was supported by the National Foundation for Scientific Research (FNRS); the Foundation for Training in Industrial and Agricultural Research (FRIA); the Université catholique de Louvain (Fonds Spéciaux de Recherche); the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme); and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y. F. D. and D.A. are Senior Research Associate and Postdoctoral Researcher of the FRS-FNRS, respectively. Work in the P. N. L. laboratory was supported by National Institutes of Health [grant number SC1 GM083756]. Work on CWI sensors in the lab of J. J. H. is supported by a grant from the Deutsche Forschungsgemeinschaft in the frame of the SFB944. Deposited in PMC for release after 12 months.
References
- Alon R., Dustin M. L. (2007). Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity 26, 17–27 10.1016/j.immuni.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Alsteens D., Dupres V., Klotz S. A., Gaur N. K., Lipke P. N., Dufrêne Y. F. (2009). Unfolding individual Als5p adhesion proteins on live cells. ACS Nano 3, 1677–1682 10.1021/nn900078p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsteens D., Garcia M. C., Lipke P. N., Dufrêne Y. F. (2010). Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 107, 20744–20749 10.1073/pnas.1013893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhaus K., Heilmann C. J., Sorgo A. G., Purschke G., de Koster C. G., Klis F. M., Heinisch J. J. (2010). A systematic study of the cell wall composition of Kluyveromyces lactis. Yeast 27, 647–660 10.1002/yea.1781 [DOI] [PubMed] [Google Scholar]
- Bershadsky A., Kozlov M., Geiger B. (2006). Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr. Opin. Cell Biol. 18, 472–481 10.1016/j.ceb.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Binnig G., Quate C. F., Gerber C. (1986). Atomic force microscope. Phys. Rev. Lett. 56, 930–933 10.1103/PhysRevLett.56.930 [DOI] [PubMed] [Google Scholar]
- Brown A. E. X., Discher D. E. (2009). Conformational changes and signaling in cell and matrix physics. Curr. Biol. 19, R781–R789 10.1016/j.cub.2009.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H. J., Wolff E. K., Gould S. A. C., Dixon Northern B., Peterson C. M., Hansma P. K. (1990). Imaging cells with the atomic force microscope. J. Struct. Biol. 105, 54–61 10.1016/1047-8477(90)90098-W [DOI] [PubMed] [Google Scholar]
- Chtcheglova L. A., Waschke J., Wildling L., Drenckhahn D., Hinterdorfer P. (2007). Nano-scale dynamic recognition imaging on vascular endothelial cells. Biophys. J. 93, L11–L13 10.1529/biophysj.107.109751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva M. B., McClelland A. C., Kayser M. S. (2007). Cell adhesion molecules: signaling functions at the synapse. Nat. Rev. Neurosci. 8, 206–220 10.1038/nrn2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D. E., Janmey P., Wang Y. L. (2005). Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- Dranginis A. M., Rauceo J. M., Coronado J. E., Lipke P. N. (2007). A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71, 282–294 10.1128/MMBR.00037-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne Y. F. (2008a). Atomic force microscopy and chemical force microscopy of microbial cells. Nat. Protoc. 3, 1132–1138 10.1038/nprot.2008.101 [DOI] [PubMed] [Google Scholar]
- Dufrêne Y. F. (2008b). Towards nanomicrobiology using atomic force microscopy. Nat. Rev. Microbiol. 6, 674–680 10.1038/nrmicro1948 [DOI] [PubMed] [Google Scholar]
- Dufrêne Y. F. (2011). Life at the Nanoscale: Atomic Force Microscopy of Live Cells. Singapore: Pan Stanford Publishing [Google Scholar]
- Dufrêne Y. F., Evans E., Engel A., Helenius J., Gaub H. E., Müller D. J. (2011). Five challenges to bringing single-molecule force spectroscopy into living cells. Nat. Methods 8, 123–127 10.1038/nmeth0211-123 [DOI] [PubMed] [Google Scholar]
- Dupres V., Alsteens D., Wilk S., Hansen B., Heinisch J. J., Dufrêne Y. F. (2009). The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 5, 857–862 10.1038/nchembio.220 [DOI] [PubMed] [Google Scholar]
- Dupres V., Dufrêne Y. F., Heinisch J. J. (2010). Measuring cell wall thickness in living yeast cells using single molecular rulers. ACS Nano 4, 5498–5504 10.1021/nn101598v [DOI] [PubMed] [Google Scholar]
- Dupres V., Heinisch J. J., Dufrêne Y. F. (2011). Atomic force microscopy demonstrates that disulfide bridges are required for clustering of the yeast cell wall integrity sensor Wsc1. Langmuir 27, 15129–15134 10.1021/la203679s [DOI] [PubMed] [Google Scholar]
- Engel A., Gaub H. E. (2008). Structure and mechanics of membrane proteins. Annu. Rev. Biochem. 77127–148 10.1146/annurev.biochem.77.062706.154450 [DOI] [PubMed] [Google Scholar]
- Garcia M. C., Lee J. T., Ramsook C. B., Alsteens D., Dufrêne Y. F., Lipke P. N. (2011). A role for amyloid in cell aggregation and biofilm formation. PLoS ONE 6, e17632 10.1371/journal.pone.0017632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray–Owen S. D., Blumberg R. S. (2006). CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 6, 433–446 10.1038/nri1864 [DOI] [PubMed] [Google Scholar]
- Heinisch J. J., Dupres V., Alsteens D., Dufrêne Y. F. (2010a). Measurement of the mechanical behavior of yeast membrane sensors using single-molecule atomic force microscopy. Nat. Protoc. 5, 670–677 10.1038/nprot.2010.19 [DOI] [PubMed] [Google Scholar]
- Heinisch J. J., Dupres V., Wilk S., Jendretzki A., Dufrêne Y. F. (2010b). Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS ONE 5, e11104 10.1371/journal.pone.0011104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S. W. (2009). Microscopy and its focal switch. Nat. Methods 6, 24–32 10.1038/nmeth.1291 [DOI] [PubMed] [Google Scholar]
- Henderson E., Haydon P. G., Sakaguchi D. S. (1992). Actin filament dynamics in living glial cells imaged by atomic force microscopy. Science 257, 1944–1946 10.1126/science.1411511 [DOI] [PubMed] [Google Scholar]
- Hinterdorfer P., Dufrêne Y. F. (2006). Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 3, 347–355 10.1038/nmeth871 [DOI] [PubMed] [Google Scholar]
- Jena B. P., Horber J. K. H. (2002). Atomic Force Microscopy in Cell Biology, Methods in Cell Biology. San Diego, CA: Academic Press [Google Scholar]
- Kasas S., Ikai A. (1995). A method for anchoring round shaped cells for atomic force microscope imaging. Biophys. J. 68, 1678–1680 10.1016/S0006-3495(95)80344-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y. S., Robinson D. N. (2008). Motor proteins: myosin mechanosensors. Curr. Biol. 18, R860–R862 10.1016/j.cub.2008.07.071 [DOI] [PubMed] [Google Scholar]
- Kim H., Arakawa H., Hatae N., Sugimoto Y., Matsumoto O., Osada T., Ichikawa A., Ikai A. (2006). Quantification of the number of EP3 receptors on a living CHO cell surface by the AFM. Ultramicroscopy 106, 652–662 10.1016/j.ultramic.2005.12.007 [DOI] [PubMed] [Google Scholar]
- Kim J., Zhang C. Z., Zhang X., Springer T. A. (2010). A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature 466, 992–995 10.1038/nature09295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Greenberg E. P. (2006). Microbial sciences: the superficial life of microbes. Nature 441, 300–302 10.1038/441300a [DOI] [PubMed] [Google Scholar]
- Lecuit T., Lenne P. F. (2007). Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644 10.1038/nrm2222 [DOI] [PubMed] [Google Scholar]
- Levin D. E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291 10.1128/MMBR.69.2.262-291.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Cao Y. (2010). Protein mechanics: from single molecules to functional biomaterials. Acc. Chem. Res. 43, 1331–1341 10.1021/ar100057a [DOI] [PubMed] [Google Scholar]
- Lipke P. N., Garcia M. C., Alsteens D., Ramsook C. B., Klotz S. A., Dufrêne Y. F. (2012). Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 20, 59–65 10.1016/j.tim.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky J., Opekarová M., Tanner W. (2010). The lateral compartmentation of the yeast plasma membrane. Yeast 27, 473–478 10.1002/yea.1772 [DOI] [PubMed] [Google Scholar]
- Marszalek P. E., Dufrêne Y. F. (2012). Stretching single polysaccharides and proteins using atomic force microscopy. Chem. Soc. Rev. 41, 3523–3534 10.1039/c2cs15329g [DOI] [PubMed] [Google Scholar]
- McIntosh B. E., Hogenesch J. B., Bradfield C. A. (2010). Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645 10.1146/annurev-physiol-021909-135922 [DOI] [PubMed] [Google Scholar]
- Morgan M. R., Humphries M. J., Bass M. D. (2007). Synergistic control of cell adhesion by integrins and syndecans. Nat. Rev. Mol. Cell Biol. 8, 957–969 10.1038/nrm2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D. J., Dufrêne Y. F. (2011). Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 21, 461–469 10.1016/j.tcb.2011.04.008 [DOI] [PubMed] [Google Scholar]
- Müller D. J., Helenius J., Alsteens D., Dufrêne Y. F. (2009). Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 5, 383–390 10.1038/nchembio.181 [DOI] [PubMed] [Google Scholar]
- Novak P., Li C., Shevchuk A. I., Stepanyan R., Caldwell M., Hughes S., Smart T. G., Gorelik J., Ostanin V. P., Lab M. J., et al. (2009). Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nat. Methods 6, 279–281 10.1038/nmeth.1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchner E. M., Gaub H. E. (2009). Force and function: probing proteins with AFM-based force spectroscopy. Curr. Opin. Struct. Biol. 19, 605–614 10.1016/j.sbi.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Radmacher M., Tillamnn R. W., Fritz M., Gaub H. E. (1992). From molecules to cells: imaging soft samples with the atomic force microscope. Science 257, 1900–1905 10.1126/science.1411505 [DOI] [PubMed] [Google Scholar]
- Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997). Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 10.1126/science.276.5315.1109 [DOI] [PubMed] [Google Scholar]
- Rodicio R., Heinisch J. J. (2010). Together we are strong—cell wall integrity sensors in yeasts. Yeast 27, 531–540 10.1002/yea.1785 [DOI] [PubMed] [Google Scholar]
- Roduit C., van der Goot F. G., De Los Rios P., Yersin A., Steiner P., Dietler G., Catsicas S., Lafont F., Kasas S. (2008). Elastic membrane heterogeneity of living cells revealed by stiff nanoscale membrane domains. Biophys. J. 94, 1521–1532 10.1529/biophysj.107.112862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D., Aguilar C., Losick R., Kolter R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 107, 2230–2234 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. A. (2009). Cell biology. The force is with us. Science 323, 588–589 10.1126/science.1169414 [DOI] [PubMed] [Google Scholar]
- Sheetz M. P. (2001). Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2, 392–396 10.1038/35073095 [DOI] [PubMed] [Google Scholar]
- Shibata M., Yamashita H., Uchihashi T., Kandori H., Ando T. (2010). High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat. Nanotechnol. 5, 208–212 10.1038/nnano.2010.7 [DOI] [PubMed] [Google Scholar]
- Sokurenko E. V., Vogel V., Thomas W. E. (2008). Catch-bond mechanism of force-enhanced adhesion: counterintuitive, elusive, but ... widespread? Cell Host Microbe 4, 314–323 10.1016/j.chom.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. (2006). Pili in gram-positive pathogens. Nat. Rev. Microbiol. 4, 509–519 10.1038/nrmicro1443 [DOI] [PubMed] [Google Scholar]
- Verstrepen K. J., Reynolds T. B., Fink G. R. (2004). Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2, 533–540 10.1038/nrmicro927 [DOI] [PubMed] [Google Scholar]
- Vogel V., Sheetz M. (2006). Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 10.1038/nrm1890 [DOI] [PubMed] [Google Scholar]
- Weber C., Fraemohs L., Dejana E. (2007). The role of junctional adhesion molecules in vascular inflammation. Nat. Rev. Immunol. 7, 467–477 10.1038/nri2096 [DOI] [PubMed] [Google Scholar]
- Wilk S., Wittland J., Thywissen A., Schmitz H-P., Heinisch J. J. (2010). A block of endocytosis of the yeast cell wall integrity sensors Wsc1 and Wsc2 results in reduced fitness in vivo. Mol. Genet. Genomics 284, 217–229 10.1007/s00438-010-0563-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


